Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
2
This product is intended for research use only. This product is not intended for diagnostic purposes.
This document and its contents are proprietary to Arima Genomics, Inc (“Arima Genomics”). Use of this
document is intended s
olely for Arima Genomics customers for use with the Arima-HiC Kit, PN A510008, and
for no other purpose. This document and its contents shall not be used, distributed or reproduced in whole
or in part and/or otherwise communicated or disclosed without the prior written consent of Arima Genomics.
This user manual must be read in advance of using the product and strictly followed by qualified and properly
trained personnel to ensure proper use of the Arima-HiC kit. Failure to do so may result in damage to the
product, injury to persons, and/or damage to other property. Arima Genomics does not assume any liability
resulting from improper use of its products or others referenced herein.
U.S. Patent No. US 9,434,985 pertains to the use of this product.
TRADEMARKS
Illumina
Ò
, MiSeq
Ò
, NextSeq
Ò
, HiSeq
Ò
, and NovaSeq
Ô
are trademarks of Illumina, Inc.
AMPure
Ò
is a trademark of Beckman Coulter, Inc.
KAPA
Ò
is a trademark of Roche Molecular Systems, Inc.
Qubit
Ò
is a trademark of Molecular Probes, Inc.
SSIbio
Ò
is a trademark of Scientific Specialties, Inc.
Bio-Rad
Ò
is a trademark of Bio-Rad Laboratories, Inc.
Fisher Scientific
Ò
is a trademark of Fisher Scientific Company, LLC.
NEB
Ò
is a trademark of New England Biolabs, Inc.
©
2019, Arima Genomics, Inc. All rights reserved.
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
3
Revision History
Document
Date
Description of Change
Material Part Number:
A510008
Document Part Number:
A160134 v00
November
2018
Initial Release
Material Part Number:
A510008
Document Part Number:
A160134 v01
October
2019
• Added BSA to crosslinking buffer in Crosslinking – Low Input
section
.
• Added a step to Crosslinking – Low Input section.
• Added Crosslinking – Cryopreserved Cells section.
• Added guidance for optional overnight Enzyme D incubation
in Arima-HiC Protocol section.
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
4
Table of Contents
Introduction ........................................................................................................................................... 5
Arima-HiC Quick Reference Protocol .................................................................................................... 6
Arima-HiC Kit Contents and Storage Info .............................................................................................. 7
Getting Started ................................................................................................................................... 8-9
Crosslinking – Standard Input .............................................................................................................. 10
Crosslinking – Low Input ...................................................................................................................... 11
Crosslinking – Cryopreserved Cells ..................................................................................................... 12
Estimating Input Amount – Standard Input .................................................................................... 13-14
Arima-HiC Protocol ......................................................................................................................... 15-17
Arima-QC1 Quality Control ................................................................................................................. 18
Warranty and Contact Info ................................................................................................................... 19
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
5
Introduction
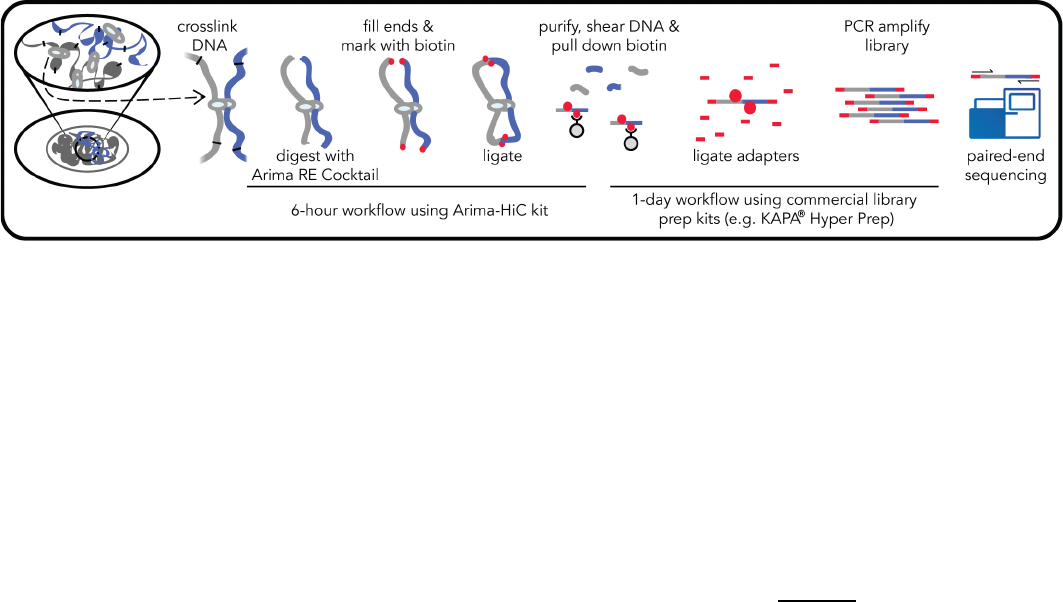
1.1 Arima-HiC Workflow Overview
Arima-HiC is an experimental workflow that captures the sequence and structure (three-dimensional
conformation) of genomes. Arima-HiC has been successfully performed on a wide-range of species
from the plant and animal kingdoms. As illustrated in the Arima-HiC workflow schematic above,
chromatin from a sample source (tissues, cell lines, or blood) is first crosslinked to preserve the
genome sequence and structure. The crosslinked chromatin is then digested using a restriction
enzyme (RE) cocktail. The 5’-overhangs are then filled in, causing the digested ends to be labeled
with a biotinylated nucleotide. Next, spatially proximal digested ends of DNA are ligated, capturing
the sequence and structure of the genome. The ligated DNA is then purified, producing pure
proximally-ligated DNA. The proximally-ligated DNA is then fragmented, and the biotinylated
fragments are enriched. The enriched fragments are then subjected to a
custom
library preparation
protocol utilizing a range of supported commercially available library prep kits. Depending on the
choice of library prep kit, a separate Arima-HiC Library Prep
user guide is provided that contains a
custom protocol for converting proximally-ligated DNA to Arima-HiC libraries.
