
The Arima-HiC Kit
and Service for
Chromosome Spanning
Assemblies
Introduction
The availability of high-quality reference genomes has had a profound
impact on the understanding of genome function and species evolution.
Recent years have seen a rapid expansion of long-read and long-range
methods, including Hi-C
1,2,
a NGS-based assay that preserves chromo-
some-range information during sample preparation.
Hi-C sequencing data uses the preserved chromosome-range information
to transform contigs to chromosomes. Despite its utility, broad adoption
of Hi-C has been plagued by labor-intensive complex protocols, prolonged
workflow durations, inconsistent experimental results, excessive sequencing
requirements, and expensive Bioinformatics analyses/services.
The Arima-HiC Kit overcomes these technical and economical limitations
with the development of a highly simplified and robust protocol that
streamlines Hi-C to a 6-hour, 8-step procedure, followed by library prep
and NGS. The Arima-HiC sequencing data is then used to scaffold contigs
to generate chromosome-spanning assemblies via open-source software
such as SALSA
3,4
and 3D-DNA
5
.
These key advancements have persuaded researchers, including
the Vertebrate Genomes Project (Fig.1), to use Arima-HiC to generate
high-quality assemblies of hundreds of species across plant and
animal kingdoms.
PRODUCT DATASHEET
GENOME ASSEMBLY
Fast and Easy Workflow
The Arima-HiC workflow was optimized to enable first time Hi-C users to
generate high-quality data with ease (Fig.2). The rapid 6-hour protocol limits
prolonged exposure of chromatin to external agents, leading to significant
enrichment of inter-contig signal.
The use of a unique combination of multiple 4-base cutting restriction
enzymes (RE) for chromatin digestion results in greater spread (per-base uni-
formity) of inter-contig signal to assemble all contigs regardless of gap-sizes.
Overall, the Arima-HiC Kit was designed to maximize the ease-of-use, with
minimal total time and hands-on steps, compatibility with downstream
library prep kits for Illumina NGS, 96-well plate compatible design,
and support for a broad range of sample types and species (Table 1). Upon
sequencing of the Arima-HiC libraries, open source Bioinformatics tools such
as SALSA
3,4
and 3D-DNA
5
can be used to map Arima-HiC data to contigs to
scaffold them into accurate chromosome-spanning assemblies. In addition
to scaffolding, Arima-HiC data has also been used to polish individual bases
of the contigs as Arima-HiC is sequenced on an Illumina NGS3 (Table 1).
Proven Performance
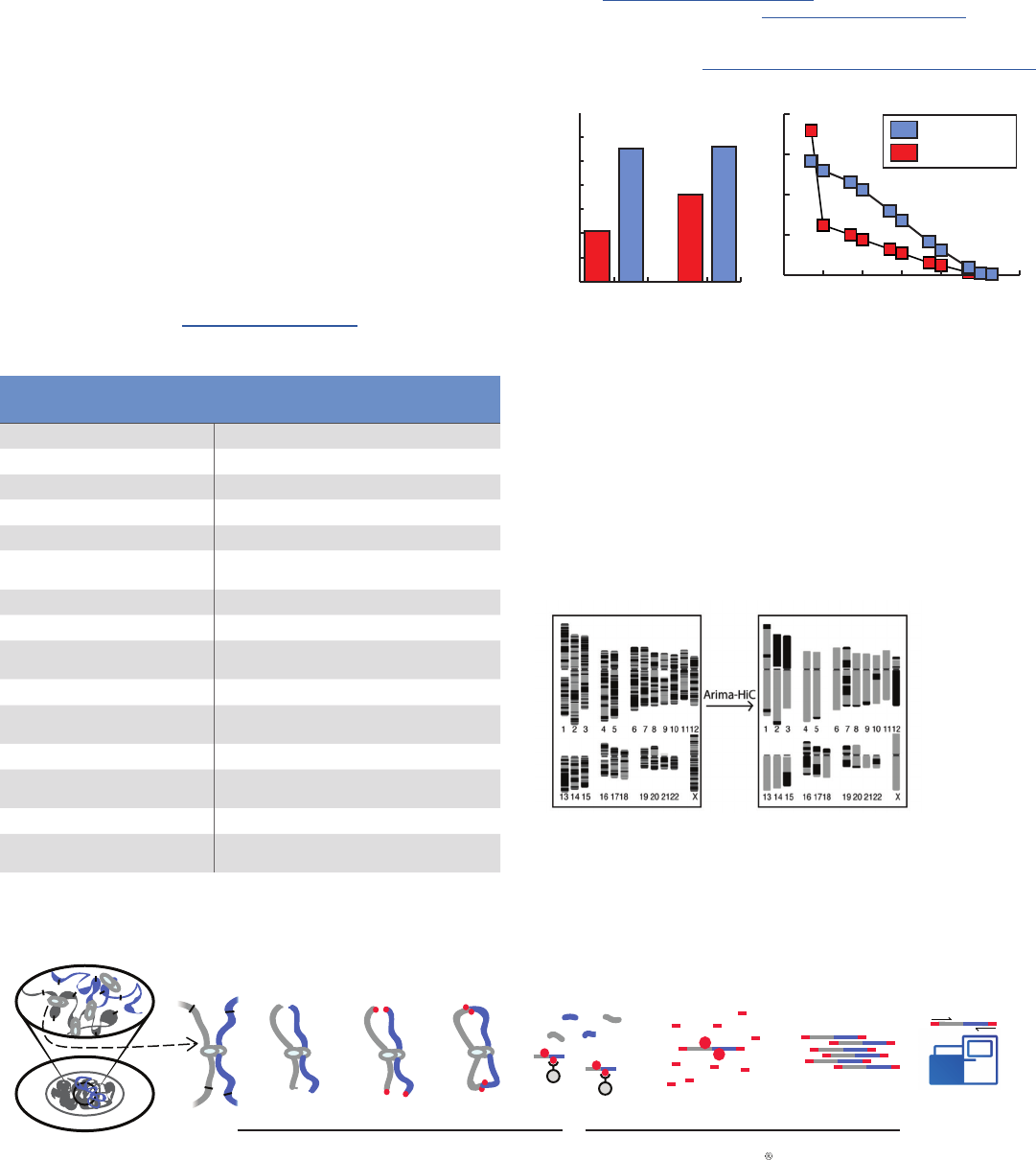
Rigorous external and internal testing resulted in an Arima-HiC Kit with
robust performance. When key opinion leaders compared Arima-HiC data
with Hi-C data generated by competitors, the Arima-HiC data manifested
higher inter-contig signal (Fig.3A). Importantly, the competitor’s Hi-C
data manifested a rapid drop in inter-contig signal with increase in contig
