

Emerging Infectious Diseases
Emerging Infectious Diseases is published month-
ly by the National Center for Infectious Diseases,
Centers for Disease Control and Prevention (CDC),
1600 Clifton Road, Mailstop D61, Atlanta, GA 30333,
USA. Telephone 404-371-5329, fax 404-371-5449,
email [email protected].
The opinions expressed by authors contributing to
this journal do not necessarily reflect the opinions of
the Centers for Disease Control and Prevention or the
institutions with which the authors are affiliated.
All material published in Emerging Infectious
Diseases is in the public domain and may be used
and reprinted without special permission; proper cita-
tion, however, is required.
Use of trade names is for identification only and
does not imply endorsement by the Public Health
Service or by the U.S. Department of Health and
Human Services.
∞ Emerging Infectious Diseases is printed on acid-
free paper that meets the requirements of
ANSI/NISO 239.48-1992 (Permanence of Paper)
Electronic Access
Retrieve the journal electronically on the World Wide Web (WWW) at
http://www.cdc.gov/eid or from the CDC home page (http://www.cdc.gov).
Announcements of new table of contents can be automatically emailed to
you. To subscribe, send an email to [email protected] with the following
in the body of your message: subscribe EID-TOC.
Editors
D. Peter Drotman, Editor-in-Chief
Atlanta, Georgia, USA
David Bell, Associate Editor
Atlanta, Georgia, USA
Vincent Deubel, Associate Editor
Lyon, France
Stephanie James, Associate Editor
Bethesda, Maryland, USA
Brian W.J. Mahy, Associate Editor
Atlanta, Georgia, USA
Takeshi Kurata, Associate Editor
Tokyo, Japan, USA
Martin I. Meltzer, Associate Editor
Atlanta, Georgia, USA
David Morens, Associate Editor
Washington, DC, USA
Tanja Popovic, Associate Editor
Atlanta, Georgia, USA
Patricia M. Quinlisk, Associate Editor
Des Moines, Iowa, USA
Gabriel Rabinovich, Associate Editor
Buenos Aires, Argentina
Didier Raoult, Associate Editor
Marseilles, France
Pierre Rollin, Associate Editor
Atlanta, Georgia, USA
Mario Raviglione, Associate Editor
Geneva, Switzerland
Polyxeni Potter, Managing Editor
Atlanta, Georgia, USA
Joseph E. McDade, Founding Editor
Rome, Georgia, USA
Mary Anne Castranio
Carolyn P. Collins
Teresa M. Hood
Ann Kitchen
Anne D. Mather
Carol D. Snarey
Reginald S. Tucker
Cathy E. Young
Dennis Alexander, Addlestone Surrey, United Kingdom
Ban Allos, Nashville, Tennessee, USA
Michael Apicella, Iowa City, Iowa, USA
Ben Beard, Atlanta, Georgia, USA
Barry J. Beaty, Ft. Collins, Colorado, USA
Martin J. Blaser, New York, New York, USA
David Brandling-Bennet, Washington, D.C., USA
Donald S. Burke, Baltimore, Maryland, USA
Charles H. Calisher, Ft. Collins, Colorado, USA
Arturo Casadevall, New York, New York, USA
Thomas Cleary, Houston, Texas, USA
Anne DeGroot, Providence, Rhode Island, USA
Vincent Deubel, Providence, Rhode Island, USA
Ed Eitzen, Washington, D.C., USA
Duane J. Gubler, Fort Collins, Colorado, USA
Scott Halstead, Arlington, Virginia, USA
David L. Heymann, Geneva, Switzerland
Sakae Inouye, Tokyo, Japan
Charles King, Cleveland, Ohio, USA
Keith Klugman, Atlanta, Georgia, USA
S.K. Lam, Kuala Lumpur, Malaysia
Bruce R. Levin, Atlanta, Georgia, USA
Myron Levine, Baltimore, Maryland, USA
Stuart Levy, Boston, Massachusetts, USA
John S. MacKenzie, Brisbane, Australia
Tom Marrie, Edmonton, Alberta, Canada
John E. McGowan, Jr., Atlanta, Georgia, USA
Stephen S. Morse, New York, New York, USA
Philip P. Mortimer, London, United Kingdom
Fred A. Murphy, Davis, California, USA
Barbara E. Murray, Houston, Texas, USA
P. Keith Murray, Ames, Iowa, USA
Stephen Ostroff, Atlanta, Georgia, USA
Rosanna W. Peeling, Geneva, Switzerland
David H. Persing, Seattle, Washington, USA
Gianfranco Pezzino, Topeka, Kansas, USA
Richard Platt, Boston, Massachusetts, USA
Leslie Real, Atlanta, Georgia, USA
David Relman, Palo Alto, California, USA
Nancy Rosenstein, Atlanta, Georgia, USA
Connie Schmaljohn, Frederick, Maryland, USA
Tom Schwan, Hamilton, Montana, USA
Ira Schwartz, Valhalla, New York, USA
Tom Shinnick, Atlanta, Georgia, USA
Robert Shope, Galveston, Texas, USA
Bonnie Smoak, Bethesda, Maryland, USA
Rosemary Soave, New York, New York, USA
P. Frederick Sparling, Chapel Hill, North Carolina, USA
Jan Svoboda, Prague, Czech Republic
Bala Swaminathan, Atlanta, Georgia, USA
Robert Swanepoel, Johannesburg, South Africa
Phillip Tarr, Seattle, Washington, USA
Timothy Tucker, Cape Town, South Africa
Elaine Tuomanen, Memphis, Tennessee, USA
David Walker, Galveston, Texas, USA
Mary E. Wilson, Cambridge, Massachusetts, USA
Editorial Board
The print journal is available at no charge to public health professionals
YES, I would like to receive Emerging Infectious Diseases.
Please print your name and business
address in the box and return by fax to
404-371-5449 or mail to
EID Editor
CDC/NCID/MS D61
1600 Clifton Road, NE
Atlanta, GA 30333
Moving? Please give us your new address (in the box) and print the number of your old
mailing label here_________________________________________
Full text free online at
www.cdc.gov/eid
Production Editors

Perspectives
Mycobacterial Aerosols and Respiratory Disease . . . .763
J.O. Falkinham III
Global Screening for Human Viral Pathogens . . . . . . .768
N.G. Anderson et al.
Synopses
Salmonella Control Programs in Denmark . . . . . . . . . .774
H.C. Wegener et al.
Disease Surveillance and the Academic,
Clinical, and Public Health Communities . . . . . . . . . . .781
R.W. Pinner et al.
Research
Acute Flaccid Paralysis
and West Nile Virus Infection . . . . . . . . . . . . . . . . . . . .788
J.J. Sejvar et al.
West Nile Virus in Farmed Alligators . . . . . . . . . . . . . .794
D.L. Miller et al.
Emergence and Global Spread of a
Dengue Serotype 3, Subtype III Virus . . . . . . . . . . . . .800
W.B. Messer et al.
Molecular Epidemiology of O139
Vibrio cholerae: Mutation, Lateral
Gene Transfer, and Founder Flush . . . . . . . . . . . . . . .810
P. Garg et al.
Amoeba-Resisting Bacteria and
Ventilator-Associated Pneumonia . . . . . . . . . . . . . . . . .815
B. La Scola et al.
Antimicrobial Resistance Markers of Class 1
and Class 2 Integron-Bearing Escherichia coli
from Irrigation Water and Sediments . . . . . . . . . . . . . .822
M.T. Roe et al.
Hantavirus Prevalence in the IX Region of Chile . . . . .827
M. Täger Frey et al.
Antimicrobial Susceptibility Breakpoints
and First-Step parC Mutations in
Streptococcus pneumoniae: Redefining
Fluoroquinolone Resistance . . . . . . . . . . . . . . . . . . . . .833
S. Lim et al.
Mutations in Putative Mutator
Genes of Mycobacterium tuberculosis
Strains of the W-Beijing Family . . . . . . . . . . . . . . . . . .838
M.E. Rad et al.
Yellow Pigmy Rice Rat
(Oligoryzomys flavescens) and Hantavirus
Pulmonary Syndrome in Uruguay . . . . . . . . . . . . . . . .846
A. Delfraro et al.
Dispatches
Serologic Evidence of West Nile Virus
Infection in Horses, Coahuila State, Mexico . . . . . . . . .853
B.J. Blitvich et al.
Serologic Evidence of West Nile Virus
Infection in Horses, Yucatan State, Mexico . . . . . . . . .857
M.A. Loroño-Pino et al.
Serologic Evidence of West Nile
Virus Transmission, Jamaica, West Indies . . . . . . . . . .860
A.P. Dupuis II et al.
A Peer-Reviewed Journal Tracking and Analyzing Disease Trends Vol.9, No.7, July 2003
On the Cover:
Henri Rousseau—known as
Le Douanier Rousseau
(1844–1910).
The Snake Charmer (1907).
Oil on canvas,
169 cm x 189.5 cm.
Musée d’Orsay, Paris,
France. Credit: Réunion des
Musées Nationaux/Art
Resource, NY
About the Cover, pg. 901
All material published in Emerging Infectious Diseases is in the public
domain and may be used and reprinted without special permission; proper
citation, however, is appreciated.

Sulfa Resistance and Dihydropteroate
Synthase Mutants in Recurrent
Pneumocystis carinii Pneumonia . . . . . . . . . . . . . . . . .864
A. Nahimana et al.
VIM- and IMP-Type Metallo-β-lactamase–
Producing Pseudomonas spp. and
Acinetobacter spp. in Korean Hospitals . . . . . . . . . . . .868
K. Lee et al.
Leishmaniasis in Germany . . . . . . . . . . . . . . . . . . . . . .872
G. Harms et al.
Probable Dengue Virus Infection among
Italian Troops, East Timor, 1999–2000 . . . . . . . . . . . . .876
M.S. Peragallo et al.
HIV Epidemic among Young
Thai Men, 1991–2000 . . . . . . . . . . . . . . . . . . . . . . . . . .881
K. Torugsa et al.
Letters
Taenia solium Cysticercosis,
Irian Jaya, Indonesia . . . . . . . . . . . . . . . . . . . . . . . . . .884
T. Wandra et al.
Recombinant Vaccine–Derived
Poliovirus in Madagascar . . . . . . . . . . . . . . . . . . . . . . .885
D. Rousset et al.
West Nile Virus Infection in Crocodiles . . . . . . . . . . . .887
A. Steinman et al.
Rickettsia aeschlimannii in Spain:
Molecular Evidence in Hyalomma
marginatum and Five Other Tick
Species that Feed on Humans . . . . . . . . . . . . . . . . . . .889
P. Fernández-Soto et al.
Hantaviruses in São Paulo State, Brazil . . . . . . . . . . . .891
L.T.M. Figueiredo et al.
Israeli Spotted Fever Rickettsia
in Sicilian Rhipicephalus sanguineus Ticks . . . . . . . . .892
G.M. Giammanco et al.
Co-feeding Transmission and Its
Contribution to the Perpetuation of the
Lyme Disease Spirochete Borrelia afzelii
(In Reply) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .893
S. Randolph and L. Gern
Co-feeding Transmission and Its
Contribution to the Perpetuation of the
Lyme Disease Spirochete Borrelia afzelii
(In Reply to Randloph and Gern) . . . . . . . . . . . . . . . . .895
D. Richter et al.
News & Notes
Conference Summaries
West Nile Virus Southeast Conference . . . . . . . . . . . .897
D. Rimland et al.
West Nile Virus and Wildlife Health . . . . . . . . . . . . . . .898
P.P. Marra et al.
About the Cover . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .901
A Peer-Reviewed Journal Tracking and Analyzing Disease Trends Vol.9, No.7, July 2003

Environmental opportunistic mycobacteria, including
Mycobacterium avium, M. terrae, and the new species M.
immunogenum, have been implicated in outbreaks of
hypersensitivity pneumonitis or respiratory problems in a
wide variety of settings. One common feature of the out-
breaks has been exposure to aerosols. Aerosols have been
generated from metalworking fluid during machining and
grinding operations as well as from indoor swimming pools,
hot tubs, and water-damaged buildings. Environmental
opportunistic mycobacteria are present in drinking water,
resistant to disinfection, able to provoke inflammatory reac-
tions, and readily aerosolized. In all outbreaks, the water
sources of the aerosols were disinfected. Disinfection may
select for the predominance and growth of mycobacteria.
Therefore, mycobacteria may be responsible, in part, for
many outbreaks of hypersensitivity pneumonitis and other
respiratory problems in the workplace and home.
H
ypersensitivity pneumonitis is an occupational hazard
of workers in two different industries, automobile
manufacturing (e.g., metal working) and leisure (e.g.,
indoor swimming pools). Pulmonary illness and infection
have also been a consequence of exposure to aerosols gen-
erated by hot tubs, spas, and coolant baths. Respiratory
problems have also been associated with exposure to
water-damaged buildings during reconstruction, and
mycobacteria isolated from materials from such buildings
have been shown to provoke inflammatory reactions. The
outbreaks share the common feature of aerosol exposure
and respiratory illness. I propose that exposure to aerosols
containing mycobacteria is a common feature of the out-
breaks and that mycobacteria or their products could be
responsible for the respiratory symptoms.
Epidemiologic studies have established that the work-
ers in such outbreaks were exposed to aerosols generated
in the workplace from water that was either a work tool
(e.g., metalworking fluid) or an integral part of the work-
place or household (e.g., swimming pools and hot tubs)
(1–7). Outbreaks of respiratory disease occurred in spite of
disinfectant treatment of the waters or fluids to reduce the
number of microorganisms. Living or working in water-
damaged buildings or as a consequence of reconstruction
of water-damaged buildings has also been associated with
outbreaks of respiratory problems (8,9). Respiratory dis-
ease has been associated with mycobacteria in reservoirs,
aerosols, or structural material in a number of cases
(2,3,6,7,9).
Hypersensitivity Pneumonitis in Workers
Exposed to Metalworking Fluid
An estimated 1.2 million workers in the United States
are exposed to aerosols generated by metal grinding (10).
Metalworking fluids are widely used in a variety of com-
mon industrial metal-grinding operations to lubricate and
cool the tool and the working surface. Metalworking fluids
are oil-water emulsions that contain paraffins, pine oils,
polycyclic aromatic hydrocarbons, and heavy metals
(10,11). Exposure to metalworking fluid aerosols can lead
to hypersensitivity pneumonitis and chronic obstructive
pulmonary disease (1,6,12–14). Mycobacteria were recov-
ered significantly more frequently from metalworking
fluid samples collected from facilities where hypersensi-
tivity pneumonitis was found; compared to facilities that
did not have hypersensitivity pneumonitis (6). In one
study, exposure to metalworking fluid mist resulted in
hypersensitivity pneumonitis in 10 workers (7). Acid-fast
microorganisms identified as mycobacteria were present in
the reservoir at 10
7
CFU/mL (7). A mycobacteria in the
reservoir was considered to be a likely cause of the hyper-
sensitivity pneumonitis because one patient was infected
by a Mycobacterium sp. and had antibodies against the
reservoir fluid (7).
Hypersensitivity pneumonitis appeared in spite of dis-
infection of the metalworking fluid with morpholine,
formaldehyde, or quaternary ammonium-based disinfec-
tants (1,6,12,13), and mycobacteria were recovered from
the metal working fluid (6,14,15). Mycobacteria are resist-
ant to formaldehyde and quaternary ammonium disinfec-
tants (16) and the heavy metals in metalworking fluids
(17). Further, mycobacteria can grow on the organic com-
pounds in metalworking fluid, including the paraffins, pine
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 763
PERSPECTIVES
Mycobacterial Aerosols and
Respiratory Disease
Joseph O. Falkinham, III*
*Virginia Polytechnic Institute and State University, Blacksburg,
Virginia, USA

oils, and polycyclic aromatic hydrocarbons (18,19) and
can degrade the disinfectant morpholine (20).
Mycobacteria present in the water (21) can likely grow on
the organic compounds in metalworking fluids in the
absence of competitors after disinfection. Cleaning would
not be expected to eradicate mycobacteria because of their
ability to form biofilms (21,22). Adding disinfectant and
cleaning the reservoir in one facility did not prevent the
reappearance of mycobacteria (7 x 10
5
CFU/mL by 2
weeks [7]). Further, disinfectant treatment would likely
result in selection of mycobacteria remaining after the
cleaning.
Hypersensitivity Pneumonitis in
Swimming Pool Attendants
Granulomatous pneumonitis has been reported in life-
guards (“lifeguard lung”) who worked at an indoor swim-
ming pool that featured waterfalls and sprays (5). Affected
lifeguards with symptoms worked longer hours than unaf-
fected lifeguards (5), which demonstrated a dose-response
effect. The waterfalls and sprays increased the number of
respirable particles fivefold and the levels of endotoxin
eightfold (5). Based on the presence of endotoxin in the
aerosol samples, endotoxin exposure was suggested as the
cause of the pneumonitis in lifeguards (5). However, sub-
sequent data provided evidence of a possible second factor
resulting in hypersensitivity pneumonitis; aerosols con-
taining mycobacteria were shown to cause granulomatous
lung disease (4). Others have reported high numbers of
mycobacteria in swimming pools and whirlpools (23) and
in hot tubs (2,3,24). Further, amoebae were reported in the
indoor swimming pool where lifeguards reported pneu-
monitis (5). Mycobacteria, including M. avium and M.
intracellulare, can survive and grow in phagocytic amoe-
bae (25) and protozoa (26). In fact, M. avium grown in
amoebae or protozoa are more virulent (25; Falkinham JO,
unpub. data). Mycobacteria are resistant to chlorine (27)
and preferentially aerosolized from water (28).
Mycobacterial Disease after Exposure to
Aerosols Generated by Hot Tubs
Hypersensitivity pneumonitis and mycobacterial pul-
monary disease has been reported after exposure to hot
tubs (2,3,24). The mycobacteria isolated (e.g., M. avium)
were likely responsible for the infections based on the
identity of patient and hot tub mycobacterial isolates by
either restriction fragment length polymorphism analysis
(24) or multilocus enzyme electrophoresis (2,3). Further,
exposure was followed closely by the onset of symptoms,
and the extent of symptoms was related to the length of
exposure (i.e., time spent in the hot tub) (2,24). Although
these reports do not document the use of disinfectants in
the hot tubs, the waters had been heated. Mycobacteria are
relatively resistant to high temperature (29) and concen-
trated in hospital hot water systems (30).
Hypersensitivity Pneumonitis in
Occupants of Water-Damaged Buildings
Inflammatory reactions—including eye irritation, res-
piratory infections, wheeze, bronchitis, and asthma—in
workers in water-damaged or “moldy” buildings have been
associated with the presence of high numbers of microor-
ganisms (8). Mycobacteria were recovered from materials
collected from water-damaged buildings, as well as from
microorganisms normally associated with building materi-
als (9). During reconstruction, those mycobacteria could
be aerosolized in the dust. Although other microorganisms
could be responsible for the respiratory problems, both
saprophytic (e.g., M. terrae) and pathogenic (e.g., M.
avium) strains isolated from moldy buildings were capable
of inducing inflammatory responses in a mouse
macrophage cell line (31). The mycobacteria elicited dose-
dependent production of cytokines interleukin-6 and tumor
necrosis factor-α, nitric oxide, and reactive oxygen species
from the murine macrophage (31). Because whole
mycobacterial cells were used in the assays (31), whether
cell metabolites, which are likely easily aerosolized, were
responsible for the induction of inflammatory reactions is
not known. Heat-shock proteins from a number of
mycobacterial species have been shown to generate Th1-
type responses, airway inflammation, and airway hyperre-
sponsiveness (32). This evidence suggests that mycobacte-
ria or their metabolites are possible causes of respiratory
disease in persons exposed to water-damaged buildings.
Ecology of Mycobacteria
The unique combination of physiologic characteristics
that distinguish the environmental opportunistic mycobac-
teria make them likely agents for causing respiratory dis-
ease in these diverse settings. Mycobacteria are found in a
great variety of natural and human-influenced aquatic
environments, including treated drinking water (21) and
aerosols (33). Mycobacteria in drinking water are associat-
ed with the presence of particulates (21). Although these
microbes are grown in rich media in the laboratory, they
are oligotrophic and capable of substantial growth in low
concentrations of organic matter. For example, M. avium
and M. intracellulare can grow in natural and drinking
water over a temperature range of 10°C to 45°C (34).
Mycobacteria are relatively resistant to high temperatures.
For example, 10% of cells of a strain of M. avium survived
after 1 h at 55°C (29). Mycobacteria are slow growing as a
consequence of their fatty acid- and wax-rich impermeable
cell wall (35). The resulting cell surface hydrophobicity
permits adherence to solid substrates (e.g., pipes and
leaves) in aquatic environments, which results in
764 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
PERSPECTIVES