1.2 Sequencing and Data Analysis
Arima-HiC libraries are sequenced via Illumina
Ò
sequencers in “paired-end” mode. The resulting
data is referred to as Arima-HiC data. The tools necessary for analyzing Arima-HiC data depend on
the application. For example, for studying 3D genome conformation, Arima-HiC data can be
processed using publicly available tools such as Juicer (Durand, 2016a) or Hi-C Pro (Servant, 2015),
and genome organizational features such as compartments, TADs, and loops can be identified and
visualized using tools such as Juicebox (Durand, 2016b). These tools require usage modifications
and/or custom input files that are specific to Arima-HiC data, so please contact Technical Support for
assistance implementing these tools. Additionally, because paired-end reads of Arima-HiC data can
originate from distal sequences along the linear genome, these data capture short- and long-range
DNA contiguity information that is valuable for applications such as de novo assembly and genome
scaffolding. Therefore, Arima-HiC data can be mapped to contigs/unitigs using our mapping
pipeline (https://github .com/ArimaGenomics) or Juicer, and then the contigs/unitigs can be
scaffolded using tools such as SALSA (Ghurye, 2019) or 3D-DNA (Dudchenko, 2017). Please contact
Technical Support for more information.
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
6
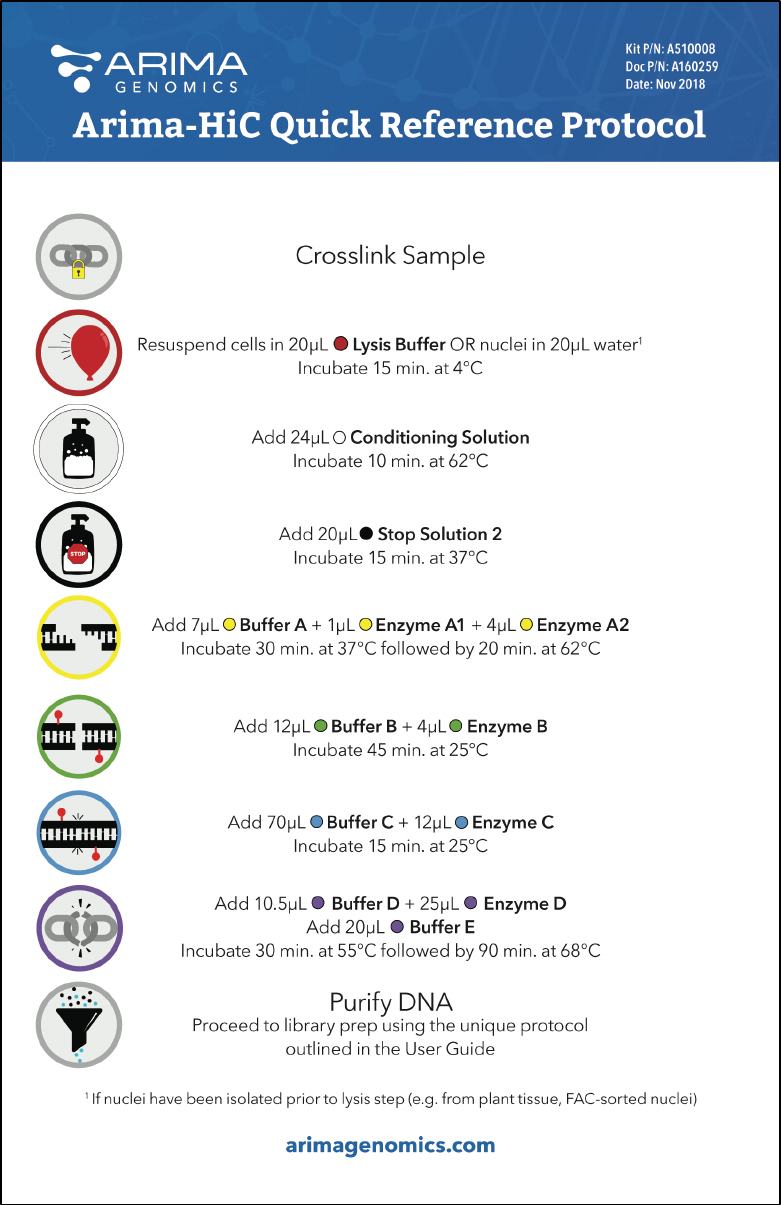
Arima-HiC Quick Reference Protocol
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
7
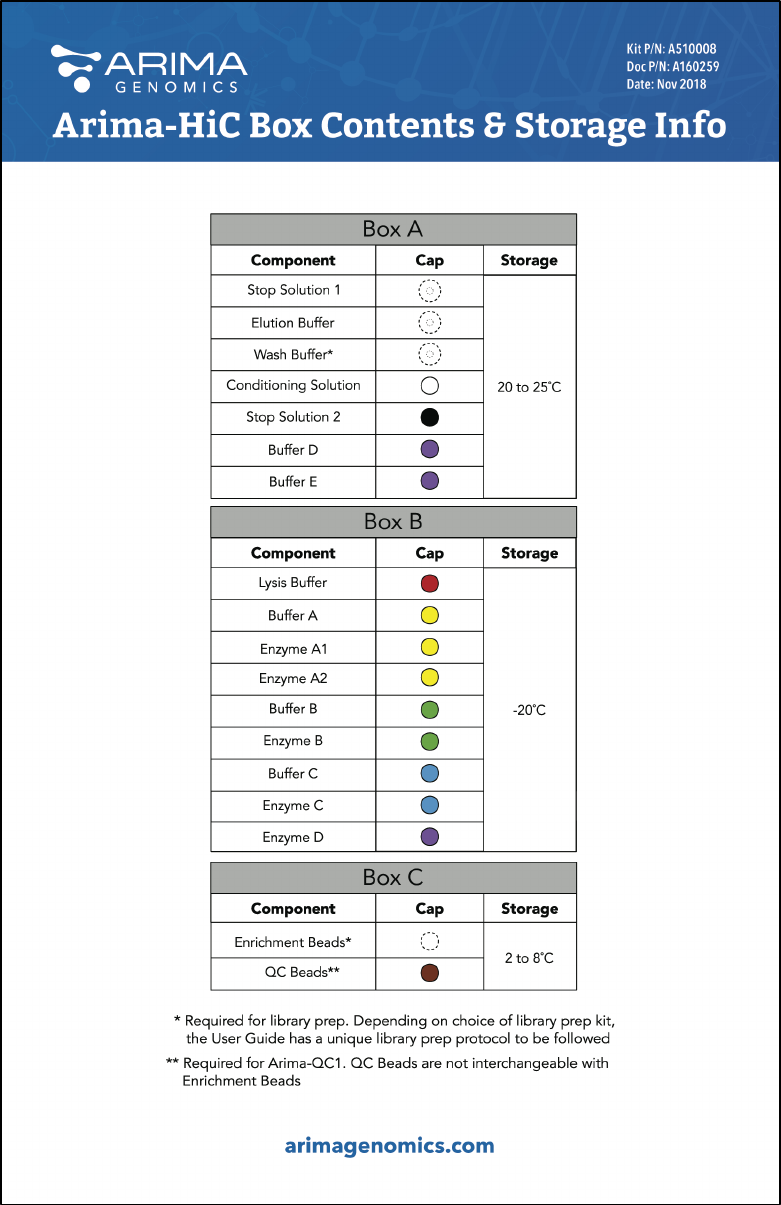
Arima-HiC Kit Contents and Storage Info

Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
8
Getting Started
2.1 Handling and Preparation
• Several steps during the Arima-HiC Protocol require preparation of a master mix. Sufficient
reagent has been included in the kit to make master mixes with 10% excess volume. Use
the master mix calculation tables provided.