gap-sizes. Arima-HiC data, on the other hand, manifested 2-3 fold higher
inter-contig signal regardless of the contig gap-size (Fig.3B).
Subsequent SALSA3,4 analyses of Arima-HiC data converted 4Mb Oxford
Nanopore contigs to 125Mb NG50 chromosome-spanning human genome
assemblies, with as little as 30X Arima-HiC sequencing depth3 (Fig.4).
This is an extraordinary performance given that even the GRCH38 reference
assembly generated by cost-prohibitive combination of multiple
technologies had an NG50 of only ~140Mb, suggesting that the high
quality of Arima-HiC libraries and sequencing data offers substantial
technical and economic benefits to the user. These benefits can be
leveraged via Arima-HiC kits or services (sample to assembly).
Highlights
Proven Performance
• Obtain best-in-class inter-contig Hi-C signal across sample types
• Innovative multi-restriction enzyme chemistry improves overall
genome coverage
• Generate uniform "spread" of inter-contig HiC signal to scaffold
contigs regardless of gap sizes
Fast and User-Friendly Workflow
• Rapid time to Hi-C libraries with minimal hands-on touchpoints,
6-hour prep time (1 hour hands-on time)
• Democratize analysis workflow by using well-documented open
source tools such as SALSA (https://github.com/machinegun/SAL-
SA) for chromosome-spanning assemblies
Assured Quality
• Arima-HiC is validated in a wide-range of sample preservation and
transportation conditions through its selection by the Genome10K
consortium to generate high-quality assemblies of 260 species
• Assured library quality to ensure sequencing and assembly success
with quantitative and predictive and QC steps
• Obtain both long-range sequence and chromatin conformation
information with one assay
PHASED CONTIGS
SCAFFOLDS 1
SCAFFOLDS 2
SCAFFOLDS 3
GAP FILLING
ERROR CORRECTION ANNOTATION
10X LINKED READSPACBIO LONG READS BIONANO MAPS ARIMA-HIC PACBIO ISO-SEQ
Figure 1: Arima Genomics is a technology
partner of the VGP Phase 1. Long-read
PacBio sequencing is performed to generate
the initial contigs, followed by long-range
scaffolding approaches to assemble contigs
into chromosomes.
(Image courtesy of Vertebrate Genomes Project)
Additionally, computational estimations suggest that about one-third of
the genome (estimated from chr1 of the human genome) is inaccessible
when Hi-C is performed with a 4-base restriction enzyme (RE), resulting in
potentially limited interaction signal for one-third of genomes. Arima-HiC
overcomes this limitation by using a unique RE cocktail and generates near
complete genome accessibility.
Demonstrated Utility
The ease-of-use, consistency, and proven performances of Arima-HiC
workflow, and, the open-source Bioinformatics strategy have made the
Arima-HiC kits and services as a highly popular choice for generating
chromosome-spanning assemblies. Indeed, Arima-HiC has been selected
by the Genome10K consortium to generate high-quality assemblies and
base-polishing of >260 vertebrate species
6
.