mycobacteria’s persistence and resistance to being washed
away at high flow rates (21,22). Further, hydrophobicity is
undoubtedly associated with the ability of these bacteria to
metabolize a wide variety of nonpolar organic compounds
(18–20) that are constituents of metal working fluids
(15,16).
Resistance of Mycobacteria to Disinfection
Mycobacteria are very resistant to the disinfectants
used in water treatment, including chlorine and ozone (27).
For example, M. avium is almost 500 times more resistant
to chlorine than is Escherichia coli (27). Mycobacteria are
also quite resistant to agents used for surface and instru-
ment disinfection, including quaternary ammonium com-
pounds, phenolics, iodophors, and glutaraldehyde
(16,22,23,36) and can degrade the disinfectant morpholine
(20). Hydrophobicity and impermeability are undoubtedly
factors contributing to the disinfection resistance of
mycobacteria (35). Chemical or enzymatic removal of sur-
face lipid, while not reducing viability, reduces surface
hydrophobicity and alters cell charge (37). Because of
their inherent impermeability, mycobacteria grow relative-
ly slowly compared to other bacteria. The slow growth is
not necessarily a disadvantage because it correlates with
increased resistance to antimicrobial agents (35), including
chlorine (Falkinham JO, unpub data).
Exposure of a mixed microbial population to disinfec-
tants results in selection of a disinfectant-resistant or toler-
ant population (38). The persistence and growth of
mycobacteria in drinking water systems (21) are due, in
part, to their disinfectant-resistance (27) and ability to
grow under oligotrophic conditions (21). Disinfection of
swimming pools, therapy pools, and spas or hot tubs with
chlorine is expected to kill nonmycobacterial flora and to
permit the growth of even the slowly growing mycobacte-
ria in the absence of competitors for nutrients. High tem-
perature would also be expected to result in enrichment of
mycobacteria (29,30). Resistance to disinfectants could
also lead to the proliferation of mycobacterial populations
in metal working fluid and coolants after disinfection
(6,12,13).
Aerosolization of Mycobacteria
Although M. tuberculosis is transmitted between
patients through aerosols, little information exists on
aerosolization of the environmental opportunistic
mycobacteria (e.g., M. avium and M. intracellulare).
Patient-to-patient transmission of environmental oppor-
tunistic mycobacteria does not occur (39). M. avium and M.
intracellulare are readily aerosolized from aqueous suspen-
sion (28,33). Transfer of mycobacteria occurs as a result of
binding of mycobacterial cells to air bubbles and ejection
of water droplets after the air bubbles reach the liquid sur-
face (28). Aerosolization can result in >1,000-fold increase
in numbers of viable mycobacterial cells per milliliter of
water droplets ejected from water (28). Mycobacteria in
natural aerosols are found in particles and droplets (i.e., <5
µm) that can enter the alveoli of the human lung (28,33).
Cell surface hydrophobicity, not surface charge, is a major
determinant of enrichment in ejected droplets (28).
Transfer of mycobacteria from water to air is subject to
prevailing physiochemical conditions and can be manipu-
lated. Salts (e.g., NaCl) or detergents reduce the rate of
transfer of mycobacteria from water to air by ejected
droplets (28). The influence of the components of metal-
working fluid or of chlorine or other disinfectants in water
upon aerosolization mycobacteria is unknown.
Mycobacteria and Immune Responses
and Airway Inflammation
Mycobacterial cells and cellular components provoke
inflammatory responses. Cells of mycobacterial strains
isolated from material collected from water-damaged
buildings provoke inflammatory responses in
macrophages (31). Mycobacterial heat-shock proteins gen-
erate Th1-type responses, airway inflammation, and hyper-
responsiveness (32). The mycolic acid-containing glycol-
ipids, mannose-containing phospholipids, glycopepti-
dolipid mycosides, phenolglycolipid mycosides, and sul-
fatides that are unique to mycobacteria have all been
reported to stimulate immune responses in animals (40).
Further, mycobacteria produce a variety of extracellular
primary and secondary metabolites (19) that could be
aerosolized and trigger immune responses, including
hypersensitivity pneumonitis. Some of these immunostim-
ulatory compounds are produced in response to growth on
polycyclic aromatic hydrocarbons (18). Unfortunately, the
studies of inflammatory responses provoked by mycobac-
teria have been limited to whole cells grown under a single
condition (31) or single proteins (32). The influence of
growth conditions (e.g., growth in metalworking fluid or
chlorinated water) or cell fractions (e.g., membranes) or
metabolites to stimulate inflammatory responses has not
been measured.
Conclusion
Contemporary reviews of airway dysfunction all
describe the need for information concerning microbial
agents of workplace and household exposure (41).
Although many more studies are needed, the evidence
points to a role of environmental opportunistic mycobacte-
ria in provoking hypersensitivity pneumonitis, respiratory
disease, and respiratory infection in both the workplace
and home. In addition to the recovery of identical species
and types of mycobacteria from reservoirs and patients,
physiologic characteristics of mycobacteria are consistent
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 765
PERSPECTIVES

with their presence in the sources, transmission by means
of aerosols, and illnesses. Identifying the factors that influ-
ence the presence of mycobacteria in aerosols in these
workplaces would have an impact on workers in a variety
of occupational settings.
On the basis of several physiologic and ecologic char-
acteristics of mycobacteria, several approaches to reduce
the impact of mycobacteria in these settings are possible.
Because mycobacteria are associated with particulates
(21), their numbers in reservoirs can be reduced by
removal of particular matter (e.g., filtration). UV light can
be used to reduce mycobacterial numbers. Disinfection of
mycobacteria at high temperatures (e.g., 40°C) is more
effective at reducing numbers, especially if cells were
grown at lower temperatures (e.g., 30°C). Agents or com-
binations with surfactant or detergent-like and disinfectant
activity would increase permeation in cells and biofilms
and kill more mycobacteria. Finally, aerosolized or water-
borne mycobacteria may be trapped in filters coated with
hydrophobic compounds (e.g., paraffin) and thereby inter-
cepted before inhalation or ingestion.
Dr. Falkinham is a professor of microbiology in the
Department of Biology at Virginia Polytechnic Institute and State
University. His research interests include identifying the genes
and physiologic characteristics of Mycobacterium avium that are
responsible for its ecology, transmission, and virulence.
References
1. Bernstein DI, Lummus ZL, Santilli G, Siskosky J, Bernstein IL.
Machine operator’s lung. A hypersensitivity pneumonitis disorder
associated with exposure to metalworking fluid aerosols. Chest
1995;108:636–41.
2. Embil J, Warren P, Yakrus M, Stark R, Corne S, Forrest D. Pulmonary
illness associated with exposure to Mycobacterium avium complex in
hot tub water: hypersensitivity pneumonitis or infection? Chest
1997;111:813–6.
3. Kahana LM, Kay JM, Yakrus M, Waserman S. Mycobacterium avium
complex infection in an immunocompetent young adult related to hot
tub exposure. Chest 1997;111:242–5.
4. Martyny JW, Rose CS. Nontuberculous mycobacterial bioaerosols
from indoor warm water sources cause granulomatous lung disease.
Indoor Air 1999;9:1–6.
5. Rose CS, Martyny JW, Newman LS, Milton DK, King TE Jr, Beebe
JL, et al. “Lifeguard lung”: endemic granulomatous pneumonitis in
an indoor swimming pool. Am J Public Health 1998;88:1795–800.
6. Shelton BG, Flanders WD, Morris GK. Mycobacterium sp. as a pos-
sible cause of hypersensitivity pneumonitis in machine workers.
Emerg Infect Dis 1999;5:270–3.
7. Muilenberg ML, Burge HT, Sweet T. Hypersensitivity pneumonitis
and exposure to acid-fast bacilli in coolant aerosols. J Allergy Clin
Immunol 1998;91:311.
8. Brunekreef B. Damp housing and adult respiratory symptoms.
Allergy 1992;47:498–502.
9. Andersson MA, Nikulin M, Koljalg U, Andersson MC, Rainey F,
Reijula K, et al. Bacteria, molds, and toxins in water-damaged build-
ing materials. Appl Environ Microbiol 1997;63:387–92.
10. Eisen EA, Tolbert PE, Smith TJ, Monson RR, Hallock M, Woskie SR,
et al. Mortality studies of machining fluids; an exposure-response
analysis of respiratory and digestive cancers. Proceedings of the 9th
International Symposium on Epidemiology in Occupational Health,
Sept 23-25, 1992, Cincinnati, Ohio. DHHS (NIOSH) pub no. 94-112.
Cincinnati (OH): National Institute for Occupational Safety and
Health; 1994. p. 113–7.
11. Howell JK, Lucke WE, Steigerwald JC. Metalworking fluids: compo-
sition and use. The Industrial Metalworking Environment:
Assessment and Control (Symposium). Nov. 13–16, 1995. Detroit:
Automobile Manufacturers Association; 1996. p. 13–20.
12. Centers for Disease Control and Prevention. Biopsy-confirmed
hypersensitivity pneumonitis in automobile production workers
exposed to metalworking fluids—Michigan, 1994–1995. MMWR
Morb Mortal Wkly Rep 1996;45:606–10.
13. Centers for Disease Control and Prevention. Respiratory illness in
workers exposed to metalworking fluid contaminated with nontuber-
culous mycobacteria—Ohio, 2001. MMWR Morb Mortal Wkly Rep
2002;51:349–52.
14. Wilson, RW, Steingrube VA, Bottger EC, Springer B, Brown-Elliot
BA, Vincent V, et al. Mycobacterium immunogenum sp. nov., a novel
species related to Mycobacterium abscessus and associated with clin-
ical disease, pseudo-outbreaks and contaminated metalworking flu-
ids: an international cooperative study on mycobacterial taxonomy.
Int J Syst Evol Microbiol 2001;51:1751–64.
15. Moore JS, Christensen M, Wilson RW, Wallace RJ Jr, Zhang, Y, Nash
DR, et al. Mycobacterial contamination of metal working fluids:
involvement of a possible new taxon of rapidly growing mycobacte-
ria. American Industrial Hygiene Association Journal
2000;61:205–13.
16. van Klingeren B, Pullen W. Comparative testing of disinfectants
against Mycobacterium tuberculosis and Mycobacterium terrae in a
quantitative suspension test. J Hosp Infect 1987;10:292–8.
17. Falkinham JO III, George KL, Parker BC, Gruft H. In vitro suscepti-
bility of human and environmental isolates of Mycobacterium avium,
M. intracellulare, and M. scrofulaceum to heavy-metal salts and
oxyanions. Antimicrob Agents Chemother 1984;25:137–9.
18. Guerin WF, Jones GE. Mineralization of phenanthrene by a
Mycobacterium sp. Appl Environ Microbiol 1988;54:937–44.
19. Krulwich TA, Pelliccione NJ. Catabolic pathways of coryneforms,
nocardias, and mycobacteria. Annu Rev Microbiol 1979;33:95–111.
20. Combourieu B, Besse P, Sancelme M, Veschambre H, Delort AM,
Poupin P, et al. Morpholine degradation pathway of Mycobacterium
aurum MO1: direct evidence of intermediates by in situ
1
H nuclear
magnetic resonance. Appl Environ Microbiol 1998;64:153–8.
21. Falkinham JO III, Norton CD, Le Chevallier MW. Factors influenc-
ing numbers of Mycobacterium avium, Mycobacterium intracellu-
lare, and other mycobacteria in drinking water distribution systems.
Appl Environ Microbiol 2001;67:1225–31.
22. Schulze-Röbbecke R, Fischeder R. Mycobacteria in biofilms.
Zentralblatt fur Hygiene und Unweltmedizin 1989;188:385–90.
23. Havelaar AH, Berwald LG, Groothuis DG, Baas JG. Mycobacteria in
semi-public swimming pools and whirlpools. Zentralblatt fur
Hygiene und Unweltmedizin 1985;180:505–14.
24. Mangione EJ, Huitt G, Lenaway D, Beebe J, Bailey A, Figoski M, et
al. Nontuberculous mycobacterial disease following hot tub expo-
sure. Emerg Infect Dis 2001;7:1039–42.
25. Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of
Mycobacterium avium with environmental amoebae enhances viru-
lence. Infect Immun 1997;65:3759–67.
26. Strahl ED, Gillaspy GE, Falkinham JO III. Fluorescent acid-fast
microscopy for measuring phagocytosis of Mycobacterium avium,
Mycobacterium intracellulare, and Mycobacterium scrofulaceum by
Tetrahymena pyriformis and their intracellular growth. Appl Environ
Microbiol 2001;67:4432–9.
766 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
PERSPECTIVES

27. Taylor RH, Falkinham JO III, Norton CD, LeChevallier MW.
Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of
Mycobacterium avium. Appl Environ Microbiol 2000;66:1702–5.
28. Parker BC, Ford MA, Gruft H, Falkinham JO III. Epidemiology of
infection by nontuberculous mycobacteria. IV. Preferential
aerosolization of Mycobacterium intracellulare from natural water.
Am Rev Respir Dis 1983;128:652–6.
29. Schulze-Röbbecke R, Buchholtz K. Heat susceptibility of aquatic
mycobacteria. Appl Environ Microbiol 1992;58:1869–73.
30. duMoulin GC, Stottmeier KD, Pelletier PA, Tsang AY, Hedley-Whyte
J. Concentration of Mycobacterium avium by hospital hot water sys-
tems. JAMA 1988;260:1599–601.
31. Hüttunen K, Ruotsalainen M, Iivanainen E, Torkko P, Katila M-L,
Hirvonen M-R. Inflammatory responses in RAW264.7 macrophages
caused by mycobacteria isolated from moldy houses. Environ Toxicol
Pharmacol 2000;8:237–44.
32. Rha Y-H, Taube C, Haczku A, Joetham A, Takeda K, Duez C, et al.
Effect of microbial heat shock proteins on airway inflammation and
hyperresponsiveness. J Immunol 2002;169:5300–7.
33. Wendt SL, George KL, Parker BC, Gruft H, Falkinham JO III.
Epidemiology of infection by nontuberculous mycobacteria. III.
Isolation of potentially pathogenic mycobacteria in aerosols. Am Rev
Respir Dis 1980;122:259–63.
34. George KL, Parker BC, Gruft H, Falkinham JO III. Epidemiology of
infection by nontuberculous mycobacteria. II. Growth and survival in
natural waters. Am Rev Respir Dis 1980;122:89–94.
35. Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev
Biochem 1995;64:29–63.
36. Collins FM. Bactericidal activity of alkaline glutaraldehyde solution
against a number of atypical mycobacterial species. J Appl Bacteriol
1986;61:247–51.
37. George KL, Pringle AT, Falkinham JO III. The cell surface of
Mycobacterium avium – intracellulare and M. scrofulaceum: effect of
specific chemical modifications on cell surface charge. Microbios
1986;45:199–207.
38. Young H-K. Antimicrobial resistance spread in aquatic environments.
J Antimicrob Chemother 1993;31:627–35.
39. Wolinsky E. Nontuberculous mycobacteria and associated diseases.
Am Rev Resp Dis 1979;119:107–59.
40. Clark-Curtiss JE. Identification of virulence determinants in patho-
genic mycobacteria. Curr Top Microbiol Immunol 1998;225:57–121.
41. Speizer FE. Occupational and environmental lung diseases: an
overview. Environ Health Perspect 2000;108(Suppl 4):603–4.
Address for correspondence: J.O. Falkinham, Fralin Biotechnology
Center, West Campus Drive, Virginia Tech, Blacksburg, VA 24061-0346,
USA; fax: (540) 231-7126; email:[email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 767
PERSPECTIVES
Search ppast iissues oof EEID aat wwww.cdc.gov/eid
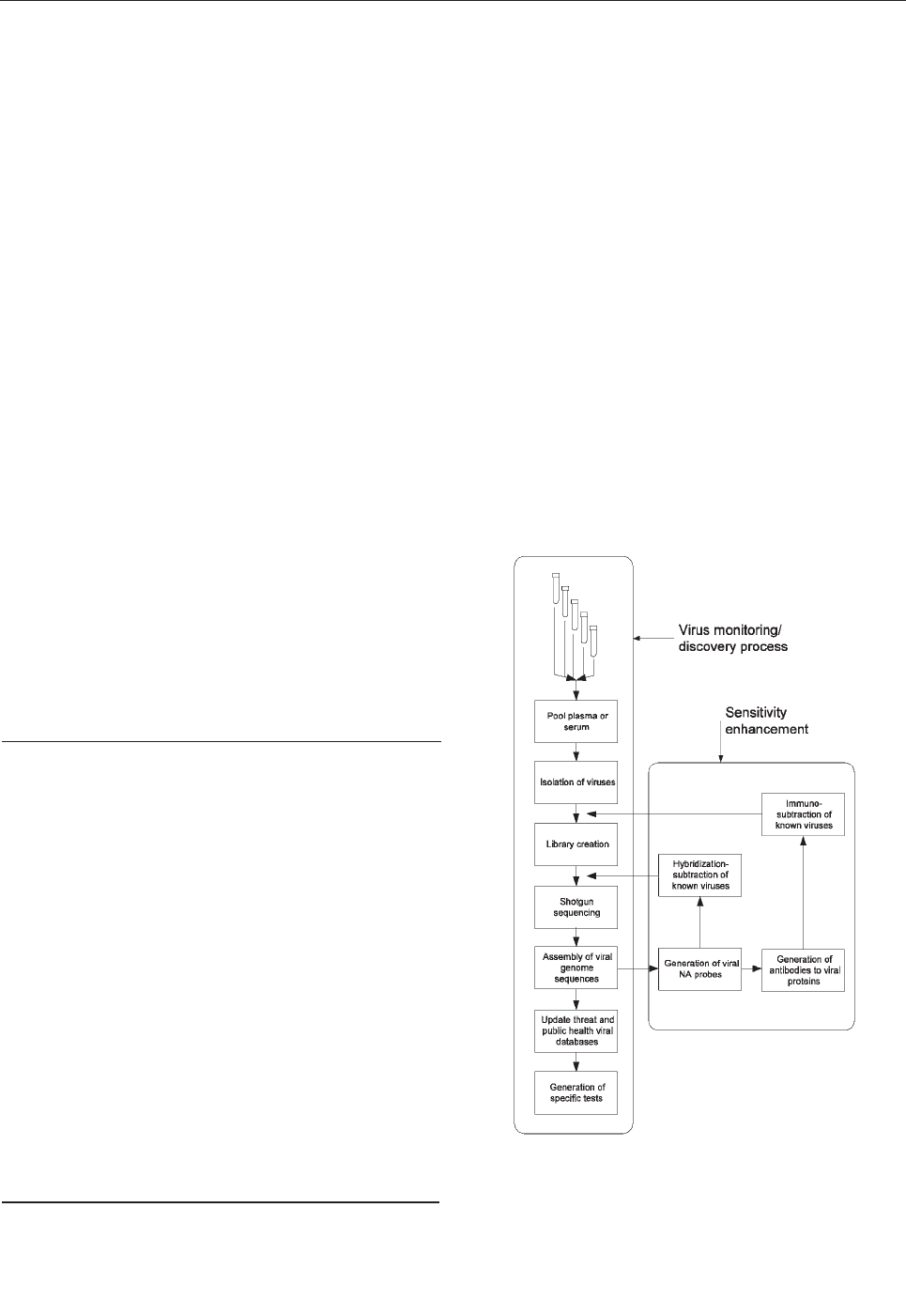
We propose a system for continuing surveillance of
viral pathogens circulating in large human populations. We
base this system on the physical isolation of viruses from
large pooled samples of human serum and plasma (e.g.,
discarded specimens from diagnostic laboratories), fol-
lowed by shotgun sequencing of the resulting genomes.
The technology for concentrating virions from 100-L vol-
umes was developed previously at Oak Ridge National
Laboratory, and the means for purifying and concentrating
virions from volumes in microliters have been developed
recently. At the same time, marine virologists have devel-
oped efficient methods for concentrating, amplifying, and
sequencing complex viral mixtures obtained from the
ocean. Given this existing technology base, we believe an
integrated, automated, and contained system for surveil-
lance of the human “virome” can be implemented within 1
to 2 years. Such a system could monitor the levels of
known viruses in human populations, rapidly detect out-
breaks, and systematically discover novel or variant human
viruses.
T
he traditional process of discovering previously
unknown human viruses, or variants of known viruses,
is neither rapid nor thoroughly systematic. The time
between back-calculated initial infection and final identifi-
cation is often many weeks, months, or even years. For a
totally new agent, the estimated interval between initial
infection and detailed characterization is variable and
depends on the presence of unusual symptoms, the failure
to identify a virus after using all available specific tests,
the recognition of a unique problem, and, in the past, the
ability to grow the agent in culture.
The idiosyncratic nature of virus discovery contrasts
with the broad survey approaches characteristic of
genomics and proteomics. Only in the relatively small
field of ocean viruses has a more inclusive, cataloging
approach been tested. Facilitated by the relative ease with
which viruses can be isolated from seawater (using com-
mercial filters), investigators in this area have examined a
broad and essentially unbiased population of viral agents at
the genome sequence level (including phage) and estimat-
ed the number of different genomes present (~5,000)
(1–3). One would expect that a comprehensive survey of
human viruses, defining what we might term the human
“virome” would be, at least conceptually, even more
straightforward.
Our proposed approach (Figure), in which large popu-
lations are continually monitored for new human-infective
768 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
PERSPECTIVES
Global Screening for
Human Viral Pathogens
Norman G. Anderson,* John L. Gerin,† and N. Leigh Anderson‡
*Viral Defense Foundation, Kensington, Maryland, USA;
†Georgetown University Medical Center, Rockville, Maryland,
USA; and ‡Plasma Proteome Institute, Washington, D.C., USA
Figure. Schematic representation of a process for systematic dis-
covery of human viruses. The basic process (left vertical series of
steps) depends on physical isolation and shotgun sequencing to
obtain sequences of frequent and rare viruses. A series of addi-
tional steps (right box) can be added to deplete known viruses at
two levels, thereby enhancing sensitivity for novel agents.