• When handling reagents, room temperature (RT) is defined as 20 to 25°C.
• If the Arima-HiC Protocol is performed in PCR plates or PCR tubes, ensure to have a total
volume capacity of at least 320µL. See Section 2.2 for recommended PCR plates and PCR
tubes. Also, ensure that plates and/or tubes are compatible with thermal cyclers and other
required equipment. Using seals and caps for PCR plates and tubes is required.
• All kit reagents should be fully thawed and thoroughly mixed before use.
•
Stop Solution 1
,
Conditioning Solution
,
and
Buffer D
from
Box A
may contain
precipitates. If present, these precipitates must be dissolved before use. Heating these
reagents at 37°C for 5-15 minutes may be necessary to dissolve precipitates.
• During handling and preparation, reagents from
Box A
should be kept at RT.
• During handling and preparation, reagents from
Box B
should be kept on ice, except for
Enzyme D,
which should be kept on ice but warmed to room temperature just before use.
• Enzyme solutions from
Box B
are viscous and require special attention during pipetting.
2.2 User-supplied reagents, consumables, and equipment checklist
1X PBS, pH 7.4 (e.g. Fisher Scientific Cat # 50-842-949)
37% Formaldehyde (e.g. Fisher Scientific Cat # F79-500)
Freshly prepared 80% Ethanol
DNA Purification Beads (e.g. Beckman Coulter Cat # A63880)!
Qubit
Ò
Fluorometer, dsDNA HS Assay Kit and consumables (e.g. Thermo Fisher Scientific
Cat # 32851, 32856)
Liquid Nitrogen or dry ice
15mL conical tubes
1.7mL microcentrifuge tubes, PCR tubes (SSIbio
Ò
Cat #
3247-00), or PCR plates (Bio-Rad
Ò
Cat # HSS9641) and magnetic rack compatible with tube selection.
Centrifuge
Thermal cycler (if performing Arima-HiC in PCR tubes or PCR plate)
Thermomixer (if performing Arima-HiC in 1.7mL microcentrifuge tubes)
BSA (e.g. NEB
Ò
Cat # B9000S)
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
9
2.3 Optimal read length, sequencing depth, and number of Arima-HiC reactions per sample
Arima-HiC libraries must be sequenced in paired-end mode, and are compatible with most Illumina
Ò
sequencing machines (e.g. MiSeq
Ò
, NextSeq
Ò
, HiSeq
Ò
, NovaSeq
Ô
) and a variety of read lengths. We
generally recommend 2x150bp read length on the HiSeq
Ò
or NovaSeq
Ô
instruments to optimize for
sequencing throughput and Arima-HiC data alignment quality, although shorter read lengths (e.g.
2x50bp, 2x100bp) and lower throughput instruments can certainly be used for certain applications of
Arima-HiC data such as 3D genome conformation analysis and genome scaffolding.
The optimal sequencing depth for Arima-HiC libraries also depends on the application. For studying
3D genome conformation, the ability to detect certain genome organization features depends on
the sequencing depth. For ~3Gb genomes such as mouse and human, we generally recommend
obtaining at least 600 million read-pairs per biological condition for high-resolution analyses of A/B
compartments, TADs, and chromatin loops. One way of obtaining at least 600 million read-pairs is
by combining at least 300 million read-pairs from 2 biological replicates. In doing so, you will be
able to assess the overall reproducibility of the Arima-HiC data across replicates, and then used the
combined replicate Arima-HiC dataset for high-resolution chromatin conformation analyses.
Alternatively, one can obtain at least 600 million read-pairs per biological replicate and then use the
common set of identified genome conformational features across replicates as a “high confidence”
set of structural features supported by their observation in both replicates. For lower resolution
analyses of A/B compartments and TADs, we generally recommend obtaining at least 300 million
read-pairs per biological condition. For help estimating the optimal sequencing depth for different
genome sizes or analysis goals, please contact Technical Support.
For applications such as de novo assembly and genome scaffolding, the required sequencing depth
can vary depending on the quality of contig/unitigs that are being scaffolded using Arima-HiC data.
For a 3Gb genome, we recommend obtaining up to 600M read-pairs, as this is the amount of
sequencing that is currently utilized from Arima-HiC libraries for genome scaffolding by the
Vertebrate Genome Project (VGP) consortia. The amount of sequencing required scales linearly with
the genome size (e.g. up to 200M read-pairs for a 1Gb genome).
Lastly, it is important to note that each Arima-HiC library should pass the Arima-QC2 assay and be
evaluated for library complexity prior to deep sequencing. As a general rule, each Arima-HiC library
should be complex enough to sequence up to ~600M read-pairs without reaching saturation. If
>600M read-pairs of Arima-HiC data are needed, it may be more efficient to sequence a second
Arima-HiC library than sequence deeper into the first Arima-HiC library.
2.4 How to cite Arima-HiC in publications
When citing the Arima-HiC protocol or kit, one may write: “Hi-C data was generated using the
Arima-HiC kit, according to the manufacturers protocols”. Please reference the catalog number
found on the kit packaging.