An additional benefit is that the same Arima-HiC data generated for genome
scaffolding can then be used to investigate the 3D conformation of the
genome. Chromatin conformation information is useful for many studies
including cell development, gene regulation, and pathogen resistance.
Additional Details
Please refer to the Genome Conformation and Low Input Application Notes,
available by contacting info@arimagenomics.com.
Learn more online at arimagenomics.com
crosslink
DNA
digest with
Arima RE Cocktail
fill ends &
mark with biotin
purify, shear DNA &
pull down biotin
ligate
ligate adapters
PCR amplify
library
paired-end
sequencing
6-hour workflow using Arima-HiC kit
1-day workflow using commercial library
prep kits (e.g. KAPA Hyper Prep)
Figure 1: The Arima-HiC workflow results in ligated and biotinylated DNA that is prepared as a library and amplified using
pre-validated library prep kits, and then subject to paired-end Illumina sequencing.
Figure 2: The Arima-HiC workflow results in ligated and biotinylated DNA that is prepared as a library and amplified using pre-validated library prep kits,
and then subject to paired-end Illumina sequencing.
Figure 4: 125 Mb NG50 Human chromosome-spanning assemblies,
generated by Arima-HiC data 4Mb contigs from Oxford Nanopore converted to
125Mb assemblies via Arima-HiC data, using SALSA open-source tool. Charge in
color in the ideogram represents a contig gap or error. Post Arima-HiC assemblies
have minimal color change, suggesting long (NG50 125Mb and chromosome-span-
ning) & accurate assemblies. Figure from Ref (3), shared with permission.
References
1. Lieberman-Aiden E, et al “Comprehensive Mapping of Long-Range Interactions Reveals Folding
Principles of the Human Genome” Science 326, 289-293 (2009)
2. Rao SP, et al “A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of
Chromatin Looping” Cell 159, 1665-1680 (2014)
3. Ghurye J, et al “Integrating Hi-C links with assembly graphs for chromosome-scale assembly”
BioRxiv, doi: http://dx.doi.org/10.1101/261149 (2018)
4. SALSA Bioinformatics Tool for Arima-HiC. https://github.com/machinegun/SALSA
5. Dudchenko O, et al "De novo assembly of the Aedes aegypti genome using Hi-C yields chromo-
some-length scaffolds" Science 356, 92-95 (2017)
6. Genome10K Press Release (1). https://www.businesswire.com/news/home/20180117006561/en
Table 1: Arima-HiC Specifications
Total Time 6 hours
Hands-on Time 1 hour
Number of Steps 8
Automation Capability Single–tube, 96-well plate compatible
Restriction Enzymes (RE) RE cutting at GATC and GANTC
Per-base Genome Uniformity “Spread”
(fraction of genome with avg. seq. depth)
~90%, as estimated from Human genome. Similar
performance expected in all species.
Sample Types Seeds, Tissue, blood, cell lines, whole insects
Sample Storage Conditions Fresh/frozen, Cross-linked, Ethanol
Input Quantity Amount of sample manifesting ~1ug DNA, low input
protocols also available
Species Plants, Invertebrates, Vertebrates
Library Prep Compatibility KAPA Hyperprep, Swift Accel NGS 2S, Illumina TruSeq,
NEBNext Ultra II, others
NGS Compatibility Illumina NGS paired end reads
Library Complexity 1 reaction provides libraries complex enough for 600M
reads
Sequencing Depth (X), recommended Arima-HiC: 15-60X, depending on the contig NG50
Data Analysis SALSA tool (Ref 3) and other open source tools
recommended
Figure 3: Arima-HiC data generates higher inter-contig signal strength
and spread, critical features that enable chromosome-spanning
assemblies at reduced sequencing. (A) Arima-HiC data and Competitor’s Hi-C
(data generated by Company D) is mapped to Hummingbird and Zebrafinch
contigs generated by Pacific Biosciences Sequencers. Regardless of the species,
Arima-HiC consistently generates higher inter-contig signal. (B) Hummingbird
Hi-C datasets from Arima Genomics and Competition analyzed in the context of
insert-sizes. That is, when Hi-C reads are categorized by insert-sizes, Arima-HiC
maintains high signal (2-3 fold) & coverage regardless of insert-size. In the
context of assembly, this “spread” of signal can enable contigs of all gap-sizes to
be well-assembled, to generate accurate assemblies at reduced sequencing cost.
To enable an unbiased analyses, these were performed with 2M Hi-C reads.
Analyses performed by Arang Rhie (NIH). Shared with permission.
A
Inter-contig signal
0
20
40
60
Hummingbird Zebrafinch
B
Per-base coverage
5
10
15
20
0
>100 >10,000 >1,000,000
Inserts or Gap-Size
Company D
Arima-HiC