viruses, has not been considered technically feasible or
medically necessary in the past. For purposes of broad sur-
veillance, we propose using pools of serum or plasma from
large numbers of persons, the most likely source of which
is excess material collected for routine clinical purposes.
These samples would be pooled and processed by using
available technology to isolate virus particles en masse,
recover viral nucleic acids, produce amplified shotgun
libraries, carry out shotgun sequencing of the mixture of
viral genomes, and reconstruct these genomes in silico
with the techniques originally developed to sequence the
entire human genome from random fragments. A central
objective is to continually repeat this monitoring process to
determine which agents change in abundance over time,
find undiscovered agents already present, and detect new
viruses when they appear. If successful, we will have for
the first time a comprehensive picture of “what is going
around.” Surprisingly, most of the systems and technology
to carry out this process exist in a basic form and have
been successfully employed to survey the extremely varied
DNA virus population of the oceans. What remains to be
done, to create a system applicable to humans, is primari-
ly its integration, optimization, and implementation in a
safely contained environment. We briefly explore the com-
ponents of this process here and suggest that it can be
made operational in less than a year.
Availability of Large Pooled Samples
The major commercial diagnostic laboratories in the
United States discard approximately 500 L of excess
human serum or plasma each week. This material repre-
sents a broad cross-section of patients and illnesses.
Plasma viral loads as a function of time after onset of ill-
ness are not known for most viral diseases, but they appear
to be highest in the initial febrile stages. Since one of the
first steps in treating a febrile illness of unknown origin is
obtaining a blood sample, we expect that current diagnos-
tic networks contain appreciable quantities of virus.
Samples from subpopulations enriched for potential viral
illness could also be selected. For those viral diseases in
which viremia precedes major illness, the inclusion of
large numbers of randomly acquired specimens in the pool
(i.e., an unselected pool) offers the best chance of detec-
tion. Analysis of pooled samples from a large number of
persons should raise minimal privacy concerns.
Virus Isolation, Sequencing, and Assembly
Methods are required for the routine isolation of all
classes of viruses from a pooled sample and for concentrat-
ing them by factors of over a million while ensuring that
all nonviral nucleic acids have been removed. The concen-
trates may be dangerously infectious, and sophisticated
containment systems will be needed.
The preferred technology for virus concentration from
large volumes was developed by the Joint NIH-AEC Zonal
Centrifuge Development Project at the Oak Ridge
National Laboratory, Oak Ridge, Tennessee, USA, and the
Oak Ridge Gaseous Diffusion Plant in the 1960s (4). At the
outset, researchers wanted to determine whether viruses as
a class differed in a systematic way from all other small
particles in nature. When the sedimentation coefficients of
then-known viruses were plotted against their isopycnic
banding densities, nearly all viruses fell into an otherwise
essentially vacant area in the center of the plot, surround-
ed at higher or lower density and higher or lower sedimen-
tation coefficients by various subcellular organelles and
macromolecules. This area was termed “the virus window”
(5). Thus, viruses exhibit a unique size and density range
and have banding densities that reflect their combined pro-
tein and nucleic acid contents. In addition, viral nucleic
acids are shielded from attack by nucleases so that contam-
inating nucleic acid-containing particles (primarily
genomic DNA from apoptotic or disrupted cells) can be
selectively destroyed by added nucleases (6). The rules of
virus isolation are the following: 1) the sedimentation rate
(based largely on particle size) falls in a specific range; 2)
the banding density in a gradient falls in a specific range;
3) the genome is protected from nuclease attack until the
protein (+/- lipid) coat is disrupted; and 4) the major pro-
teins present have sequences that agree with at least part of
the genome.
Exploiting the virus window required a two-dimension-
al separation based on sedimentation rate (S) in one dimen-
sion and banding density (rho) in the other, usually carried
out in the order: S-ρ. For large-scale isolation and purifica-
tion, the challenge was to perform these separations con-
tinuously and simultaneously in large continuous-flow
centrifuge rotors spinning at high speed in a vacuum.
In this scheme, a flowing stream passes inboard of a
thin, nonflowing density gradient, held in place against the
rotor wall by centrifugal force. The recovered virus forms
a narrow band in the gradient, which is recovered after
reorienting the gradient to rest at the end of the run. If the
flow through two centrifuges is cascaded, the first operat-
ing at lower speed than the second, particles having a high-
er S rate than viruses could be removed from the flowing
stream, and the viruses then concentrated and banded in
the second higher speed centrifuge, thus providing a large-
volume S-ρ separation.
The end result of this work included the design and
construction of the K-II large-scale ultracentrifuge (7–10).
This device was designed to recover virus in a high state of
purity from 100-L batches of crude influenza vaccine in an
8-hour day (11,12) and subsequently was used for the
large-scale purification of the hepatitis B (Australia) sur-
face antigen (13,14) from human serum for use as a vac-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 769
PERSPECTIVES

cine and for mass isolation of polyhedral inclusion bodies
(15). K centrifuges have come into worldwide use for
large-scale virus isolation and have been commercially
available with little rotor modification for the last 35 years
(16,17). Approximately 200 such systems have been con-
structed.
Methods are now available to further purify compo-
nents of complex viral mixtures, to sediment the viruses
through gradients containing nonsedimenting zones (e.g.,
of nucleases), and to concentrate them down to tens of
microliters (18,19) for sequencing or mass spectrometric
analysis.
Ocean Viruses
During the Oak Ridge centrifuge development project,
large volumes of test material were required, and seawater,
among other sources, was examined as a possible source of
virus. The ocean was found to contain examples of almost
every known viral form at high titers (20), initiating the
exploration of marine virology (1–3,21–30). Recent data
in this field suggest that the oceans of the world contains
approximately 10
31
phage particles or virions (27) (c. 22
million metric tons), much of it turning over once per day
and including some human pathogens (28). This vast
mutation engine, even if one assumes a minimal mutation
rate, generates the equivalent of hundreds of new complete
human genomes per day. That viruses are ubiquitous in the
ocean has been demonstrated by studies on samples recov-
ered in widely separated locations from filtration systems
installed on surface ships (29), nuclear submarines (3), and
remotely operated vehicles (30). Indeed, the entire ocean
has an average viral content in the lower range of the viral
loads reported for human plasma from viremic patients.
Marine virologists have in fact come closest to implement-
ing a surveillance system such as we propose for humans.
On limited budgets, these researchers have developed the
means of recovering marine viruses from large volumes by
filtration (especially well-suited to such a dilute sample),
and for producing shotgun libraries from them by random
amplification (1). Marine virologists have also begun to
estimate the diversity of marine viruses (1–3,27,30) and
are reconstructing large numbers of complete viral
genomes. In one study (2), a 200-L sample of surface sea-
water was concentrated; ~2 x 10
12
viral particles were
recovered; the DNA was randomly sheared and cloned;
and 1,934 fragments were sequenced. Data analysis
showed that most of the sequences were from previously
unknown viruses. Approximately 3.5% of the total
sequence samples overlapped, suggesting that the marine
viral community was highly diverse. A unique mathemati-
cal analysis (2) further suggested that less than 10
4
differ-
ent viral types were present and that shotgun sequencing of
the most abundant could be done, with existing facilities,
in 1 month. Although efforts to date have focused on virus-
es with DNA genomes, most human viral pathogens have
RNA genomes. Genomic sequencing libraries will there-
fore have to be prepared from mixtures of both single- and
double-stranded DNA and RNA viruses (the latter generat-
ed by reverse transcription).
The feasibility of the genomics assembly and annota-
tion components of this project derives from the demon-
stration that the entire human genome could be fragment-
ed, the fragments sequenced, and the original sequence
reconstructed from overlaps; that abundant sequencing
capacity now exists in search of high-value projects; and
that marine virologists have succeeded in parallel ventures.
The challenge is to shorten the time of the entire process so
useful epidemiologic and viroterrorism response data can
be rapidly obtained.
Remaining Challenges
Recovering viruses from large pools of human serum
and plasma and routinely cloning and sequencing the viral
nucleic acids (using established shotgun approaches)
appears technically feasible. Consequently, the titers of
many known human viral pathogens may be estimated rou-
tinely, and new viruses (both pathogenic and nonpathogen-
ic) may be discovered systematically.
The initial choice is between filtration and centrifuga-
tion. Seawater contains little contaminating material of the
size and density of the virus particles, filtration is simple
and efficient, and no free nucleic acids have been report-
ed. Plasma and serum present different problems, compli-
cated by the presence of large amounts of protein, some
nonviral particles in the virus window, and variable
amounts of soluble nonviral nucleic acids (31) that must
be eliminated. An advantage of centrifugal methods is that
all separations, down to banding in microliter gradients,
can be (and have been) done with the virions in suspen-
sion, a process that avoids aggregation that may occur on
filter surfaces.
Several key questions remain to be addressed in a prac-
tical project: 1) Can this process be carried out rapidly
enough to support a timely therapeutic or prophylactic
response to a new agent (natural or engineered)? 2) Will
the novel virions that originated from one or a very few
infected persons be recovered and detected? 3) Can the
affected persons be located? 4) Will the prescence of anti-
bodies in the starting samples against most known viruses
affect the separations?
Speed of Operation
Samples can be collected weekly, and materials rapidly
transported to one or more processing sites. Virus isola-
tion, library construction, and preparation of clones for
sequencing require <7 days but the time may be com-
770 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
PERSPECTIVES

pressed into <4 days with a 24-hour per day operation.
Sequencing time will be determined by available capacity,
but since capacity is abundant, with large increases in
sight, extensive library sequencing (e.g., 10–100 megabas-
es) could be carried out in 2 to 3 days. Less than 1 day
would be required to assemble viral genomes as contigs in
silico. By the conducting of each step in sequence, turn-
around (serum pool to raw sequence data) could be com-
pleted in approximately 10 operating days. Additional time
would be required for bioinformatics analysis of the data
and annotation, but prevalence and novelty conclusions
should be available almost immediately. These estimates
assume an integrated and fully developed system in contin-
uous operation, analogous in some respects to those moni-
toring computer viruses.
Sensitivity
The mass of virus ultimately recovered from pooled
human plasma is difficult to estimate in advance. If the
average virus has a mass of 1.0 x 10
-15
g; if the average titer
of an infected person is 10
6
virions/mL, and if 0.1% of the
samples are from viremic patients, then ~0.5 µg of virus
would be recovered from each 500-L pool, substantially
more than the few nanograms required to make a large
library with current technology (1,2). If the average sam-
ple contributed to the pool was 1 mL, and if the final con-
centrated virus were in 1 mL, the final concentration of a
totally new virus would be close to that in the original indi-
vidual sample. The possibility of detecting all viruses for
which polymerase chain reactions (PCR) primers are
available, down to contributions from single patients,
therefore exists.
Dynamic Range
The problem of dynamic range can be addressed in
three ways. First, given large sequencing capacity, one
could sequence deeply into the libraries (millions of clones
instead of a few thousand), thereby detecting parts-per-
million sequences. Second, one could apply antibody-
based affinity methods to deplete known viral particles
from the initial concentrated viral sample. Third, one could
use subtractive hybridization to remove known viral
genomic sequences to further enrich libraries in novel
genomes. The last two approaches can be progressively
extended as viruses are characterized to provide a continu-
ous increase in sensitivity to new agents (Figure).
Identification of Viral Sources
Two approaches can be used, if necessary, to link virus-
es to patients. In the first approach, viruses would be
tracked geographically, first in terms of large regions, and
then, sequentially, in terms of smaller areas. Detecting a
new agent in large pooled samples would thus be repeated
in smaller, localized pools that had been combined hierar-
chically to generate the larger pool (32).
A potentially more efficient approach involves overlap-
ping subpools designed such that a new viral sequence can
be assayed (e.g., by PCR) in the subpools and the affected
persons identified in one step. To achieve this result, each
sample is added to a series of different pools, the identity
of these subpools providing an “address” of the sample
(33). This process can be visualized by analogy to a 3-D
chessboard, where each position represents a sample, and
the subpools are the various planes parallel to the top,
front, and side: each sample would contribute to three sub-
pools. In practice, additional pools would be created to
provide a relatively unambiguous means of backtracking
from the pattern of subpools positive for a specific
sequence to one or a few persons.
Viral Pathogens That May Be Missed
Not all human viral pathogens will be detected easily
by analyzing plasma or serum samples. Neurotropic virus-
es such as rabies, for example, are found in cells and tis-
sues and do not appear free in serum or plasma in appre-
ciable amounts. Thus, these viruses would escape the
screening system described to this point. Although using
rabies for viroterrorism would be unlikely, such viruses are
of great public health interest, and efforts should ultimate-
ly be made to include them in any global screening system.
The rapid turnover of viruses found in plasma suggests
that they are removed into cells, and that appears to be gen-
erally true. Centrifugal S-ρ technology was originally
developed for cell fractionation with the aim of isolating
viruses from tumors, cells, and tissues (5). Trace quantities
of virus could be added to tissue homogenates and recov-
ered in a high state of purity. The basic technology there-
fore exists for isolating viruses from lymphocytes and a
variety of different tissues. At a later stage, the proposed
approach should be applied to whole blood (with cells
lysed before virus recovery), nasal washings, tissues, and
other potentially virus-laden samples.
Automation and Containment
To routinely detect new and potentially lethal viruses,
researchers may need to create completely automated and
contained laboratories that continually search for and
sequence viruses from a wide variety of sources to hone
skills; demonstrate efficiency; and develop improved sys-
tems, methods, and reagents.
Containment was of great concern in the original
Manhattan Project to deal with radiologic hazards, and in
the Oak Ridge centrifuge project (34) to contain infectious
agents. Containment systems have since evolved in two
directions. In biological sciences, interest has centered on
schemes to allow investigators to work in a safe environ-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 771
PERSPECTIVES