!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
10
Crosslinking – Standard Input
Input:
Cells collected from cell culture
Output:
Crosslinked cells
Before you begin:
The Arima-HiC workflow for mammalian cell lines begins with the
harvesting and crosslinking of at least 1 million cells, but performs optimally with 5-10 million
mammalian cells. If fewer than 1 million cells are available, please follow the Crosslinking –
Low Input protocol in the following section. The crosslinking protocol below involves several
cell pelleting centrifugations. During these centrifugations, pellet your specific cell types at a
speed and duration as you normally would. Alternatively, we generally recommend
centrifuging for 5 min at 500 x G.
1. Harvest cells from cell culture using standard protocols and pellet cells by centrifugation.
2. Resuspend in cell culture media, obtain a cell count by hemocytometer or automated cell
counting methods.
3. Transfer 5-10 million cells to be crosslinked into a new 15mL conical tube, pellet cells by
centrifugation and remove supernatant.
4. Resuspend cells in 5mL of RT
1X PBS
.
5. Add 286μL of
37% formaldehyde
, bringing the final formaldehyde concentration to 2%.
6. Mix well by inverting 10 times and incubate at RT for 10 min.
7. Add 460μL of
Stop Solution 1,
mix well by inverting 10 times and incubate at RT for 5
min.
8. Place sample on ice and incubate for 15 min.
9. Pellet cells by centrifugation.
10. Discard supernatant.
11. Resuspend cells in 5mL
1X PBS
.
12. Aliquot cells into several new tubes, with 1 x 10
6
cells per aliquot. Mix sample by inversion
between aliquots to ensure all aliquots are equally homogeneous.
13. Pellet cells in all aliquots by centrifugation.
14. Discard supernatant leaving only the crosslinked cell pellet and no residual liquid.
15. Freeze samples on dry ice or liquid nitrogen, and store at -80°C until ready to proceed to
the Estimating Input Amount – Standard Input
protocol in a following section.
!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
11
Crosslinking – Low Input
Input:
Cells collected from cell culture, cell sorting, or other sources
Output:
Crosslinked cells
Before you begin:
The Arima-HiC workflow for low input mammalian cells begins with the
collection and crosslinking of fewer than 1 million cells. The crosslinking protocol below
involves several cell pelleting centrifugations. During these centrifugations, pellet your
specific cell types at a speed and duration that is higher than you normally would to ensure
minimal sample loss. Alternatively, we generally recommend centrifuging for a minimum of 5
min at 2500 x G, and faster speeds can be used to ensure maximum cell pelleting efficiency.
The cell pellets may be very difficult or impossible to see by eye, so we advise to make
special note of where the cell pellet would be expected in the bottom side of the sample tube
after centrifugation and to avoid that region when pipetting.
Note: Steps 2-4 and Step 8 involve the addition of reagents pertaining to crosslinking or
washing. Please note that each step involves mixing by inversion. Do not mix by pipetting to
ensure minimal sample loss during the crosslinking workflow.
1. Collect cells in a 1.7mL microfuge tube and pellet cells by centrifugation.
2. Add 1mL of RT
1X PBS containing 3% BSA (v/v)
and mix by inverting 5 times.
3. Add 57μL of
37% formaldehyde
,
mix well by inverting 10 times and incubate at RT for 10
min. with occasional inversion.
4. Add 91.9μL of
Stop Solution 1,
mix well by inverting 10 times and incubate at RT for 5
min. with occasional inversion.
5. Place sample on ice and incubate for 15 min.
6. Pellet cells by centrifugation.
7. Discard supernatant.
8. Add 1mL of RT
1X PBS containing 3% BSA (v/v)
and mix by inverting 5 times.
9. Pellet cells by centrifugation.
10. Discard supernatant leaving only the crosslinked cell pellet and no residual liquid.
11. Proceed directly to the Arima-HiC Protocol section, or freeze samples on dry ice or liquid
nitrogen and store at -80°C until ready to proceed to the Arima-HiC Protocol section.

!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
12
Crosslinking – Cryopreserved Cells
Input:
Cryopreserved cells
Output:
Crosslinked cells
Before you begin:
We recommend that the Arima-HiC workflow for mammalian cell lines
begin with the crosslinking of cells harvested from cell culture, however, under certain
circumstances one can also crosslink cells preserved in a cryogenic “freeze” media such as a
mixture of complete cell culture media, FBS, and DMSO. A typical example would be cells
that were once cultured and then collected at 5 million cells per mL in cryogenic “freeze”
media, and stored in a liquid nitrogen tank. The crosslinking protocol below involves several
cell pelleting centrifugations. During these centrifugations, pellet your specific cell types at a
speed and duration as you normally would. Alternatively, we generally recommend
centrifuging for 5 min at 500 x G.
1. Fill a 15mL conical tube with 4mL of
1X PBS
.
2. Thaw the cryopreserved cells in a 37°C water bath.
Note: In the following step, the entire contents of the cryopreserved cell sample (i.e. cells and
the cryogenic media) are transferred into the conical tube containing PBS. Do not centrifuge
the cells to try and remove the cryogenic freeze media. The following step also assumes the
cells are preserved in 1mL of cryogenic freeze media, and transferring the cells into the PBS
will bring the total volume to 5mL. If the cells are not frozen in 1mL of cryogenic freeze
media, adjust the volume of PBS so that the total sample volume after Step 3 will be 5mL.
3. Gently transfer cells, including the cryogenic freeze media, into the conical tube
containing 4mL of
1X PBS
, bringing the total volume to 5mL.
4. Add 286μL of
37% formaldehyde
, bringing the final formaldehyde concentration to 2%.
5. Mix well by inverting 10 times and incubate at RT for 10 min.
6. Add 460μL of
Stop Solution 1,
mix by inverting 10 times and incubate at RT for 5 min.
7. Place sample on ice and incubate for 15 min.
8. Pellet cells by centrifugation and discard supernatant.
9. Resuspend cells in 5mL
1X PBS
.
10. Aliquot cells into several new tubes, with 1 x 10
6
cells per aliquot. Mix sample by inversion
between aliquots to ensure all aliquots are equally homogeneous.
11. Pellet cells in all aliquots by centrifugation.
12. Discard supernatant leaving only the crosslinked cell pellet and no residual liquid.
13. Freeze samples on dry ice or liquid nitrogen, and store at -80°C until ready to proceed to
the Estimating Input Amount – Standard Input
protocol in a following section.
!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
13
Estimating Input Amount – Standard Input
Input:
Crosslinked cells
Output:
Purified genomic DNA
Before you begin:
The Estimating Input Amount protocol is required if one does not know
how many crosslinked cells will comprise 500ng-5µg of DNA, and if sufficient cells are
available to perform this protocol. Arima-HiC reactions are optimally performed on
crosslinked cells comprising ~500ng-5µg of DNA. The Estimating Input Amount protocol
measures the amount of DNA obtained per 1 x 10
6
crosslinked cells, which guides the
calculation of the optimal cellular input for an Arima-HiC reaction. The Arima-HiC kit contains
enough reagents to perform this protocol on 8 samples. This protocol concludes with a
descriptive example of how to estimate the optimal number of crosslinked cells to use per
Arima-HiC reaction.
Note: Step 2 requires addition of several reagents in the same step. These reagents should
be combined into master mixes with 10% excess volume before use.
1. Thaw one aliquot of 1 x 10
6
cells prepared during the Crosslinking – Standard Input
or
Crosslinking – Cryopreserved Cells protocol.
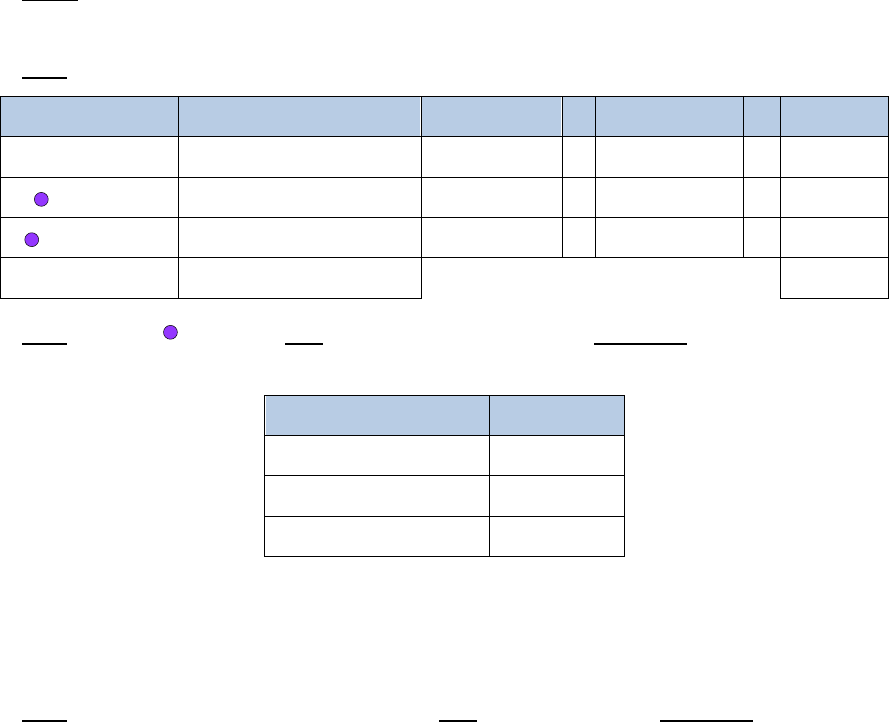
2. Add 209.5µL of a master mix containing the following reagents:
Reagent
Volume per reaction
10% extra
# reactions
Final
Elution Buffer
174µL
191.4µL
x
2
=
382.8µL
Buffer D
10.5µL
11.55µL
x
2
=
23.1µL
Enzyme D
25µL
27.5µL
x
2
=
55µL
Total
209.5
µ
L
460.9µL
3. Add 20µL of
Buffer E
, mix gently by pipetting, and incubate as follows. If using a
thermal cycler, set the lid temperature to 85°C.
Temperature
Time
55°C
30 min.
68°C
90 min.
4°C
∞
Note: DNA Purification Beads (e.g. AMPure
Ò
XP Beads) should be warmed to RT and
thoroughly mixed before use. The DNA Purification Beads are a user-supplied reagent and
should not be mistaken for the Enrichment Beads or QC Beads provided in the Arima-HiC kit.
4. Add 150µL of
DNA Purification Beads
, mix thoroughly, and incubate at RT for 5 min.

!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
14
5. Place sample against magnet, and incubate until solution is clear.
6. Discard supernatant. While sample is still against magnet, add 400µL of 80% ethanol, and
incubate at RT for 1 min.
7. Discard supernatant. While sample is still against magnet, add 400µL of 80% ethanol, and
incubate at RT for 1 min.
8. Discard supernatant. While sample is still against magnet, incubate beads at RT for 3 – 5
min. to air-dry the beads.
9. Remove sample from magnet, resuspend beads thoroughly in 20µL of
Elution Buffer
, and
incubate at RT for 5 min.
10. Place sample against magnet, incubate until solution is clear, and transfer supernatant to
a new tube.
11. Quantify sample using Qubit
Ò
. The total DNA yield corresponds to the amount of DNA
obtained from 1 x 10
6
mammalian cells.
12. Estimate how many mammalian cells to use per Arima-HiC reaction. See the example
description below:
Example:
In the following Arima-HiC Protocol, it is recommended to use crosslinked cells
corresponding to at least 500ng of DNA per Arima-HiC reaction, but no more than 5µg of
DNA. If 250ng of DNA was obtained per 1 x 10
6
mammalian cells as calculated in step 11,
one can estimate that at least 2 x 10
6
crosslinked cells should be used per Arima-HiC
reaction (~500ng of DNA). More crosslinked cells should be used if available, as long as
the total DNA per reaction is not more than 5µg. If possible, we recommend aiming to
use crosslinked cells comprising 3µg of DNA per Arima-HiC reaction. Additionally, please
note that the crosslinked cell pellet for one Arima-HiC reaction should occupy no more
than 20µL of volume in the sample tube. If the crosslinked cell pellet comprises 500ng-
5µg of DNA but occupies greater than 20µL of volume, aliquot the cells into multiple
Arima-HiC reactions such that the sum of the DNA input from all reactions is at least
500ng and each cell pellet occupies no more than 20µL of volume, or contact Technical
Support for additional guidance.
Recommended HiC Input Amount Explanation:
The recommendation to use crosslinked
cells comprising at least 500ng of DNA is only a general recommendation. If crosslinked
cells comprising at least 500ng of DNA cannot be obtained, one should proceed with the
Arima-HiC Protocol as described in this user guide and then use our validated low-input
library prep protocol.