ment using essentially the same tools they would use on an
open bench. As a result, designs has evolved in which
human operators are contained in “space suits.” In nuclear
programs, in contrast, (where containment systems actual-
ly originated), operators are completely isolated from the
contents of “hot cells,” and operations in these cells are
done remotely with specially designed equipment. Given
the national urgency for automated systems for virologic
studies, completely robotic automated systems should be
developed, analogous to those used in nuclear research,
because 1) the concentrated samples to be analyzed are
potentially extremely dangerous, 2) work must be done
without interruption, and 3) speed and precision depend on
automation. Although the K-II centrifuge has not thus far
been automated for totally remote operation, including
cleaning between runs, this is not an overwhelming prob-
lem and should involve cascaded centrifuges, as was done
for the mass isolation of Tussock moth polyhedral inclu-
sion bodies (15).
False Positives, False Negatives,
and the Price of Errors
All current diagnostic tests have the potential for false-
negative and false-positive results. In the atmosphere of an
actual terrorist attack with a biological agent, however, the
consequences of these false outcomes place an enormous
strain on public services, as demonstrated by the recent
anthrax episodes. A false positive triggers highly disrup-
tive responses, whereas a false-negative result exposes the
population to the obvious health concern. The approach
described here reduces the possibility of such outcomes
and only assumes that the virus has the expected biophys-
ical properties of size and mass and an internalized
genome. If a particle with the appropriate biophysical
properties coincides with an internal genome that codes for
structural proteins that are also found in the same fraction,
the possible number of false-positive error is acceptably
small and few mechanisms exist by which false evidence
for a truly nonexistent viral sequence might emerge from
the process described. The level of false negatives depends
not only on the overall quality of the analysis but also on
its sensitivity to rare events, i.e., the dynamic range. For
sequence-based analyses, sensitivity depends on the fre-
quency with which a sequence appears in a fragment
library, the number of clones produced, the efficiency of
known virus subtraction (if applied), and the number of
different clones sequenced.
The number of intentional false positives (duping) is
another matter and one that has two aspects. First, inten-
tional introduction of unexpected pathogens or their genes
into a global analytical system is itself a terrorist act and
one that should be detected and known. To insert a substan-
tial amount of recoverable viral particles into the sample
collection system, a person trying to deceive the system
would have to engineer and grow these viruses—an act
sufficiently close to actual bioterrorist use that it requires
detection, whether the agent is a serious human pathogen
or not. The second aspect concerns the best response to the
suspicion that such duping has occurred. Complete
sequencing of the agent(s) involved would be important
since one cannot initially distinguish a genuine sample
from one used in duping. Only after extensive further stud-
ies and the demonstration that an outbreak has not occurred
may the sample be determined not to be of patient origin.
Forewarned is forearmed. Given advance notice, even
by weeks, of an impending viral outbreak, the hope exists
that the tools and imaginations of molecular biology will
find the means to prepare some effective biological
defense.
Medical Contributions of Global Surveillance
The problem of developing new antiviral agents, espe-
cially those specific for one or only a few viral diseases, is
circular. Without such treatments rapid agent identification
is not necessary, but without such identification no press-
ing commercial justification for developing specific antivi-
ral agents exists (except for HIV) because they will not be
widely used. To be successful, diagnosis and therapy must
be linked. This project would assist in forging that link.
Conclusion
Isolating and sequencing the genomes of a wide variety
of viruses from pools of the excess human serum and plas-
ma currently collected and discarded by large diagnostic
laboratories is now technically feasible. This collection
and analysis process could allow new or unknown
pathogens to be identified in the first, or at most second,
round of infection. Not all human viral pathogens will be
present in such mixtures, but they will include a large frac-
tion of all known highly infectious viral agents. Since the
core technologies, though varied, are highly developed, we
believe that the initial feasibility studies could be complet-
ed in 1 year.
Former Senator Sam Nunn and William H. Wulf, pres-
ident of the National Academy of Engineering, have both
proposed setting up a project concerned with bioterrorism,
modeled after the Manhattan Project. We believe that the
project described could form the nucleus of such an effort
and suggest that lessons learned in the Oak Ridge cen-
trifuge project may apply. As noted by Alvin Weinberg,
that project was the first (and hopefully not the last) large-
scale project in the biological sciences in which facile
access to a wide range of technologies was provided, on
the model of the original Manhattan Project (35).
In separate articles, we will discuss the possibility of
linking rapid detection to rapid responses, including vac-
772 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
PERSPECTIVES
cine and therapeutic antibody development, in an attempt
to abort epidemics caused by new viruses while they are in
progress.
Acknowledgments
We acknowledge with gratitude the continuing support and
encouragement of Alvin Weinberg, who was director of the Oak
Ridge National Laboratory when the original development was
done.
Dr. Anderson was director of the Molecular Anatomy
Program at the Oak Ridge National Laboratory, held a similar
position at the Argonne National Laboratory, was chief scientist
at Large Scale Biology Corporation, and is now president of the
Viral Defense Foundation. His chief interests include proteomics,
virology, and biophysical separations.
References
1. Rohwer F, Seguritan V, Choi DH, Segall AM, Azam F. Production of
shotgun libraries using random amplification. Biotechniques
2001;31:108–17.
2. Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead,
D, et al. Genomic analysis of uncultured marine viral communities.
Proc Natl Acad Sci U S A 2002;99:14250–55.
3. Steward GF, Montiel JL, Azam F Genome size distributions indicate
variability and similarities among marine viral assemblages from
diverse environments. Limnology and Oceanography
2000;45:1697–706.
4. Anderson NG, editor. The development of zonal centrifuges and
ancillary systems for tissue fractionation and analysis. Natl Cancer
Inst Monogr 1966;21:526.
5. Anderson NG, Harris WW, Barber AA, Rankin CT, Candler EL.
Separation of sub-cellular components and viruses by combined rate-
and isopycnic-zonal centrifugation. Natl Cancer Inst Monogr
1966;21:253–83.
6. Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. A virus dis-
covery method incorporating DNase treatment and its application to
the identification of two bovine parvovirus species. Proc Natl Acad
Sci U S A 2001;98:11609–14.
7. Anderson NG, Waters DA, Nunley CE, Gibson RF, Schilling RM,
Denny C, et al. K-Series centrifuges. I. Development of the K-II con-
tinuous-sample-flow-with-banding centrifuge system for vaccine
purification. Anal Biochem 1969:32:460–94.
8. Perardi TE, Leffler RA, Anderson, NG. K-Series centrifuges. II.
Performance of the K-II rotor. Anal Biochem 1969;32:495–511.
9. Perardi TE, Anderson NG. K-Series centrifuges. III. Effect of core
taper on particle capture efficiency. Anal Biochem 1970;34:112–22.
10. Brantley JN, Willis DD, Breillatt JP, Gibson RF, Patrick LC,
Anderson NG. K-Series centrifuges IV. Measurement and control of
temperature. Anal Biochem 1970;36:434–42.
11. Gerin JL, Anderson, NG. Purification of influenza virus in the K-II
zonal centrifuge. Nature 1969;221:1255–6.
12. Reimer CB, Baker RS, Van Frank RM, Newlin TE, Cline GB,
Anderson NG. Purification of large quantities of influenza virus by
density gradient centrifugation. J Virol 1967;1:1207–6.
13. Gerin JL, Holland PV, Purcell RH. Australia antigen: large-scale
purification from human serum and biochemical studies of its pro-
teins. J Virol 1971;7:569–76.
14. Hilleman MR, McAleer WJ, Buynak EB, McLean AA. The prepara-
tion and safety of hepatitis B vaccine. J Infect 1983(Suppl 1);7:3–8.
15. Breillatt JP, Brantley JN, Mazzone HM, Martignoni ME, Franklin JE,
Anderson, NG. Mass purification of nucleopolyhedrosis virus inclu-
sion bodies in the K-Series centrifuge. Appl Microbiol
1972;23:923–30.
16. Alfa Wassermann Inc. Available from: URL: www.smartcentrifuge.com
17. Kendro Laboratory Products. Available from: URL: www.kendro.com
18. Anderson NG, Anderson NL. Detection and characterization of
microorganisms. U.S. Patent Application publication no.
2002/0137026 A1, Sept. 26, 2002.
19. Anderson NG, Anderson NL, Della-Cioppa, G. Method for discover-
ing new infectious particles. U.S. Patent Application publication no.
2003/0044771 A1, March 6, 2003.
20. Anderson NG, Cline GB, Harris WW, Green JG. Isolation of viral par-
ticles from large fluid volumes. In: Berg G, editor. Transmission of
viruses by the water route. New York: Interscience Publishers; 1967.
p. 75–88.
21. Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of
viruses found in aquatic environments. Nature 1989;340:467–8.
22. Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosys-
tems. Microbiol Mol Biol Rev 2000:64:69–114.
23. Noble RT, Fuhrman JA. Rapid virus production and removal as meas-
ured with fluorescently labeled viruses as tracers. Appl Environ
Microbiol 2000;66:3790–7.
24. Brussaard CP, Marie D, Bratbak G. Flow cytometric detection of
viruses. J Virol Methods 2000;85:175–82.
25. Chen F, Suttle CA, Short SM. Genetic diversity in marine algal virus
communities as revealed by sequence analysis of DNA polymerase
genes. Appl Environ Microbiol 1996;62:2869–74.
26. Fuhrman JA. Marine viruses and their biogeochemical and ecological
effects. Nature 1999;399:541–8.
27. Rohwer F, Edwards R. The phage proteomic tree: a genome-based
taxonomy for phage. J Bacteriol 2002;184:4529–5.
28. Griffin DW, Donaldson KA, Paul JH. Pathogenic human viruses in
coastal waters. Clin Microbiol Rev 2003;16:129–43.
29. Hara S, Koike I, Terauchi K, Kamiya H, Tanoue E. Marine Ecology
Progress Series 1996;145:269–77.
30. Steward GF, DeLong EF, Preston CM Assessing viral diversity in the
sea. Eos Trans 2002(Suppl);83:OS19.
31. Lee TH, Montalvo L, Chrebtow V, Busch MP. Quantitation of genom-
ic DNA in plasma and serum samples: higher concentrations of
genomic DNA found in serum than in plasma. Transfusion
2001;41:276–82.
32. Quinn TC, Brookmeyer R, Kline R, Shepherd M, Paranjape R,
Mehendale, S, et al. Feasibility of pooling sera for HIV-1 viral RNA
to diagnose acute primary HIV-1 infection and estimate HIV inci-
dence. AIDS 2000;14:2751–7.
33. Lefkovits I, Kettman JR, Frey JR. Proteomic analysis of rare molec-
ular species of translated polypeptides from a mouse fetal thymus
cDNA library. Proteomics 2001;1:560–73.
34. Cho N, Barringer HP, Amburgey JW, Cline GB, Anderson NG,
McCauley LL, et al. Problems in biocontainment. Natl Cancer Inst
Monogr 1966;21:485–502.
35. Weinberg AM The birth of big biology. Nature 1999;401:738.
Address for correspondence: Norman G. Anderson, Viral Defense
Foundation, Box 2126, Kensington, MD 20891, USA; fax: 301-770-
9091; email: [email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 773
PERSPECTIVES

We describe Salmonella control programs of broiler
chickens, layer hens, and pigs in Denmark. Major reduc-
tions in the incidence of foodborne human salmonellosis
have occurred by integrated control of farms and food pro-
cessing plants. Disease control has been achieved by mon-
itoring the herds and flocks, eliminating infected animals,
and diversifying animals (animals and products are
processed differently depending on Salmonella status) and
animal food products according to the determined risk. In
2001, the Danish society saved U.S.$25.5 million by con-
trolling Salmonella. The total annual Salmonella control
costs in year 2001 were U.S.$14.1 million (U.S.$0.075/kg
of pork and U.S.$0.02/kg of broiler or egg). These costs are
paid almost exclusively by the industry. The control princi-
ples described are applicable to most industrialized coun-
tries with modern intensive farming systems.
S
almonellosis is one of the most common causes of
foodborne diarrheal disease worldwide. Most of these
infections are zoonotic and are transmitted from healthy
carrier animals to humans through contaminated food. The
main reservoir of zoonotic Salmonella is food animals, and
the main sources of infections in industrialized countries
are animal-derived products, notably fresh meat products
and eggs. In developing countries, contaminated vegeta-
bles, water, and human-to-human transmission are
believed to contribute to a comparatively larger proportion
of the human cases than those in industrialized countries
(1). However, the incidence of human salmonellosis
increased in most industrialized countries in the 1980s and
1990s. Rapid spread of a limited number of successful
Salmonella clones in different sectors of food animal pro-
duction (swine, broiler chickens, and particularly layer
hens) has been suggested as the most important cause of
this increase (2).
Despite much research and many national and interna-
tional attempts to implement control strategies, the inci-
dence of human salmonellosis in most countries remains
high. One notable exception is Sweden, which remains
essentially free from the Salmonella problems typical for
most other industrialized countries. The background for
the Swedish success has been described (3). Unfortunately,
other countries cannot apply the Swedish model of
Salmonella control, which requires near freedom from
Salmonella in domestic food animal production from the
onset. In the European Union, the Zoonosis Directive (4)
was an attempt to initiate a European Union–wide control
effort against foodborne zoonoses, particularly Salmonella
in broiler chickens and layer hens. Most European Union
countries found that they either could not or would not
implement the directive, which did not permit use of vac-
cines, antimicrobial drugs, or both as elements in the con-
trol program of Salmonella in broiler chickens or layer
hens. This constraint was seen as an obstacle by some
countries. Recently a new directive has been formulated,
which is awaiting final approval by the European Union
Parliament.
In Denmark, the incidence in human salmonellosis
increased rapidly in the second half of the 1980s because
of the spread of Salmonella in broiler chickens. This
increase led to the initiation of a targeted national control
program (5). Subsequent spread of Salmonella in swine
and layer hens has also led to increases in human disease
incidence and subsequently to the development and imple-
mentation of targeted control efforts (6–8). We review
Denmark’s Salmonella control programs and the effect on
Salmonella in food animals, food, and humans. We also
evaluate and discuss control costs and public health econ-
omy aspects.
Control of Salmonella in Broiler Chickens
Objectives, Program, and Effects
The initial aim of the program was that <5% of broiler
flocks would be infected with Salmonella. The program
was successful and was gradually revised towards assur-
ance of complete freedom from Salmonella in broiler pro-
duction.
The program is based on the principle of top-down
eradication, ensuring freedom from Salmonella from the
top of the broiler-breeding pyramid down. Infected flocks
of breeding animals are destroyed, and infected birds are
774 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES
Salmonella Control Programs
in Denmark
Henrik C. Wegener,* Tine Hald,* Danilo Lo Fo Wong,* Mogens Madsen,* Helle Korsgaard,*
Flemming Bager,* Peter Gerner-Smidt,† and Kåre Mølbak†
*Danish Veterinary Institute, Copenhagen, Denmark; and †Statens
Serum Institut, Copenhagen, Denmark