!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
15
Arima-HiC Protocol
Input:
Crosslinked cells containing ~500ng-5µg of DNA
Output:
Proximally-ligated DNA
Before you begin:
The cell pellet for one Arima-HiC reaction should occupy no more than
20µL of volume and should be devoid of any residual liquid. If the cell pellet occupies greater
than 20µL of volume, aliquot the cells such that the sum of the DNA input from all reactions is
between 500ng-5µg and each cell pellet occupies no more than 20µL of volume, or contact
Technical Support for additional guidance. Note that steps 2 – 3 require consecutive heated
incubations. Make sure your thermal device(s) are set to 62°C and 37°C for these incubations.
The safe stopping point in this section is after completing Step 21.
Note: Choose to perform either Step 1a if the input sample type is crosslinked cells, or Step
1b only if the input sample type is crosslinked nuclei that have been previously purified from
cells.
1a. Resuspend one reaction of crosslinked cells in 20µL of
Lysis Buffer
in a tube or a well of
a PCR plate, and incubate at 4°C for 15 min.
1b. Resuspend one reaction of purified crosslinked nuclei in 20µL of
Water
in a tube or a well
of a PCR plate and procced to the next step.
2. Add 24µL of
Conditioning Solution
, mix gently by pipetting, and incubate at 62°C for
10 min. If using a thermal cycler, set the lid temperature to 85°C.
3. Add 20µL of
Stop Solution 2
, mix gently by pipetting, and incubate at 37°C for 15 min.
If using a thermal cycler, set the lid temperature to 85°C.
Note: Steps 4, 6, 8 and 10 require addition of several reagents in the same step. These
reagents should be combined into master mixes following the master mix tables.
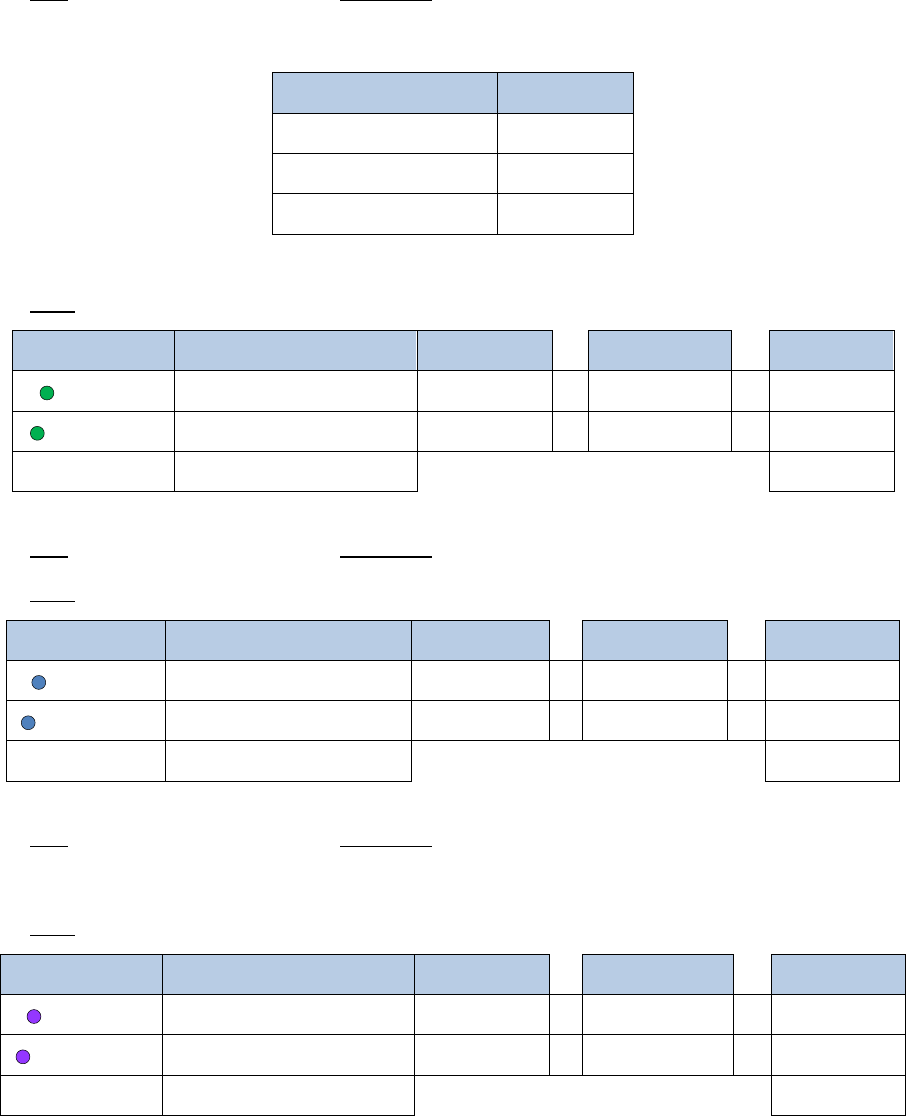
4. Add 12µL of a master mix containing the following reagents:
Reagent
Volume per reaction
10% extra
# reactions
Final
Buffer A
7µL
7.7µL
x
2
=
15.4µL
Enzyme A1
1µL
1.1µL
x
2
=
2.2µL
Enzyme A2
4µL
4.4µL
x
2
=
8.8µL
Total
12
µ
L
26.4µL
!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
16
5. Mix gently by pipetting, and incubate as follows. If using a thermal cycler, set the lid
temperature to 85°C. Note that there are sequential incubations at different
temperatures:
Temperature
Time
37°C
30 min.*
65°C
20 min.
25°C
10 min.
* Optimal performance can be achieved with incubation durations between 30 and 60min.
6. Add 16µL of a master mix containing the following reagents:
Reagent
Volume per reaction
10% extra
# reactions
Final
Buffer B
12µL
13.2µL
x
2
=
26.4µL
Enzyme B
4µL
4.4µL
x
2
=
8.8µL
Total
16
µ
L
35.2µL
7. Mix gently by pipetting, and incubate at room temperature (RT) for 45 min.
8. Add 82µL of a master mix containing the following reagents:
Reagent
Volume per reaction
10% extra
# reactions
Final
Buffer C
70µL
77µL
x
2
=
154µL
Enzyme C
12µL
13.2µL
x
2
=
26.4µL
Total
82
µ
L
180.4µL
9. Mix gently by pipetting, and incubate at RT for 15 min.
Note: Enzyme D should be warmed to RT to prevent precipitation in the below master mix.
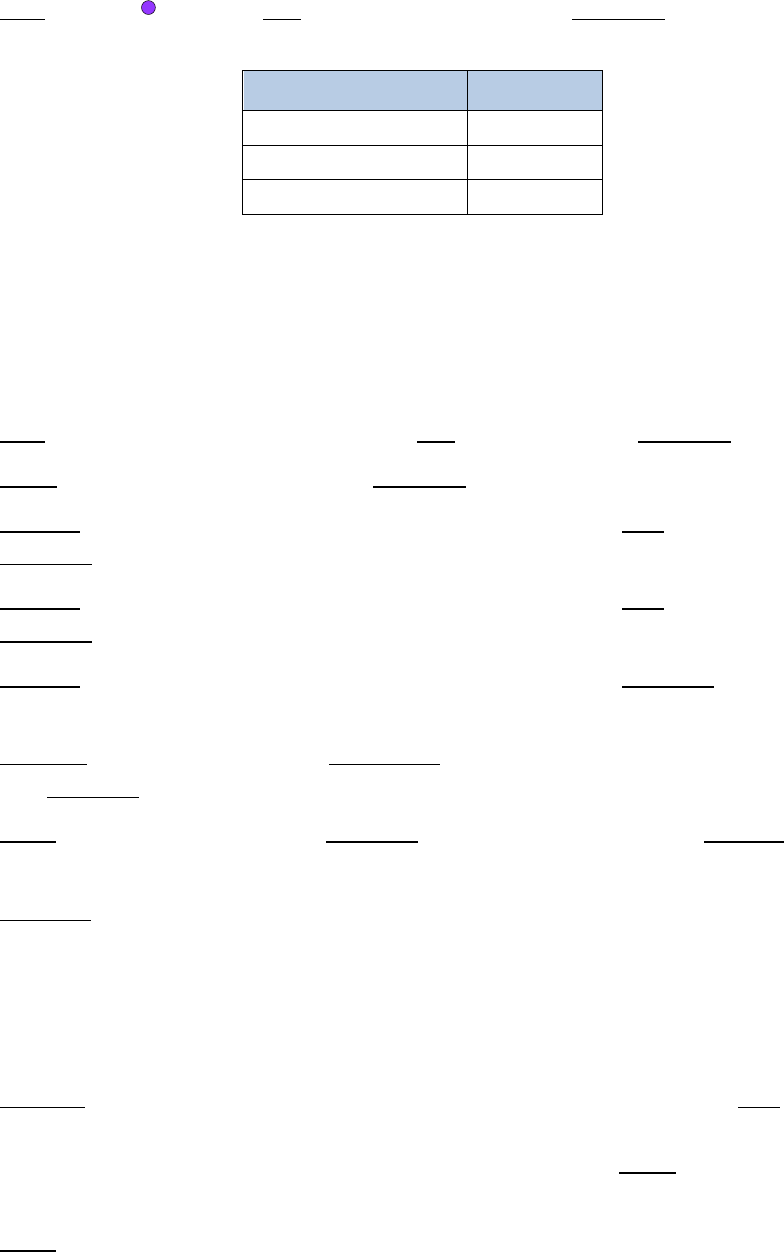
10. Add 35.5µL of a master mix containing the following reagents:
Reagent
Volume per reaction
10% extra
# reactions
Final
Buffer D
10.5µL
11.55µL
x
2
=
23.1µL
Enzyme D
25µL
27.5µL
x
2
=
55µL
Total
35.5
µ
L
78.1µL
!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
17
11. Add 20µL of
Buffer E
, mix gently by pipetting, and incubate as follows. If using a
thermal cycler, set the lid temperature to 85°C.
Temperature
Time
55°C
30 min.
68°C
90 min.
25°C*
10 min.*
* To provide flexibility, this incubation can also be held overnight at 4°C. Do not incubate at
68°C for longer than 90 min. unless doing so using a thermal cycler with a heated lid.
Note: DNA Purification Beads (e.g. AMPure
Ò
XP Beads) should be warmed to RT and
thoroughly mixed before use. The DNA Purification Beads are a user-supplied reagent and
should not be mistaken for the Enrichment Beads or QC Beads provided in the Arima-HiC kit.
12. Add 100µL of
DNA Purification Beads
, mix thoroughly, and incubate at RT for 5 min.
13. Place sample against magnet, and incubate until solution is clear.
14. Discard supernatant. While sample is still against magnet, add 300µL of 80% ethanol, and
incubate at RT for 1 min.
15. Discard supernatant. While sample is still against magnet, add 300µL of 80% ethanol, and
incubate at RT for 1 min.
16. Discard supernatant. While sample is still against magnet, incubate beads at RT for 3 – 5
min. to air-dry the beads.
17. Remove sample from magnet, resuspend beads thoroughly in 100µL of
Elution Buffer
,
and incubate at RT for 5 min.
18. Place sample against magnet, incubate until solution is clear, and transfer supernatant to
a new tube.
19. Quantify sample using Qubit
Ò
.
Note: If the proximally-ligated DNA yield is less than 275ng, we recommend skipping the
Arima-QC1 assay mentioned in Step 20 and described in the following Arima-QC1 Quality
Control section, and strongly recommend performing the Arima-QC2 assay described in our
Arima-HiC Library Preparation user guide for low input samples.
20. Transfer 75ng of sample into a new tube labelled “Arima-QC1
”,
and add
Elution Buffer
to
Arima-QC1 to bring the volume to 50µL. The “Arima-QC1” sample should now contain
75ng of proximally-ligated DNA in 50µL of
Elution Buffer
. Store at -20°C until use in the
following Arima-QC1 Quality Control protocol.
21. Store all remaining samples at -20°C until ready to proceed to library preparation
following an accompanying Arima-HiC Library Preparation
user guide
.