processed for slaughter. The testing program has devel-
oped gradually to adjust to higher food safety objectives.
As progress stalled, more intensive serologic and bacterio-
logic testing was developed and applied (5,9–11). The cur-
rent testing scheme is shown in Table 1.
Birds from infected flocks are slaughtered on separate
slaughter lines or late in the day to avoid cross-contamina-
tion. Farmers get a better price for birds from Salmonella-
free flocks, and slaughterhouses can use the label
“Salmonella-free” for birds that meet criteria determined
by the authorities. No decontaminants, such as organic
acids or chlorine, are used during carcass processing.
The proportion of Salmonella-infected broiler flocks
has been markedly reduced since the initiation of the con-
trol program. Figure 1 shows that >65% of broiler flocks
tested positive for Salmonella during the first year of the
program, 1988–89, versus <5% in 2000. This decrease in
Salmonella has led to a concomitant reduction in the pro-
portion of infected broiler carcasses after slaughter and at
retail.
The Danish government and the European Union equal-
ly compensate owners of destroyed breeding stock for their
losses. In 1993, a major Danish retailer (COOP-Denmark)
stopped the marketing of broiler chicken, which exceeded
a 5% target. Danish chicken could not meet this target at
that time, so producers suffered severe losses because they
had to export their chicken to lower priced markets.
Salmonella can be effectively reduced (nearly eliminat-
ed) from broiler chickens by intensive flock-level testing
and top-down eradication. Essential to success is a suffi-
ciently sensitive testing program in the breeding and rear-
ing flocks as well as in the hatcheries, i.e., one that
involves intensive sampling and a combination of serolog-
ic and bacteriologic testing methods (Table 1).
Bacteriologic testing alone is not sufficiently sensitive to
achieve control, especially if S. Enteritidis infections are
present. Removal of all organic material, thorough clean-
ing and disinfection of the poultry house, and an empty
resting period of 10–14 days between flocks can effective-
ly eliminate residual infections. In Denmark, most infec-
tions appear to be vertically transmitted (nearly always
traceable to an infected hatchery or parent flock), whereas
horizontal transmission from the environment and wild
fauna appear to play a minor role. Competitive exclusion
cultures, vaccines, or antibiotics have not been used in the
Danish control program.
Control of Salmonella in Layer Hens
Objectives, Program, and Effects
All shell eggs from commercial layer flocks should be
free from S. Enteritidis and S. Typhimurium. Control of
layer breeders in Denmark is essentially identical to the
control program for broiler breeders (Table 1). Blood and
fecal samples of rearing flocks are tested (8,11), and
infected flocks are destroyed. All commercial flocks of
layer hens in production are tested routinely every 9 weeks
by a combination of serologic testing of egg yolk and bac-
teriologic testing of environmental samples (Table 1,
Figure 2).
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 775
SYNOPSES
Table 1. Salmonella surveillance of the broiler and egg production, Denmark, 2000
Stage of production
Age or frequency
Samples taken
Method
Day-old chickens
10 samples of crate material, 20 dead or destroyed chickens
a
Bacteriologic
1 wk
40 dead chickens
Bacteriologic
2 wks
2 pairs of sock samples
Bacteriologic
4 wks
60 fecal samples
a
Bacteriologic
8 wks
2 pairs of sock samples
Bacteriologic
Central rearing stations,
broiler and egg sector
2 weeks before moving
60 fecal samples and 60 blood samples
ab
Bacteriologic, serologic
Every 2 wks
50 dead chickens or meconium from 250 chickens
taken from the hatchery
ac
Bacteriologic
Breeders (hatching egg
production)-broiler and
egg sector
Every wk
2 pairs of sock samples
d
Bacteriologic
Hatchery
After each hatching
Wet dust
Bacteriologic
Day-old chickens
10 samples of crate material and 20 dead chickens
Bacteriologic
3 wks
5x2 sock samples in floor production units
or 300 fecal samples
Bacteriologic
Rearing egg production
12 weeks
5x2 sock samples in floor production units
or 300 fecal samples, and 60 blood samples
b
Bacteriologic, serologic
Every 9th wk for eggs sold to
authorized egg-packing centers
2 pairs of sock samples in floor production
units or fecal samples and egg samples
Bacteriologic, serologic
Egg production
Every 6 mo for eggs sold at
barnyard sale
2 pairs of sock samples or fecal samples and egg samples
Bacteriologic, serologic
a
Requirements of the European Union Zoonosis Directive (92/117/EEC).
b
Samples taken by the district veterinary officer.
c
Samples taken by the district veterinary officer every 8 weeks.
d
Samples taken by the district veterinary officer every 3 months.
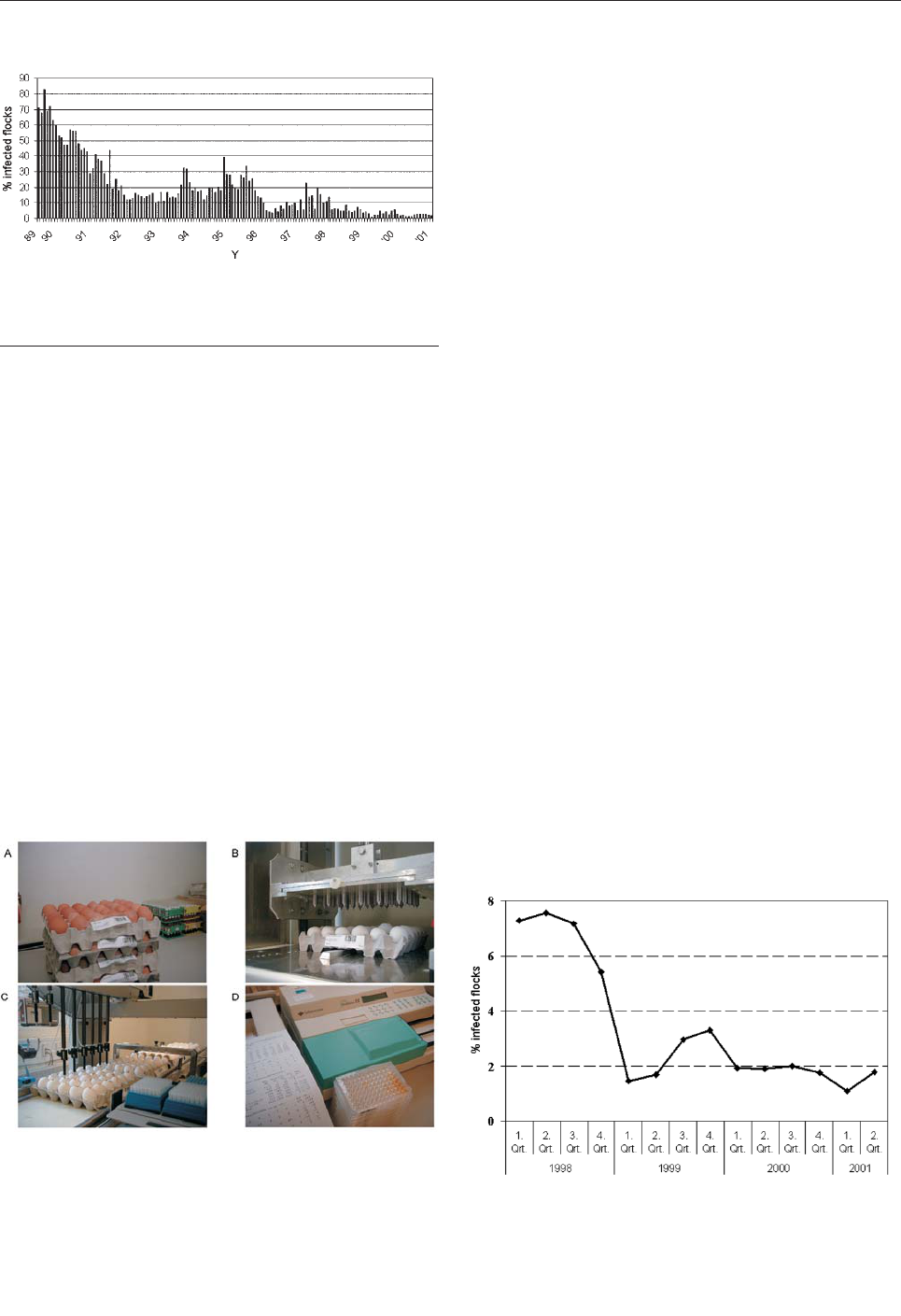
All eggs from suspect or confirmed-positive layer
flocks are pasteurized. All shell eggs are distributed in a
cold chain (not exceeding 12°C) and kept refrigerated at
retail; eggs are generally refrigerated in private homes.
The government and the European Union equally com-
pensate owners of destroyed breeding stock for their loss-
es. The proportion of layer flocks infected with
Salmonella, notably S. Enteritidis, has been markedly
reduced since the initiation of the control program. Figure
3 shows that >7% of layer flocks tested positive for
Salmonella in the first year of the program, 1998, versus
<2% in 2001. The level of Salmonella-contaminated shell
eggs has not been measured from the initiation of the con-
trol program. However, a year before the program began, a
study of 13,000 eggs from different types of production
determined the level to be 1 per 1,000 eggs (20% of the
contaminated eggs harbored S. Enteritidis) (12).
Top-down eradication of S. Enteritidis has effectively
reduced the level of Salmonella, notably S. Enteritidis, in
Danish commercial layer flocks. The program has been
effective in free range, deep litter, organic, and caged birds.
Frequent testing by a combination of serologic and bacteri-
ologic testing methods is essential to achieve adequate sen-
sitivity in the monitoring program. Control of residual
infections in poultry houses can be conducted with a suc-
cess rate of nearly 70% by thorough cleansing and disinfec-
tion of the depopulated house (removal of all organic mate-
rial, disinfection of surfaces, and resting of the empty
house for 2 weeks). Day-old chicks for rearing must be
antibiotic free. Competitive exclusion cultures and vacci-
nation are not used in the Danish program. Vaccination
cannot, at present, be used in combination with serologic
testing because of problems of cross-reaction.
Control of Salmonella in Pork
Objectives, Program, and Effects
Denmark is the only country with a nationwide control
program of Salmonella in pork that is integrated from
“feed-to-food.” The program is based on routine testing
and classification of slaughter pig herds and subsequent
slaughter of pigs according to the inherent risk, as meas-
ured by the continual test program (Figure 2; Table 2). The
program has been described in detail elsewhere (7,8,13).
Pre-Harvest Control
Pigs from breeding and multiplying herds are tested
monthly by serologic testing of blood samples. If a specif-
ic cutoff level is reached, bacteriologic confirmatory test-
ing is carried out. Further, if the serologic reactions exceed
a specific high level, all movement of animals is restricted.
Slaughter pig herds are monitored continuously by sero-
logic testing of “meat juice” (drip fluid released from meat
after freezing and thawing) (14). Meat samples for testing
are collected at the slaughter line, and the number of sam-
ples and frequency of sampling are determined by the size
of the herd. Approximately 700,000 slaughter pigs are cur-
776 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES
Figure 1. Salmonella in Danish broiler flocks as determined by
bacteriologic testing of every flock 2–3 weeks before slaughter
(N>4,000 flocks/year).
Figure 2. A) Receipt of 60 eggs per producer every 9 weeks (bar-
code indicating producer is shown). B) The “eggbreaker” punches
a hole in 30 eggs at a time. C) Withdrawal of egg yolk from 30
eggs and transfer to microtiter tray. D) Enzyme-linked immunosor-
bent assay analysis, reading, and transfer of results to central
database.
Figure 3. Salmonella in Danish layer flocks as determined by sero-
logic and bacteriologic testing of each commercial flock in week 9
of production.
rently tested each year (Figure 4). Herds sending <200 pigs
to slaughter each year are not tested, leaving 1.6% of the
slaughter pigs outside the monitoring scheme (13). The
herds are categorized in three levels based on the propor-
tion of seropositive meat juice samples during the last 3
months. Owners in level 2 and 3 are encouraged to seek
advice on how to reduce the Salmonella problem in the
herd (e.g., feeding, hygiene, and management).
Furthermore, payment from the slaughterhouse is reduced
by 2% and 4%, respectively.
The postharvest surveillance program has been
described (8,15,16). Pigs from herds in levels 1 and 2 are
slaughtered traditionally without any special precautions.
Pigs from level 3 herds can only be slaughtered in special
slaughterhouses under special hygienic precautions.
Carcasses from level 3 herds are tested for bacteria after
slaughter, and if the level of contamination exceeds a cer-
tain limit all carcasses from the particular herd have to
undergo heat treatment or other risk-reducing processing.
All slaughterhouses do routine bacteriologic testing of car-
casses according to a sampling plan, which ensures that
testing is random and representative of the national swine
production (>30,000 samples/year). Slaughterhouses that
exceed a certain predetermined level of Salmonella in the
routine monitoring of carcasses are obliged to investigate
and reduce the contamination problem to an acceptable
level.
The prevalence of swine herds in level 2 and 3 respec-
tively, has been steadily reduced since the program began
(Figure 5). Bacteriologic testing has indicated that the herd
infection level was reduced by 50% (from 14.7% to 7.2%
in small herds and 22.2% to 10.4% in large herds) from
1993 (when the program was implemented) to 1998 (17).
In the same period, the level of Salmonella contamination
in pork products, as determined by the routine monitoring
program, was reduced from 3% to <1% (Figure 6).
As Salmonella eradication in swine herds is difficult
because of the continual nature of the production system,
reducing the infection level should be the aim of a control
strategy. The low infection level in the herds and contami-
nation in the products can likely be reduced further in
Denmark. As the contamination level goes below 1%, test-
ing for contamination requires increasingly large numbers
of samples and consequently becomes very expensive,
which is one reason the sampling plan of herds and prod-
ucts has become more sensitive in recent years. This
change in testing sensitivity makes it difficult to compare
current and past levels of infection and contamination but
is nevertheless a necessity for the continued improvement
of the program.
A combination of serologic and bacteriologic testing is
essential for the success of the program. Nearly 1 million
serologic samples are tested each year. Testing this large
number of samples would not be possible because of finan-
cial and logistic constraints if the program were to rely on
bacteriologic testing alone.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 777
SYNOPSES
Table 2. Salmonella surveillance in pig and pork production, Denmark, 2001
Type of production
Sample
No. and frequency
Response
Breeding and multiplying
herds
Blood
10 times per mo
Confirmatory bacteriologic testing and restrictions on the movement of
animals if above predetermined level
Pig herds
Feces
100 in 20 pools of five
collected on indication
Salmonella reduction plan implemented
Slaughter pig producers
producing >200 pigs per year
Meat juice
Depending on herd
size (60–100 samples
per yr). Samples are
collected continuously
and semi-randomly
Confirmatory bacteriologic testing (20 pools of 5 fecal samples). Herds are
assigned to one of three levels depending on serology. Level 1: no sanctions;
level 2: implementation of Salmonella reducing actions in the herd; and level
3: same as level 2 and obligatory slaughter of pigs under special hygienic
precautions, including postslaughter microbial testing and potential heat
treatment of all meat products
Carcass after slaughter
Surface
swab
Swabs of five
carcasses are pooled
into one sample. One
sample per day in each
slaughterhouse.
Slaughterhouses exceeding a predetermined number of positive swabs in a 3-
months period are obliged to implement corrective actions
Figure 4. A) Receipt of pork samples from the slaughterhouse.
Each tube is labeled with a barcode, indicating herd of origin.
Samples are frozen overnight. B) The tube is entered in a rack with
the barcode facing outward. Meat juice is sieved into the tube from
the container during thawing. C) Withdrawal of meat juice from
tube and transfer to microtiter tray. D) Enzyme-linked immunosor-
bent assay analysis, reading, and transfer of results to central
database.
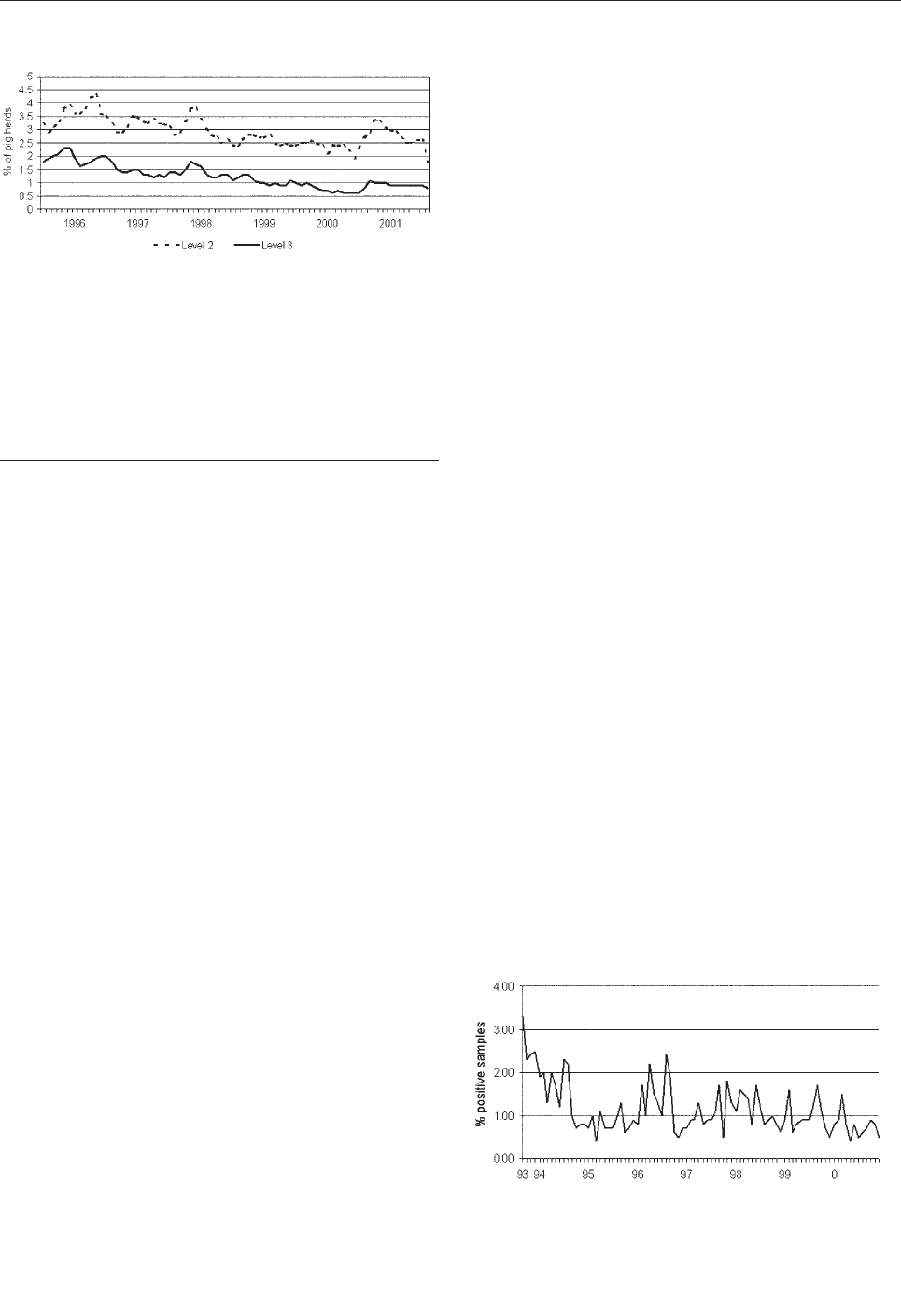
Feeding strategy (e.g., increased coarseness of feed and
wet feeding) and improved management (e.g., sectioning
and all-in all-out production) and hygiene standards are
important elements in the preharvest control efforts. Using
commercial prebiotic cultures is not necessary; natural
microflora in the feed, especially wet fermented feed,
appear to have a protective effect (18).
A reduction of Salmonella in slaughter pig herds has
been attained without Salmonella–free breeding herds.
However, to ensure the highest degree of consistency in
the program, the levels of Salmonella in breeding herds
should be kept as low as possible, and infected breeding
herds should not be sold to producers of herds of a superi-
or Salmonella status.
Determination of Public Health Impact
To better explain the mechanisms in the occurrence of
Salmonella infections in humans, the Danish Zoonosis
Centre has previously described a method that estimates
the number of human cases attributable to each of the
major animal-food sources (19,20). Using this method, we
compared Salmonella types isolated from animals and
foods with Salmonella types isolated from humans. In
brief, subtypes of Salmonella that are almost exclusively
found in a particular food animal reservoir or food type
(unique types) are used as anchor points for the distribu-
tion of subtypes occurring in several reservoirs and
sources. All human infections caused by the unique types
are associated with the indicated food type or derived from
the indicated food animal reservoir (e.g., pork, beef, chick-
ens, or eggs). Salmonella types, which occur in several
reservoirs, are distributed relative to the prevalence of
unique types in each reservoir or food type. Detailed
knowledge of the distribution of Salmonella types in all
relevant food animals and food types, generated through
intensive and continuous monitoring, is a prerequisite for
the analysis. Recently, a stochastic model based on the
principles of the previous method was developed and
applied. This model allows us to consider the uncertainty
around the estimated parameters (21).
Figure 7 shows the human salmonellosis incidence
associated with the three major sources of human salmo-
nellosis in Denmark from 1988 to 2001. The year that a
control program was launched for a specific food animal
production system is also indicated. The control programs
have been successful in achieving the main objective, a
reduction of the incidence of human salmonellosis. The
broiler-associated salmonellosis incidence
(cases/100,000) has been reduced by >95.0, from 30.8 in
1988 to 0.5 in 2001; the pork-associated salmonellosis
incidence has been reduced by >85, from 22.0 in 1993 to
3.0 in 2001; and the egg-associated salmonellosis inci-
dence has been reduced by nearly 75, from 57.7 in 1997 to
15.5 in 2001. Trends in the animal and food-specific dis-
ease incidence estimates show a high degree of agreement
with the trends in prevalence of Salmonella in specific
food animals and the corresponding animal-derived food
products. These trends serve as an indirect validation of
the estimates because these estimates do not rely on preva-
lence data.
Economy of Salmonella Control
Costs of Salmonella Control
The Audit Office of Denmark has evaluated the govern-
ment spending in relation to the national Salmonella con-
trol efforts (22). From 1994 to 1999, the control program
for broiler chickens and layer hens involved government
finances of a total of 188.1 million Danish kroner (DKK)
(U.S.$26.5 million) (U.S.$1.00 = 7.1 DKK). A total of
109.7 million DKK (U.S.$15.45 million) was paid to com-
pensate farmers for destroyed animals; most of the remain-
ing costs were associated with establishing and running the
surveillance program. These costs were highest in the ini-
tial phase of the layer hen control program in 1997 but
778 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES
Figure 5. Prevalence of Salmonella in Danish pig herds as deter-
mined by continuous serologic testing of all commercial pig herds
(N >700,000 samples tested/year). Herds are categorized in three
levels based on the proportion of seropositive meat juice samples
during the last 3 months. Owners in level 2 and 3 are encouraged
to seek advice on how to reduce the Salmonella problem in the
herd (e.g., feeding, hygiene, and management). Furthermore, pigs
from level 3 herds can only be slaughtered in special slaughter-
houses under special hygienic precautions. Data from the Danish
Veterinary and Food Administration.
Figure 6. Salmonella detected in pork, as determined by continu-
ous randomized sampling of pork end-products from all major
national pig slaughterhouses (N >30,000 samples/year).