!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
18
Arima-QC1 Quality Control
Before you begin:
The following protocol quantifies the fraction of proximally-ligated DNA
that has been labeled with biotin, and is a quality control metric after completing the Arima-
HiC Protocol but before proceeding to library preparation. The Arima-QC1 Quality Control
protocol involves using
QC Beads
to enrich an aliquot of proximally-ligated DNA, which is
then quantified using a Qubit
Ò
fluorometer. Unlike standard Qubit
Ò
readings which involve
quantifying a transparent unobstructed DNA sample, the Arima-QC1 value is obtained by
quantifying DNA that is still bound to the
QC Beads
. This protocol can be performed in either
plates or tubes. Set your thermal device (thermal cycler or thermomixer) to hold at 55°C. After
completing the Arima-QC1 Quality Control protocol, use the provided
Arima-HiC QC
Worksheet
to determine the Arima-QC1 values.
1. If necessary, thaw the “Arima-QC1” samples prepared during Step 20 of the Arima-HiC
Protocol in the previous section.
2. Add 50µL of
QC Beads
, mix thoroughly by pipetting, and incubate at RT for 15 min.
3. Place sample against magnet, and incubate until solution is clear.
4. Discard supernatant, and remove sample from magnet.
5. Wash beads by resuspending in 200µL of
Wash Buffer
, and incubate at 55°C for 2 min.
6. Place sample against magnet, and incubate until solution is clear.
7. Discard supernatant, and remove sample from magnet.
8. Wash beads by resuspending in 200µL of
Wash Buffer
, and incubate at 55°C for 2 min.
9. Place sample against magnet, and incubate until solution is clear.
10. Discard supernatant, and remove sample from magnet.
11. Wash beads by resuspending in 100µL of
Elution Buffer
.
12. Place sample against magnet, and incubate until solution is clear.
13. Discard supernatant, and remove sample from magnet.
14. Resuspend beads in 7µL of
Elution Buffer.
Proceed to next step with resuspended beads.
Note: The following step involves the quantification of the bead-bound DNA using the
Qubit
Ò
dsDNA HS Assay Kit
.
15. Quantify the total amount of bead-bound DNA using Qubit
Ò
. Use 2µL of thoroughly
mixed bead-bound DNA for the Qubit
Ò
assay.
16. Determine the
Arima-QC1
value by following the
Arima-HiC QC Worksheet
. High quality
Arima-QC1 values are expected to be >15%. If the Arima-QC1 value did not obtain a
‘PASS’ status, please contact Technical Support for troubleshooting assistance.
!
Arima-HiC Kit
User Guide Mammalian Cell Lines
Doc A160134 v01
19
Warranty and Contact Info
WARRANTY DISCLAIMERS
THE EXPRESS WARRANTIES AND THE REMEDIES SET FORTH ABOVE ARE IN LIEU OF, AND ARIMA
GENOMICS AND ITS LICENSORS, SUPPLIERS AND REPRESENTATIVES HEREBY DISCLAIM, ALL OTHER
REMEDIES AND WARRANTIES, EXPRESS, STATUTORY, IMPLIED, OR OTHERWISE, INCLUDING, BUT NOT
LIMITED TO, ANY WARRANTIES OF MERCHANTABILITY, SATISFACTORY QUALITY, NONINFRINGEMENT
OR FITNESS FOR A PARTICULAR PURPOSE, OR REGARDING RESULTS OBTAINED THROUGH THE USE OF
ANY PRODUCT OR SERVICE (INCLUDING, WITHOUT LIMITATION, ANY CLAIM OF INACCURATE, INVALID
OR INCOMPLETE RESULTS), IN EACH CASE HOWEVER ARISING, INCLUDING WITHOUT LIMITATION
FROM A COURSE OF PERFORMANCE, DEALING OR USAGE OF TRADE, OR OTHERWISE. TO THE
MAXIMUM EXTENT PERMITTED BY APPLICABLE LAW, ARIMA AND ITS LICENSORS, SUPPLIERS AND
REPRESENTATIVES SHALL NOT BE LIABLE FOR LOSS OF USE, PROFITS, REVENUE, GOODWILL, BUSINESS
OR OTHER FINANCIAL LOSS OR BUSINESS INTERUPTION, OR COSTS OF SUBSTITUTE GOODS OR
SERVICES, OR FOR ANY SPECIAL, CONSEQUENTIAL, INCIDENTAL, EXEMPLARY OR INDIRECT DAMAGES
FOR BREACH OF WARRANTY.
WARRANTY
All warranties are personal to the Purchaser and may not be transferred or assigned to a third-party, including
an affiliate of the Purchaser. The warranty described below excludes any stand-alone third-party goods that
may be acquired or used with the Product. Arima Genomics only warrants that the kit reagents will be made
and tested in accordance with Arima Genomics manufacturing and quality control processes. Arima Genomics
makes no warranty that the reagents provided in this kit will work as intended by the Purchaser or for the
Purchaser’s intended uses. ARIMA GENOMICS MAKES NO OTHER WARRANTY, EXPRESSED OR IMPLIED.
THERE IS NO WARRANTY OF MERCHANTABILITY OR FITNESS FOR A PARTICULAR PURPOSE. The warranty
provided herein and the data and descriptions of Arima Genomics products appearing in Arima Genomics
product literature and website may not be altered except by express written agreement signed by an officer
of Arima Genomics. Representations, oral or written, which are inconsistent with this warranty or such
publications are not authorized and if given, should not be relied upon.
The foregoing warranties do not apply to the extent a non-conformance is due to (i) abuse, misuse, neglect,
negligence, accident, improper storage, or use contrary to the Documentation or Specifications, (ii) use that is
an Excluded Use, (iii) improper handling, (iv) unauthorized alterations, (v) natural disasters, or (vi) use with a
third-party’s good that is not specified in the product documentation. In the event of a breach of the
foregoing warranty, customer shall promptly contact Arima Genomics customer support to report the non-
conformance and shall cooperate with Arima Genomics in confirming or diagnosing the non-conformance.
Additionally, Arima Genomics may request return shipment of the non-conforming product at Arima
Genomics cost. Arima Genomics sole obligation shall be to replace the applicable product or part thereof,
provided the customer notifies Arima Genomics within 90 days of any such breach. If after exercising
reasonable efforts, Arima Genomics is unable to replace the product, then Arima Genomics shall refund to
the Purchaser all monies paid for such applicable product.
CONTACT US
Technical Support: tech[email protected]
Order Support: ordersupport@arimagenomics.com