have been reduced considerably since then. In 2001, all
costs associated with running the program were assumed
by the poultry industry (with the exception of govern-
ments’ compensation for flocks destroyed according to the
European Union Zoonosis Directive). The current control
costs for layer hens and broiler chickens are estimated to
be in the range of US$4.2 million per year (for 344 broiler
producers, producing 135 million broiler chickens year,
and 392 shell-egg producers, producing 1 billion shell-
eggs per year).
In the initial phase, the Salmonella control program of
pigs and pork cost the industry and government a total of
U.S.$14 million per year. With the recent revision of the
program, the responsibility has been taken over solely by
the industry, and operational costs have been reduced to
approximately U.S.$8.5 million per year (for 21,000 pro-
ducers, producing 21–22 million slaughter pigs each year)
(B. Nielsen, pers. comm.).
Public Health Economy
Direct health costs (e.g., hospitalization, consulting a
physician, and laboratory testing) as well as the costs of
lost labor (e.g., loss of production per day away from
work) in relation to a case of salmonellosis in Denmark
were evaluated as part of a multidisciplinary task force
(Korsgaard and Wegener, pers. comm.). For 2001, food-
borne salmonellosis cost the Danish U.S.$15.5 million.
The estimate is based on an incidence of 54.6 cases per
100,000, and approximately 10% of cases are laboratory
confirmed. Assuming that 5% or 20% of cases are labora-
tory-confirmed changes the estimate to U.S.$25.5 million
and U.S.$10.4 million, respectively.
Costs and Benefits
Assuming that salmonellosis associated with each of
the major sources would have remained at the precontrol
program incidence (and not increased further) (i.e., if no
action had been taken to curb the problem), we calculated
a hypothetical “no-control” salmonellosis incidence. This
incidence would have been 137.5 (pork 22, broiler chick-
ens 30.8, eggs 57.7, and average residual, 27). The socie-
tal costs, in the absence of the existing control programs,
would thus have been U.S.$41 million per year (assuming
10% of cases are laboratory confirmed). Thus, in 2001,
Denmark saved U.S.$25.5 million by controlling
Salmonella. The estimated annual Salmonella control costs
from 2000 and onwards are approximately U.S.$14.1 mil-
lion. These costs are borne almost exclusively by the ani-
mal producers and the food industry, which suggests that
the costs are passed on to consumers through higher food
prices. Based on the figures above and data on annual pro-
duction (23), control costs amount to approximately
U.S.$0.075/kg of pork and U.S.$0.02/kg of broiler or egg.
Discussion
Danish Salmonella control efforts have been successful
in achieving their objective; reduction of human salmonel-
losis. These efforts illustrate that with a focused and inte-
grated programs, including a strong element of preharvest
control, and based on a public-private partnership,
Salmonella can be reduced. At the same time, the indus-
tries involved have remained profitable and internationally
competitive (approximately 75% of the chicken products
and 85% of the pork are exported).
Initially, the programs have received some government
funding, primarily for research, development, and com-
pensation for destroyed animals. After the initial imple-
mentation and clean-up phase of the programs, the respon-
sibility for running and funding the programs have been
nearly completely taken over by the industries involved.
The government, however, maintains access to all relevant
information and data through a central database managed
by the Danish Zoonosis Centre, and food safety objectives
continue to be determined by the Danish government.
A proactive and collaborative approach to food safety
by food industry and government ensures consumers’ con-
fidence in the domestic food production. For example,
when the bovine spongiform encephalopathy (BSE) crisis
hit Europe, the beef industry in most countries was
adversely affected by reduced consumer demand. In
Denmark, the sale of beef remained nearly unaffected by
the crisis even after the first positive BSE findings
occurred in Danish cattle. These steady beef sales are like-
ly attributable to a high degree of consumer confidence in
the public and private control systems.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 779
SYNOPSES
Figure 7. Effects of Salmonella control programs as indicated by
incidence of human infection attributable to the different major
sources of human salmonellosis in Denmark; 1) Salmonella con-
trol program for broiler chickens implemented, 2) Salmonella con-
trol program implemented for pigs and pork, 3) Salmonella control
program implemented for layer hens and eggs The three sources
account for approximately 50% to 75% of Salmonella each year.
Remaining cases are attributable to beef, imported food products,
infections acquired while traveling abroad, and unknown sources.
Incidence (cases/100.000 inhabitants).

The success of the programs supports the effectiveness
of a preharvest control approach to Salmonella.
Monitoring and intervention at the farm and in the food
animal breeding systems are feasible means to achieve
lasting control of the Salmonella problem. Development
and application of a two-tiered detection system based on
a combination of serologic testing and bacteriologic con-
firmation have been essential for the success of the pro-
grams. Serologic testing enables semi-automated mass
screening of animals and eggs at a low price and with
good, and in some cases superior, sensitivity. Bacteriologic
testing serves to compensate for the sub-optimal specifici-
ty of a serologic-based monitoring system. The programs
could not have been operated solely on the basis of bacte-
riologic testing because of the higher costs involved and
logistical problems (i.e., screening nearly 2 million sam-
ples per year by bacteriologic testing is unrealistic).
Preharvest control tools, such as vaccines, antibiotics, or
competitive exclusion, are not used to control Salmonella
in Denmark; these tools might be counterproductive, as
they mask the Salmonella problem rather than aid in its
reduction or eradication.
Evaluating the costs and benefits of the national
Salmonella control efforts is difficult; estimating the pub-
lic health and societal costs in the absence of the control
program is impossible. However, this conservative esti-
mate suggests that efforts have been cost beneficial and
those benefits are likely to increase with time.
Dr. Wegener is a professor of zoonoses epidemiology and
head of the Danish Zoonosis Center. His main research interests
are the epidemiology of foodborne zoonosis and antimicrobial
resistant bacterial in the food chain. He is involved in the coordi-
nation of the World Health Organization Global Salmonella
Surveillance Program.
References
1. Salmonella. In: Acha PN, Szyfres B, editor. Zoonoses and communi-
cable diseases common to man and animals. PAHO Scientific and
Technical publications no. 580; 2001.
2. Thorns CJ. Bacterial food-borne zoonoses. Rev Sci Tech
2000;19:226–39.
3. Eld K, Gunnarsson A, Holmberg T, Hurvell B, Wierup M. Salmonella
isolated from animals and feedstuffs in Sweden during 1983-1987.
Acta Vet Scand 1991;32:261–77.
4. Directive concerning measures for protection against specified
zoonoses and specified zoonotic agents in animals and in products of
animal origin in order to prevent outbreaks of food-borne infections
and intoxications (Zoonosis Directive). Council Directive
92/117/EEC, December 1992.
5. Bisgaard M. A voluntary Salmonella control program for the broiler
industry, implemented by the Danish Poultry Council. Int J Food
Microbiol 1992;15:219–24.
6. Mousing J, Jensen PT, Halgaard C, Bager F, Feld N, Nielsen B, et al.
Nation-wide Salmonella Enterica surveillance and control in Danish
slaughter swine herds. Prev Vet Med 1997;29:247–61.
7. Nielsen B, Alban L, Stege H, Sorensen LL, Mogelmose V, Bagger J,
et al. A new Salmonella surveillance and control program in Danish
pig herds and slaughterhouses. Berl Munch Tierarztl Wochenschr
2001;114:323–6.
8. Feld NC, Ekeroth L, Gradel KO, Kabell S, Madsen M. Evaluation of
a serological Salmonella mix-ELISA for poultry used in a national
surveillance program. Epidemiol Infect 2000;125:263–8.
9. Skov MN, Carstensen B, Tornoe N, Madsen M. Evaluation of sam-
pling methods for the detection of Salmonella in broiler flocks. J Appl
Microbiol 1999;86:695–700.
10. Gradel KO, Andersen J, Madsen M. Comparisons of sampling proce-
dures and time of sampling for the detection of Salmonella in Danish
infected chicken flocks raised in floor systems. Acta Vet Scand
2002;43:21–30.
11. Skov MN, Feld NC, Carstensen B, Madsen M. The serologic
response to Salmonella Enteritidis and Salmonella Typhimurium in
experimentally infected chickens, followed by an indirect
lipopolysaccharide enzyme-linked immunosorbent assay and bacteri-
ologic examinations through a one-year period. Avian Dis
2002;46:265–73.
12. Bager F. Investigation of Salmonella prevalence in shell-eggs
(Undersøgelse af salmonella-forekomst i konsumæg) [In Danish].
Zoonose-Nyt 1996;3:7–8.
13. Alban L, Stege H, Dahl J. The new classification system for
slaughter-pig herds in the Danish Salmonella surveillance-and-
control program. Prev Vet Med 2002;53:133–46.
14. Nielsen B, Ekeroth L, Bager F, Lind P. Use of muscle fluid as a source
of antibodies for serologic detection of Salmonella infection in
slaughter pig herds. J Vet Diagn Invest 1998;10:158–63.
15. Nielsen B, Wegener HC. Public health and pork and pork products:
regional perspectives of Denmark. Rev Sci Tech 1997;16:513–24.
16. Hald T, Andersen JS. Trends and seasonal variations in the occur-
rence of Salmonella in pigs, pork and humans in Denmark,
1995–2000. Berl Munch Tierarztl Wochenschr 2001;114:346–9.
17. Christensen J, Baggesen DL, Nielsen B, Stryhn H. Herd prevalence
of Salmonella spp. in Danish pig herds after implementation of the
Danish Salmonella Control Program with reference to a pre-
implementation study. Vet Microbiol 2002;88:175–88.
18. Stege H, Jensen TK, Moller K, Baekbo P, Jorsal SE. Risk factors for
intestinal pathogens in Danish finishing pig herds. Prev Vet Med
2001;50:153–64.
19. The Ministry of Food, Agriculture and Fisheries. Annual report on
zoonoses in Denmark 1998. Copenhagen, Denmark: The Ministry.
Available from: URL: http://www.vetinst.dk
20. Hald T, Wegener H C. Quantitative assessment of the sources of
human salmonellosis attributable to pork. In Proceedings of the 3rd
International Symposium on Epidemiology and Control of
Salmonella in Pork, 4–7 Aug 1999, Washington, D.C. Urbana-
Champaign (IL): University of Illinois. p. 200–5.
21. Hald T, Vose D, Wegener HC. Quantifying the contribution of ani-
mal-food sources to human salmonellosis by a Bayesian approach.
Risk Anal. In press 2003.
22. Report from the National Audit Office of Denmark: The Danish
Governments Control of Salmonella [In Danish]: Beretning om
statens bekæmpelse af Salmonella). Rigsrevisonen. Report No. 8/99
2000. Available from: URL: http://www.rigsrevisionen.dk
23. The Ministry of Food, Agriculture and Fisheries. Annual report on
zoonoses in Denmark 2000. Copenhagen: The Ministry. Available
from: URL: www.vetinst.dk
Address for correspondence: Henrik C. Wegener, Danish Zoonosis
Centre, Danish Veterinary Institute, 27 Bülowsvej, DK-1790 Copenhagen
V, Denmark; fax: +45 35 30 03 77; email: [email protected]
780 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES

The Emerging Infections Programs (EIPs), a popula-
tion-based network involving 10 state health departments
and the Centers for Disease Control and Prevention, com-
plement and support local, regional, and national surveil-
lance and research efforts. EIPs depend on collaboration
between public health agencies and clinical and academic
institutions to perform active, population-based surveillance
for infectious diseases; conduct applied epidemiologic and
laboratory research; implement and evaluate pilot preven-
tion and intervention projects; and provide capacity for flex-
ible public health response. Recent EIP work has included
monitoring the impact of a new conjugate vaccine on the
epidemiology of invasive pneumococcal disease, providing
the evidence base used to derive new recommendations to
prevent neonatal group B streptococcal disease, measuring
the impact of foodborne diseases in the United States, and
developing a systematic, integrated laboratory and epi-
demiologic method for syndrome-based surveillance.
D
uring the 1980s, clinicians added newly recognized
infectious diseases, such as toxic shock syndrome and
AIDS, to their differential diagnoses when evaluating pre-
viously healthy young adults with severe illness. More
recently, clinicians in the United States found themselves
considering the possibility of inhalational anthrax among
patients with influenzalike illnesses and adding West Nile
virus infection to their workup of posttransfusion fevers
(1–3). The existence of these and dozens of other emerg-
ing and reemerging infectious diseases, naturally or inten-
tionally transmitted, has removed any doubt about the
interdependence of clinical medicine and public health.
Clinicians are sentinels for detection of new or reemerging
diseases and may benefit from information acquired
through public health surveillance and research projects,
which helps to place the quantitative risks of these new
diseases in perspective amidst the media attention that
often accompanies the latest medical mysteries.
In 1992, the Institute of Medicine (IOM) articulated the
concept of emerging infections, discarding the naive view
that infectious diseases were problems of the past and cau-
tioning against complacency about public health prepared-
ness for infectious diseases (4). By defining emerging
infectious diseases as “new, reemerging, or drug-resistant
infections whose incidence in humans has increased with-
in the past two decades or whose incidence threatens to
increase in the near future,” IOM recognized the broad
scope of these diseases. The IOM report also cited factors
that influence the emergence of infectious diseases:
changes in human demographics and behavior; advances
in technology and changes in industry practices; economic
development and change in land-use patterns; increased
volume and speed of international travel and commerce;
microbial adaptation and change; and breakdown of public
health capacity at the local, national, and global levels. The
intentional release of anthrax in the United States in 2001
emphasized the need to add intentionally inflicted harm to
the list of factors that influence the emergence of infec-
tious diseases and to suspect the unexpected.
In response to the IOM report, Addressing Emerging
Infectious Disease Threats to Health: A Prevention
Strategy for the United States was developed by the
Centers for Disease Control and Prevention (CDC) (5). A
key recommendation of the plan called for establishing
population-based centers to complement and support local,
regional, and national surveillance and research efforts.
This concept was realized through Emerging Infections
Programs (EIPs), a network of state health departments
(Figure 1) coordinated by CDC. EIPs are intended to be a
national resource for surveillance and epidemiologic
research by conducting work that goes beyond the routine
public health department functions; by fostering collabora-
tions between the public health, academic, and clinical
communities; and by maintaining an infrastructure flexible
enough to address new infectious diseases challenges as
they emerge. An updated plan released in 1998 described
the important role assumed by EIPs in addressing emerg-
ing infections and identified several high-priority target
areas (6), which include: antimicrobial drug resistance,
foodborne and waterborne diseases, vector-borne and
zoonotic diseases, chronic diseases caused by infectious
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 781
SYNOPSES
Disease Surveillance and the
Academic, Clinical, and Public
Health Communities
Robert W. Pinner,* Catherine A. Rebmann,* Anne Schuchat,* and James M. Hughes*
*Centers for Disease Control and Prevention, Atlanta, Georgia,
USA
agents, diseases transmitted through blood transfusions or
products, vaccine development and use, diseases of preg-
nant women and newborns, diseases of persons with
impaired host defenses, and diseases of travelers, immi-
grants, and refugees. We describe EIP accomplishments
and future directions.
EIP Methods
The principal functions of EIPs are to perform active,
population-based surveillance for infectious diseases; con-
duct applied epidemiologic and laboratory research;
implement and evaluate pilot prevention and intervention
projects; and provide capacity for flexible public health
response. EIPs also develop and evaluate public health
practice and transfer what is learned to the public health
and medical communities.
These programs are supported through cooperative
agreements between CDC and state health departments,
who engage collaborators in local health departments, hos-
pitals, and academic institutions. Additional funding for
certain EIP activities comes from other sources; for exam-
ple, the U.S. Department of Agriculture and the Food and
Drug Administration provide support for activities involv-
ing foodborne illnesses, and the National Vaccine Program
Office has provided support for postlicensure vaccine eval-
uations.
The population base for EIP activities is approximately
36 million persons, though the base varies by project. This
population represents an approximation of the U.S. popu-
lation with respect to demographic characteristics such as
age, gender, race, and urban residence, as well as health
indicators such as population density and percentage of
persons at or below the poverty level (7). EIPs are geo-
graphically dispersed throughout the country (Figure 1).
Active, laboratory-based surveillance is the foundation
of two core EIP projects conducted at all sites: Active
Bacterial Core Surveillance (ABCs) and Foodborne
Disease Active Surveillance (FoodNet) (Table 1). These
active surveillance projects generate reliable estimates of
the incidence of certain infections and provide the founda-
tion for a variety of epidemiologic studies to explore risk
factors, disease spectrum, and prevention strategies (8,9).
For example, the total impact of foodborne illnesses in the
United States has been estimated by combining FoodNet
active surveillance data with other data sources and results
from FoodNet surveys of the general population (to learn
about the frequency of diarrhea in the general population
and to determine what proportion of persons with diarrhea
seeks medical care), physicians (to determine the frequen-
cy of stool-culturing by physicians), and clinical laborato-
ries (to determine the frequency of culturing for selected
foodborne pathogens) (9–11). These data provide esti-
mates of the overall occurrence of diarrheal illness (0.7 ill-
nesses/person-year), as well as the likely degree of under-
reporting for specific infections under surveillance (10).
Other projects are conducted by EIPs, depending on
local priorities and expertise. The Unexplained Deaths and
Critical Illness (UNEX) project, a prospective study that
uses epidemiologic and laboratory methods to detect and
investigate unexplained illnesses with clinical features
suggesting infectious diseases, has been in place at four
states with EIPs since the inception of the program (12,13).
The Connecticut EIP conducts active surveillance for
emerging tick-borne diseases that are transmitted by a sin-
gle tick vector (Ixodes scapularis) in the state (14). EIPs
also strive to maintain the flexibility to meet new chal-
lenges effectively. For example, in 1996 four EIP sites con-
ducted active surveillance for variant Creutzfeldt-Jakob
782 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES
Table 1. Surveillance and focus area for two core projects conducted at all Emerging Infections Program sites
a
Projects
Type of surveillance
Focus
Active Bacterial Core Surveillance
Active, laboratory-based
Invasive disease (isolated from a normally sterile site such as
blood or cerebrospinal fluid) caused by group A streptococcus,
group B streptococcus, Haemophilus influenzae, Neisseria
meningitidis, and Streptococcus pneumoniae
FoodNet/Foodborne Disease Active Surveillance
Active, laboratory-based
Disease (first isolation from a person) caused by
Campylobacter, Listeria, Salmonella, Shigella, Yersinia,
Vibrio, Shiga toxin–producing Escherichia coli, including
O157:H7, Cryptosporidium, and Cyclospora
a
Intended to generate reliable estimates of the incidence of certain infections and provide the foundation for a variety of epidemiologic studies to explore risk factors,
disease spectrum, and prevention strategies.
Figure 1. Distribution of Emerging Infections Programs (EIPs), a
network of 10 state health departments and their collaborators in
local health departments, academic institutions, and clinical set-
tings, coordinated by the Centers for Disease Control and
Prevention. *New Mexico was added as the 10th EIP site in late
2002 and will begin EIP activities during 2003.

Disease (CJD) and physician-diagnosed CJD cases. This
study contributed to surveillance methods by confirming
that death certificate reviews are a sensitive method for
detecting CJD deaths while providing some assurance that
variant CJD was not occurring in these states (15).
Impact of a New Pneumococcal Vaccine
Through ABCs, we are evaluating the effect of the
pneumococcal conjugate vaccine on the epidemiology of
invasive pneumococcal disease in the United States.
Streptococcus pneumoniae (pneumococcus), which is an
important cause of serious illness among young children,
is the leading cause of bacterial pneumonia and meningitis
in the United States. For many years, immunization against
pneumococcus with a 23-valent polysaccharide vaccine
was recommended for persons >2 years of age who are at
high risk and for all adults >65 years of age. Although dis-
ease incidence is highest in the first 2 years of life, the
polysaccharide vaccine was poorly immunogenic in this
group. In February 2000, a protein-polysaccharide pneu-
mococcal conjugate vaccine for seven pneumococcal
serotypes (Prevnar, Wyeth Pharmaceuticals, Pearl River,
NY) was licensed for use in infants and children (16). This
conjugate vaccine is now recommended in the United
States for all children <2 years of age, with catch-up vac-
cination schedules suggested for children 2 to 4 years of
age. In clinical trials, the vaccine was efficacious against
invasive disease in infancy and reduced nasopharyngeal
colonization by vaccine-type strains, an indication of
potential for herd immunity.
One method used by ABCs is to collect available iso-
lates from identified cases. Serotyping data were analyzed
to learn about the epidemiology of S. pneumoniae in the
pre-conjugate vaccine era and to predict the potential
impact of the conjugate vaccine (17). Of pneumococcal
cases identified by ABCs from 1995 to 1998, at least 82%
in children <2 years of age were caused by serotypes
included in the 7-valent pneumococcal conjugate vaccine.
These population-based ABCs data were used to formulate
the original pneumococcal conjugate vaccine schedules
and provide recommendations for administering the vac-
cine to infants and children. When a vaccine shortage
became evident in 2001, ABCs data were again used by
public health officials to weigh alternative strategies for
delivering available doses (18). Surveillance is now
focused on evaluating changes in disease impact after the
conjugate vaccine was introduced, including whether it
interrupts transmission of antibiotic-resistant pneumococ-
ci. Analysis of ABCs data shows a substantial decline in
disease caused by serotypes in the vaccine formulation
among children in the age group for whom the vaccine is
recommended. More modest declines also occur in select-
ed adult groups (19).
ABCs will continue to evaluate the impact of the
recently introduced pneumococcal conjugate vaccine,
including whether vaccine shortages have slowed the ini-
tial steep decline in disease occurrence. Other goals are
measurement of vaccine efficacy, assessment of whether
the vaccine is interrupting transmission, and evaluation of
the distribution of serotypes causing disease (to determine
if decline in disease because of serotypes included in the
vaccine has been counterbalanced by emergence of inva-
sive disease caused by nonvaccine serotypes). While this
“replacement disease” phenomenon was recognized for
otitis media and colonization in the prelicensure vaccine
trials, no evidence of replacement invasive disease has
thus far been recognized.
Clinicians were challenged by the emergence of mul-
tidrug-resistant pneumococci during the 1990s, when new
treatment guidelines were developed for meningitis, otitis
media, and pneumonia (20). Vaccines, in concert with
campaigns to promote appropriate use of antibiotics, pro-
vide opportunities to transform the problem of drug-resist-
ant pneumococci from a treatment dilemma to a prevention
success story (21).
Revised Recommendations for Preventing Perinatal
Group B Streptococcal Disease
Data developed through ABCs provided a basis for
revising recommendations for the prevention of perinatal
group B streptococcal (GBS) disease. Since its emergence
in the 1970s, GBS disease has been the leading invasive
bacterial infection associated with illness and death among
newborns in the United States. Surviving infants may have
long-term developmental disabilities, such as mental retar-
dation or hearing and vision loss. Newborns at increased
risk for GBS disease are those born to women who are col-
onized with GBS in the genital or rectal areas. Although
the use of intrapartum prophylaxis has led to a 70%
decline in the incidence of GBS disease during the 1990s
(Figure 2) (22,23), early-onset GBS disease (in infants <7
days old) remains a leading cause of illness and death
among newborns. Guidelines issued in 1996 recommend-
ed either screening pregnant women for GBS colonization
by means of prenatal cultures (screening approach) or
assessing obstetric risk factors intrapartum (risk-based
approach) to identify candidates for intrapartum antibiotic
prophylaxis.
An EIP population-based, retrospective cohort study
compared the effectiveness of prenatal screening for GBS
with the risk-based approach for preventing early-onset
GBS sepsis (24). The analysis, which combined ABCs
population-based active surveillance data on GBS cases
with a sample survey representing >600,000 deliveries,
showed that infants born to women who had been screened
for GBS before delivering had less than half the risk for
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 783
SYNOPSES
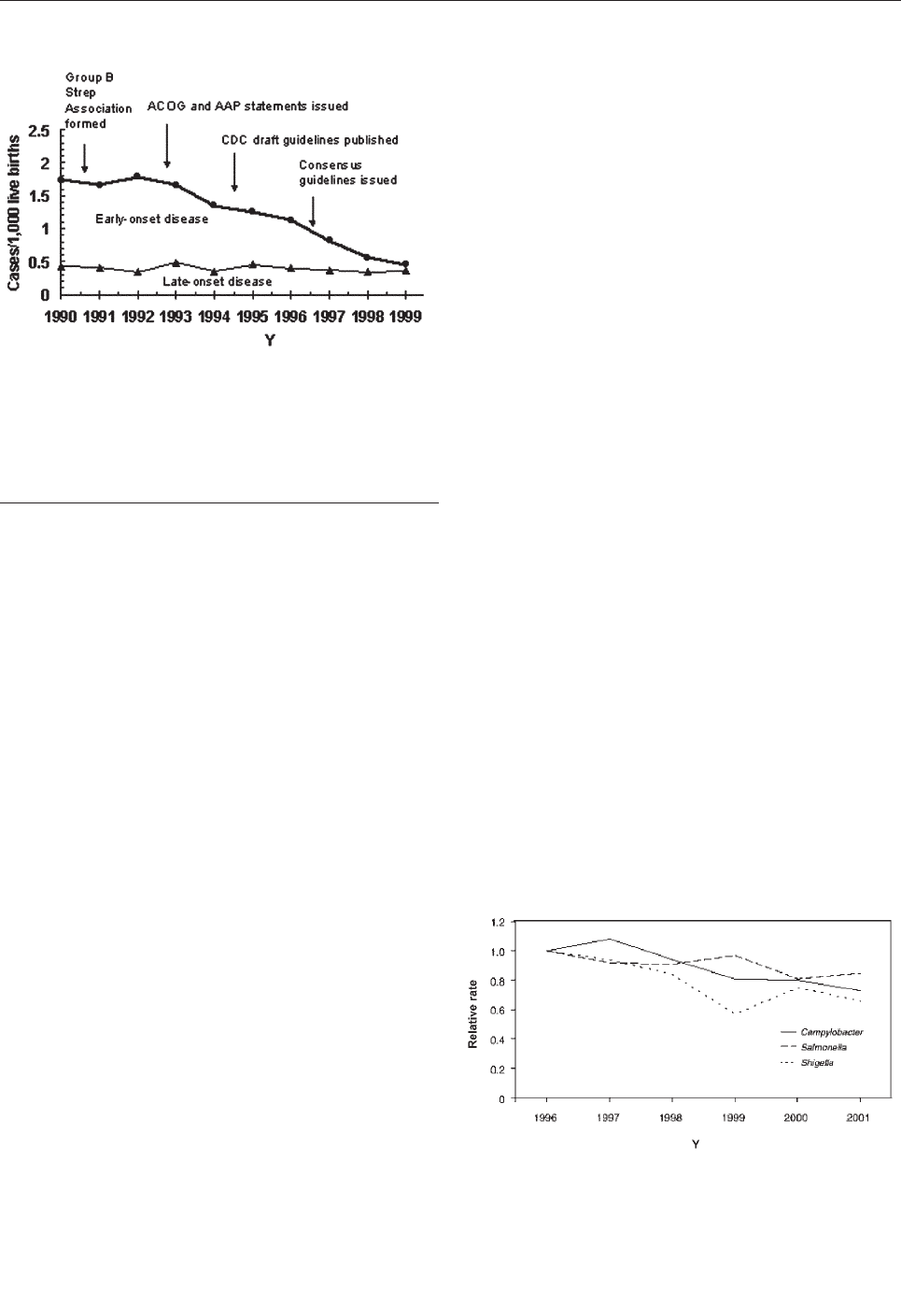
early-onset GBS compared to infants of women who had
not been screened, after adjustments were made for poten-
tial confounders. The protective effect of the screening
approach resulted mainly from broader coverage of the
population at risk because many early-onset GBS cases in
the preprevention era occurred in GBS-colonized women
without obstetric risk factors. The evidence for updated
prevention recommendations from key health organiza-
tions (i.e., American College of Obstetricians and
Gynecologists, American Academy of Pediatrics,
American College of Nurse-Midwives, and CDC) was
based on the finding that routine screening for GBS during
pregnancy more effectively prevents cases of early-onset
disease than the risk-based approach (25). Through ABCs,
CDC will continue to monitor GBS disease trends to
understand the impact of the new recommendations and
detect potential adverse consequences of intrapartum
antibiotic use such as emergence of sepsis caused by other
organisms or new patterns of antimicrobial resistance
(26,27).
Decrease in Bacterial Foodborne Diseases
FoodNet documented a decrease in bacterial foodborne
illnesses from 1996 to 2001. Many infections are transmit-
ted through food and can cause illness ranging from mild
gastroenteritis to severe illness requiring hospitalization.
Foodborne pathogens cause an estimated 76 million ill-
nesses, 325,000 hospitalizations, and 5,000 deaths in the
United States each year (11). Clinicians treating patients
with acute gastroenteritis are principally focused on
whether empiric antimicrobial agents are warranted and
the value of diagnostic evaluation. However, the task of
providing accurate information on trends in specific food-
borne pathogens capable of causing this syndrome, as well
as probable sources of infection, has historically fallen to
public health authorities.
Data from FoodNet documented recent declines in the
occurrence of several major bacterial foodborne illnesses
(9,28); preliminary surveillance data for 2001 were com-
pared with 1996–2000 data (28). Significant declines
occurred in major bacterial foodborne illnesses, including
infections caused by Yersinia (49%), Listeria (35%),
Campylobacter (27%), and Salmonella (15%) (Figure 3).
The combined estimated incidence of infections caused by
Listeria, Campylobacter, Salmonella, and E. coli O157 in
2001 was 21% lower than in 1996, on the basis of a multi-
variate regression model.
The factors influencing the occurrence of foodborne ill-
nesses are complex. However, the observed declines in
foodborne disease incidence did occur in the context of
several control measures, including the U.S. Department
of Agriculture’s Food Safety Inspection Service’s imple-
mentation of the Pathogen Reduction/Hazard Analysis and
Critical Control Point regulations in meat and poultry
slaughter and processing plants, egg-quality assurance pro-
grams for Salmonella Enteritidis, and increased consumer
education in food safety (28).
FoodNet will continue to monitor the occurrence of
foodborne diseases. In 2003, FoodNet will also conduct
studies of the consequences of and risk factors for illness
caused by S. Enteritidis, S. Newport, and illness in infants
caused by Campylobacter and Salmonella. Other activities
include a project to improve collection and transport of
specimens during outbreaks so that a cause is identified in
a higher percentage of outbreaks.
Rapid identification of a cause for cases of infectious
diarrhea and appropriate reporting of cases of foodborne
illnesses to state or local public health authorities are
important not only in identifying and controlling outbreaks
but also for more precise assessments of the local, region-
784 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES
Figure 2. Incidence of early- and late-onset invasive group B strep-
tococcal disease in three active surveillance areas (California,
Georgia, and Tennessee), 1990–1998, and activities for the pre-
vention of group B streptococcal disease (22). CDC, Centers for
Disease Control and Prevention. ACOG, American College of
Obstetricians and Gynecologists; AAP, American Academy of
Pediatrics.
Figure 3. Relative rates compared with 1996, adjusted for sites, of
laboratory-diagnosed cases of Campylobacter, Salmonella, and
Shigella, by year, FoodNet, United States, 1996–2001 (28).
Bacterial pathogens with highest incidences of the 10 studied dis-
eases are shown.

al, and national trends in foodborne illnesses (29). In turn,
such estimates can inform clinicians of likely causes, prob-
able sources, and prognostic factors for episodes of illness
in persons under their care.
Unexplained Deaths and Critical Illnesses Project
Many clinicians have treated patients with puzzling sit-
uations, in which the acute onset of a critical illness sug-
gestive of an infectious origin occurred in otherwise
healthy young people for whom diagnostic tests failed to
identify an etiologic agent. Occasionally, such episodes are
retrospectively diagnosed many years later with the recog-
nition of a new infectious disease and testing of stored
clinical specimens. For example, hantavirus pulmonary
syndrome was first recognized and described in the United
States in 1993 by an alert clinician during an outbreak in
the Southwest (30); retrospective reviews of fatal illnesses
showed that unrecognized cases of hantavirus pulmonary
syndrome had preceded the 1993 outbreak by at least 15
years (31). Similarly, cases of legionellosis and AIDS were
recognized in hindsight years after they had occurred (13).
These observations, coupled with the new laboratory tech-
niques for pathogen identification, particularly methods
that do not rely on culture, suggested that an effort to
prospectively identify pathogens causing unexplained syn-
dromes might yield useful information (12,13); this was
the beginning of the UNEX project. Laboratory evaluation
of cases includes traditional serologic and in vitro culture
diagnostic methods as well as molecular techniques. This
combined epidemiologic and laboratory approach is a hall-
mark feature of other EIP projects that study hepatitis,
acute respiratory diseases, and encephalitis (32).
The UNEX project has developed methods for evaluat-
ing severe syndromes indicating infection, including non-
culture-based methods to identify etiologic agents. From
May 1, 1995, to December 31, 1998, 137 illnesses meeting
the UNEX case definition were reported to participating
EIPs. After adjustments for age and race were made, this
number translates to an estimated 920 U.S. cases per year;
the overall annual incidence rates did not change during
this time. No differences were observed in the seasonal
distribution of cases of unexplained illnesses, nor did cases
cluster by time or place. The largest proportion of cases
was treated as a neurologic syndrome (29%), followed by
respiratory (27%) and cardiac (21%) syndromes.
Diagnostic testing through UNEX identified a cause in 34
(28%) of 122 cases from which specimens were available
(Table 2).
Two recent outbreaks demonstrate the usefulness of the
approach developed for UNEX. During a 1999 outbreak of
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 785
SYNOPSES
Table 2. Infectious causes and explanations for unexplained deaths and critical illnesses cases, 1995–1998, California, Oregon,
Connecticut, and Minnesota (n=34)
a,b
Syndrome
Etiologic agent (n)
Tests (n)
Neisseria meningitidis (4)
16S rDNA PCR (2), PCR (1), EIA IgM (1)
a
Bartonella hensaelae (1)
PCR, IFA, IgG
Bartonella spp. (2)
IFA, IgG
Chlamydia pneumoniae (1)
MIF, IgG
Mycoplasma pneumoniae (1)
EIA, IgM/IgG
Cytomegalovirus (1)
EIA and IFA,IgG
Coxsackie B virus (1)
EIA, IgM, viral culture
Enterovirus (1)
EIA, IgM
Epstein-Barr virus (1)
IFA, IgG (VCA and EA)
Human herpesvirus 6 (1)
IFA and EIA (IgM and IgG)
Neurologic (n=15)
Mumps virus (1)
IFA IgM, IFA and EIA, IgG
Chlamydia pneumoniae (2)
MIF IgG (2), IFA, IgM
Mycoplasma pneumoniae (4)
PCR (blood), EIA, IgM/IgG
Streptococcus pneumoniae (2)
16S rDNA PCR (pleural fluid)
Legionella spp. (1)
PCR (from lung)
Adenovirus (1)
EIA and IFA, IgG
Influenza B virus (1)
EIA and IFA, IgG
Influenza A virus (1)
EIA and IFA, IgM, EIA (IgG)
Respiratory (n=13)
Human parainfluenza virus types 1 and 3 (1)
EIA and IFA, IgG
Borrelia burgdorferi/Ehrlichia chaffeensis (1)
EIA/IFA flagella, IgG, Western blot (IgG and IgM)
Enterovirus (1)
EIA IgM
Cardiac (n=3)
Legionella spp. (1)
PCR (heart)
Neisseria meningitidis (1)
PCR (cerebrospinal fluid)
Adenovirus (1)
PCR (blood)
Multisystem (n=3)
Enterovirus (1)
IgM, EIA
a
PCR, polymerase chain reaction; EIA, enzyme immunosorbent assay; IFA, indirect immunofluorescent assay; Ig, immunoglobulin; EA,
early antigens; VCA, viral capsid
antigens; MIF, microimmunofluorescence.
b
Reference 12.

West Nile encephalitis in the northeastern United States,
which was recognized by an alert clinician (33), and dur-
ing an outbreak of unexplained illness among injecting
drug users in Scotland and Ireland (34), initial reports of
illness were received and initial laboratory testing per-
formed through the laboratory infrastructure established
for the UNEX project.
The frequency and distribution of the syndromes identi-
fied through this project undoubtedly reflect both the dis-
tribution of their occurrence and gaps in our ability to diag-
nose causes of neurologic and respiratory syndromes in
particular. Although novel pathogens have not yet been dis-
covered through the UNEX project, this systematic
approach improves chances of recognizing infectious dis-
ease causes earlier than in the past and lays the groundwork
for the development of improved diagnostic tools.
Moreover, concerns about bioterrorism have put a premi-
um on the early detection of an intentional release or infec-
tious or chemical agents; this syndrome-based surveil-
lance, which seeks early identification and diagnosis, can
contribute to public health preparedness for such events.
Future Directions of EIPs
Since the release of the plan that launched the EIPs,
these programs have made substantial contributions to the
practice of U.S. public health. Using domestic EIPs as a
model, CDC has begun developing a network of interna-
tional EIPs (IEIPs) in collaboration with Ministries of
Health and other international partners. The first IEIP was
established in Thailand during 2001, and a second IEIP is
being established in Kenya. Collaborations between EIPs
and IEIPs will provide valuable opportunities for training.
In addition, the new U.S. EIP in New Mexico will feature
work along the U.S.-Mexico border and also promises to
enhance international collaborations.
Opportunities presented by new laboratory and infor-
mation technologies, as well as challenges posed by poten-
tial bioterrorism, will influence the evolution of the EIPs
over the next several years. EIP work will build on experi-
ence gained through the combined epidemiologic and lab-
oratory evaluation of syndromes to enhance bioterrorism
preparedness and develop the capacity for identifying pre-
viously unrecognized pathogens. However, even as new
technologies are found, knowledgeable and engaged clini-
cians will remain a vital element in efforts to detect,
respond to, and prevent emerging infectious diseases.
Acknowledgments
We thank Cynthia Whitney for providing information about
the pneumococcal conjugate vaccine and pneumococcal disease
and Matthew Moore for providing information about foodborne
disease.
Dr. Pinner is the director of the Office of Surveillance,
National Center for Infectious Diseases, Centers for Disease
Control and Prevention. His scientific interests are public health
surveillance, especially for infectious diseases; trends in deaths
attributed to infectious diseases; and the epidemiology of several
bacterial and fungal diseases.
References
1. Gerberding JL, Hughes JM, Koplan JP. Bioterrorism preparedness
and response: clinicians and public health agencies as essential part-
ners. JAMA 2002;287:898–90.
2. Petersen LR, Marfin AA. West Nile virus: a primer for the clinician.
Ann Intern Med 2002;137:173–9.
3. Centers for Disease Control and Prevention. West Nile virus activi-
ty—United States, September 26–October 2, 2002, and investigations
of West Nile virus infections in recipients of blood transfusion and
organ transplantation. MMWR Morb Mortal Wkly Rep
2002;51:884,895.
4. Institute of Medicine. Emerging infections: microbial threats to
health in the United States. Washington: National Academy Press;
1992.
5. Centers for Disease Control and Prevention. Addressing emerging
infectious disease threats: a prevention strategy for the United States.
Atlanta: U.S. Department of Health and Human Services, Public
Health Service; 1994.
6. Centers for Disease Control and Prevention. Preventing emerging
infectious diseases: a strategy for the 21st Century. Atlanta: U.S.
Department of Health and Human Services, Public Health Service;
1998.
7. Hardnett FP, Hoekstra RM, Kennedy MH, Angulo FJ, EIP Foodnet
Working Group. Comparability of FoodNet and United States popu-
lations. Poster presented at: International Conference on Emerging
Infectious Diseases; 24–27 March 2002; Atlanta, Ga.
8. Schuchat A, Hilger T, Zell E, Farley MM, Reingold A, Harrison L, et
al. Active bacterial core surveillance of the emerging infections pro-
gram network. Emerg Infect Dis 2001;7:92–9.
9. Centers for Disease Control and Prevention. Foodborne diseases
active surveillance network. MMWR Morb Mortal Wkly Rep
1997;46:258–61.
10. Herikstad H, Yang S, Van Gilder TJ, Vugia D, Hadler J, Blake P, et al.
A population-based estimate of the burden of diarrhoeal illness in the
United States: FoodNet, 1996–7. Epidemiol Infect 2002;129:9–17.
11. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et
al. Food-related illness and death in the United States. Emerg Infect
Dis 1999;5:607–25.
12. Hajjeh RA, Relman D, Cieslak PR, Sofair AN, Passaro D, Flood J, et
al. Surveillance for unexplained deaths and critical illnesses due to
possibly infectious cases, United States, 1995–1998. Emerg Infect
Dis 2002;8:145–53.
13. Perkins BA, Flood JM, Danila R, Holman RC, Reingold AL, Klug
LA, et al. Unexplained deaths due to possibly infectious causes in the
United States: defining the problem and designing surveillance and
laboratory approaches. Emerg Infect Dis 1996;2:47–53.
14. Ijdo JW, Meek JI, Cartter ML, Magnarelli LA, Wu C, Tenuta SW, et
al. Emergence of another tickborne infection in the 12-town area
around Lyme, CT: human ehrlichiosis. J Infect Dis
2000;181:1388–93.
15. Centers for Disease Control and Prevention. Surveillance for
Creutzfeldt-Jakob disease—United States. MMWR Morb Mortal
Wkly Rep 1996;45:665–8.
16. Centers for Disease Control and Prevention. Preventing pneumococ-
cal disease among infants and young children: recommendations of
the Advisory Committee on Immunization Practices (ACIP). MMWR
Morb Mortal Wkly Rep 2000;49(RR-9):1–35.
786 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
SYNOPSES

17. Robinson KA, Baughman W, Rothrock G, Barrett NL, Pass M, Lexau
C, et al. Epidemiology of invasive Streptococcus pneumoniae infec-
tions in the United States, 1995–1998: opportunities for prevention in
the conjugate vaccine era. JAMA 2001;285:1729–35.
18. Centers for Disease Control and Prevention. Notice to readers:
decreasing availability of pneumococcal conjugate vaccine. MMWR
Morb Mortal Wkly Rep 2001;50:783.
19. Whitney CG, Farley MM, Hadler J, Harrison LH, Bennet NM,
Lynfield R, et al. Decline in invasive pneumococcal disease after the
introduction of protein-polysaccharide conjugate vaccine. N Engl J
Med 2003;348;1737–46.
20. Whitney CG, Farley MM, Hadler J, Harrison LH, Lexau C, Reingold
A, et al. Increasing prevalence of multidrug-resistant Streptococcus
pneumoniae in the United States. N Engl J Med 2000;343:1917–24.
21. Heffelfinger JD, Dowell SF, Jorgensen JH, Klugman KP, Mabry LR,
Musher DM, et al. Management of community-acquired pneumonia
in the era of pneumococcal resistance: a report from the drug-resist-
ant Pneumococcus pneumonia therapeutic working group. Arch
Intern Med 2000;160:1399–408.
22. Schrag SJ, Zywicki S, Farley M, Reingold A, Harrison L, Lefkowitz
L, et al. Group B streptococcal disease in the era of intrapartum
antibiotic prophylaxis: population-based surveillance in the United
States, 1993–1998. N Engl J Med 2000;342:15–20.
23. Centers for Disease Control and Prevention. Early-onset group B
streptococcal disease–United States, 1998–1999. MMWR Morb
Mortal Wkly Rep 2000;49:793–6.
24. Schrag SJ, Zell ER, Lynfield R, Roome A, Arnold KE, Craig AS, et
al. A population-based comparison of strategies to prevent early-onset
group B streptococcal disease in neonates. N Engl J Med
2002;347:233–9.
25. Schrag S, Gorwitz R, Fultz-Butts K, Schuchat A. Prevention of peri-
natal group B streptococcal disease. Revised guidelines from the
CDC. MMWR Morb Mortal Wkly Rep 2002;51(RR-11):1–22.
26. Hyde TB, Hilger TM, Reingold A, Farley MM, O’Brien KL,
Schuchat A. Trends in the incidence and antimicrobial resistance of
early-onset sepsis: population-based surveillance in San Francisco
and Atlanta. Pediatrics 2002;110:690–5.
27. Baltimore RS, Huie SM, Meek JI, Schuchat A, O’Brien KL. Early-
onset neonatal sepsis in the era of group B streptococcal prevention.
Pediatrics 2001:108:1094–8.
28. Centers for Disease Control and Prevention. Preliminary FoodNet
data on the incidence of foodborne illnesses—select sites, United
States, 2001. MMWR Morb Mortal Wkly Rep 2002;51:325–9.
29. Centers for Disease Control and Prevention. Diagnosis and manage-
ment of foodborne illnesses. MMWR Morb Mortal Wkly Rep
2001;50(RR-2):1–69.
30. Duchin JS, Koster FT, Peters CJ, Simpson GL, Tempest B, Zaki SR,
et al. Hantavirus pulmonary syndrome: a clinical description of 17
patients with a newly recognized disease. N Engl J Med
1994;331:546–8.
31. Zaki SR, Khan AS, Goodman RA, Armstrong LR, Greer PW,
Coffield LM, et al. Retrospective diagnosis of hantavirus pulmonary
syndrome, 1978–1993: implications for emerging infectious diseases.
Arch Pathol Lab Med 1996;120:134–9.
32. Park SY, Glaser C, Murray WJ, Kazacos KR, Rowley HA, Frederick
DR, et al. Raccoon roundworm (Baylisascaris procyonis) encephali-
tis: case report and field investigation. Pediatrics 2000;106:e56.
33. Asnis DS, Conetta R, Teixeira AA, Waldman G, Sampson BA. The
West Nile virus outbreak of 1999 in New York: the Flushing Hospital
experience. Clin Infect Dis 2000;30:413–8.
34. Centers for Disease Control and Prevention. Update: Clostridium
novyi and unexplained illness among injecting drug users—Scotland,
Ireland, and England, April–June 2000. MMWR Morb Mortal Wkly
Rep 2000;49:543–5.
Address for correspondence: Robert W. Pinner, Centers for Disease
Control and Prevention, 1600 Clifton Rd., Mailstop D59, Atlanta, GA
30333, USA; fax: 404-371-5445; email: [email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 787
SYNOPSES
All material published in Emerging Infectious Diseases is in the
public domain and may be used and reprinted without special per-
mission; proper citation, however, is appreciated.

Acute weakness associated with West Nile virus
(WNV) infection has previously been attributed to a periph-
eral demyelinating process (Guillain-Barré syndrome);
however, the exact etiology of this acute flaccid paralysis
has not been systematically assessed. To thoroughly
describe the clinical, laboratory, and electrodiagnostic fea-
tures of this paralysis syndrome, we evaluated acute flac-
cid paralysis that developed in seven patients in the setting
of acute WNV infection, consecutively identified in four hos-
pitals in St. Tammany Parish and New Orleans, Louisiana,
and Jackson, Mississippi. All patients had acute onset of
asymmetric weakness and areflexia but no sensory abnor-
malities. Clinical and electrodiagnostic data suggested the
involvement of spinal anterior horn cells, resulting in a
poliomyelitis-like syndrome. In areas in which transmission
is occurring, WNV infection should be considered in
patients with acute flaccid paralysis. Recognition that such
weakness may be of spinal origin may prevent inappropri-
ate treatment and diagnostic testing.
M
ost human infections with West Nile virus (WNV), a
flavivirus within the Japanese encephalitis virus
antigenic complex, are clinically inapparent (1,2). Mild
febrile illness develops in approximately 1 in 5 infected
persons; more severe neurologic disease, mostly meningi-
tis or encephalitis, occurs in and 1 in 150 (1–4). Less fre-
quently, acute WNV infection has been associated with
acute flaccid paralysis, which has been attributed to
Guillain-Barré syndrome, motor axonopathy, or axonal
polyneuropathy (4–6). However, these reports describe
clinical and laboratory features that seem inconsistent with
such diagnoses, and the exact cause of acute flaccid paral-
ysis has not been thoroughly assessed with rigorous elec-
trophysiologic, laboratory, and neuroimaging data. Brief
descriptions of six patients have suggested that this flaccid
paralysis is due to anterior horn cell involvement with a
resultant poliomyelitis-like syndrome (7–9). Because
understanding the clinical characteristics and underlying
etiology of WNV-induced acute flaccid paralysis is critical
for therapeutic decisions as well as prognosis, we describe
the detailed clinical, laboratory, and electrophysiologic
findings from these six patients and from one additional
patient.
Patients and Methods
Seven patients were detected through WNV surveil-
lance conducted by the Mississippi Department of Health
and the Louisiana Office of Public Health. For each
patient, a standardized questionnaire, including demo-
graphics, medical history, initial signs and symptoms, risk
factors, and treatment, was completed; a standardized neu-
rologic examination was performed by a single neurologist
(JJS). Electrodiagnostic studies were performed by neurol-
ogists (AAL and JAVG) specializing in electrodiagnostic
medicine.
Cerebrospinal fluid (CSF) and acute- or convalescent-
phase serum specimens (or both) from each patient were
tested for antibody to WNV by immunoglobulin (Ig) M
antibody-capture enzyme immunoassay (10) or plaque
reduction neutralization assay (11). The initial specimen
for one patient (patient 5, Table 1) was tested with a slight-
ly modified IgM antibody assay at a commercial laborato-
ry (12). IgM assays were considered positive if the optical
density ratio of the patient and negative control samples
(P/N ratio) was greater than three. For patient samples, a
P/N ratio for WNV at least three times that for St. Louis
encephalitis virus indicated WNV infection (13). A plaque
reduction neutralization test result of at least 10 was con-
sidered positive.
All seven patients had serologic evidence of WNV infec-
tion (Table 1). On the basis of serologic data, three of the
patients were classified as confirmed case-patients (patients
4, 6, and 7) and four as probable case-patients (patients 1–3,
5), according to the national case definition (14).
Case 1
On July 1, 2002, a previously healthy, 56-year-old,
male Mississippi resident was hospitalized with a 1-week
788 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Acute Flaccid Paralysis and West
Nile Virus Infection
James J. Sejvar,* A. Arturo Leis,† Dobrivoje S. Stokic,† Jay A. Van Gerpen,‡
Anthony A. Marfin,* Risa Webb,§ Maryam B. Haddad,* Bruce C. Tierney,* Sally A. Slavinski,§
Jo Lynn Polk,† Victor Dostrow,† Michael Winkelmann,† and Lyle R. Petersen*
*Centers for Disease Control and Prevention, Atlanta, Georgia,
USA; †Methodist Rehabilitation Center, Jackson, Mississippi,
USA; ‡Ochsner Clinic, New Orleans, Louisiana, USA; and
§Mississippi State Department of Health, Jackson, Mississippi,
USA

history of fever, chills, night sweats, myalgias, and acute
encephalopathy. Neurologic examination showed pro-
found weakness in both arms, asymmetric weakness in the
legs with a right foot drop, and acute respiratory distress
(Table 2). Sensory test results were normal. Although com-
puted tomography (CT) and magnetic resonance imaging
(MRI) of the brain showed normal results, heparin was
administered for suspected evolving stroke. Admission
laboratory values (Table 3) showed serum leukocytosis,
and cerebrospinal fluid (CSF) obtained on day 3 showed
elevated protein. By day 8, Guillain-Barré syndrome was
suspected, and intravenous immunoglobulin (IVIG) was
administered. Electrodiagnostic studies performed that day
were interpreted as showing a proximal neuropathy or
myopathy. A deltoid muscle biopsy for suspected inflam-
matory myopathy showed mild type 2 fiber atrophy but no
obvious necrosis or marked inflammatory response. On
day 30, the patient was transferred for rehabilitation with
flaccid, areflexic paralysis in the right leg and variable
weakness and diminished reflexes in all other limbs. Neck
flexors were normal in strength, and sensation was pre-
served in all limbs; loss of bladder function was evident.
MRI of the cervical spine showed normal results.
Electrodiagnostic studies showed widespread but variable
denervation, reduced compound muscle action potentials
(CMAPs), and normal sensory nerve action potentials
(SNAPs), consistent with a severe, asymmetric process
affecting anterior horn cells or motor axons. Myopathy,
demyelinating polyneuropathy, and diffuse axonal
polyneuropathy were not evident.
Case 2
On July 15, 2002, a 57-year-old male Mississippi resi-
dent with a remote history of prostate cancer and glucose
intolerance was hospitalized with a 3-day history of fever,
chills, nausea, vomiting, and headache. Neurologic exam-
ination showed encephalopathy and asymmetric weakness
in all limbs (Table 2). Results of a brain MRI weree nor-
mal. Admission laboratory studies showed a CSF pleocy-
tosis with elevated protein (Table 3). On day 5, acute res-
piratory distress developed, and the patient required
mechanical ventilation. Upon extubation 2 weeks later, the
patient had continued extremity weakness and was aspirat-
ing fluids; a modified barium swallow study showed
oropharyngeal dysphagia. Upon transfer to a rehabilitation
center on day 30, the patient had asymmetric weakness in
the legs and right arm and moderate weakness in neck flex-
ors and facial muscles. Hypotonia and areflexia were noted
in all limbs. Sensation was slightly diminished to vibration
and proprioception in toes bilaterally but preserved to light
touch, pinprick, and temperature. Sensory testing was nor-
mal in the upper limbs. Urinary incontinence was noted.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 789
RESEARCH
Table 1. Serologic results for West Nile virus (WNV)–specific antibodies in patients with acute flaccid paralysis associated with acute
WNV infection
a
IgM-capture enzyme immunoassay
Plaque reduction neutralization assay
Case no.
Onset
Collection
Sample
SLEV
WNV
SLEV
WNV
1
6/24
7/12
Serum
3.5
22.3
320
5,120
2
7/12
7/16
Serum
8.0
22.7
80
1,280
3
7/26
8/1
Serum
2.79
24.9
<10
640
4
7/29
8/3
Serum
1.1
14.1
<10
80
4
7/29
8/3
CSF
3.3
39.2
4
7/29
8/13
Serum
4.4
23.5
40
2,560
5
8/11
8/15
Serum
2.02
5
8/11
8/29
Serum
3.4
25.7
6
8/13
8/16
CSF
6.1
23.8
6
8/13
8/16
Serum
1.0
5.7
<10
40
7
9/1
10/24
Serum
2.8
10.6
10
320
7
9/1
9/6
CSF
Not performed
7.4
a
IgM, immunoglobulin M; SLEV, Saint Louis encephalitis virus; CSF, cerebrospinal fluid.
Table 2. Initial clinical signs and symptoms in patients with acute flaccid paralysis associated with acute West Nile virus infection
Case no.
Fever (>38.5°C)
Headache
Nuchal rigidity
Altered mental status
Tremor
Distribution of weakness
a
1
+
+
+
+
+
Upper and lower limbs, R > L
2
+
+
-
+
-
Upper and lower limbs, R > L
3
+
-
-
-
+
Lower limbs, R > L
4
+
+
+
+
+
R upper limb
5
-
+
-
-
-
R upper limb
6
+
+
-
-
+
Lower limbs, R > L
7
+
+
+
-
-
Upper and lower limbs, L > R;
bulbar muscles
a
R, right; L, left.

Electrodiagnostic studies showed widespread denervation,
reduced CMAP amplitudes in all nerves of the lower limbs
and right upper limb, and normal SNAP responses, consis-
tent with a severe, asymmetric process affecting anterior
horn cells or motor axons. Myopathy, demyelinating
polyneuropathy, and diffuse axonal polyneuropathy were
not apparent.
Case 3
On July 24, 2002, a low-grade fever, nausea, and vom-
iting, followed by shaking chills and sweats, developed in
a 56-year-old male Louisiana resident with a history of
hypertension and coronary artery disease. The next day,
asymmetric weakness developed in the lower extremities,
with no pain or numbness. Upper extremities were normal.
No bowel or bladder dysfunction was present. The patient
was hospitalized on July 29, and neurologic examination
showed a flaccid, areflexic right lower extremity and a
weak left lower extremity with diminished reflexes.
Results of strength and reflex testing of the upper extrem-
ities were normal. Sensory examination results were nor-
mal except for a mild decrease in sensitivity to pinprick,
temperature, touch, and vibration in a stocking-and-glove
distribution (i.e., distal arms and legs). A coarse bilateral
upper extremity action tremor was noted. The patient had
no headache, neck stiffness, or alteration of mental status
(Table 2). Admission laboratory values showed leukocyto-
sis and CSF pleocytosis (Table 3). Results of other diag-
nostic tests were unremarkable. Postviral demyelination
syndrome and viral-induced polyradicultis were consid-
ered, and IVIG, dexamethasone, and antibacterial and
antiviral medications were administered without patient
improvement. On day 15, the patient was discharged to a
skilled nursing facility for rehabilitation.
MRI of the cervical, thoracic, and lumbosacral spine
obtained during rehabilitation was notable for showing
mild cervical and lumbosacral spinal stenosis and forami-
nal restrictions from C3 through C7 and homogeneous
enhancement of the nerve roots of the cauda equina consis-
tent with meningitis. Electrodiagnostic studies showed
denervation in thoracic and lumbosacral myotomes, with
no muscle activation in the right leg and reduced muscle
activation in the left leg. CMAPs in the right leg were
absent; SNAPs were normal. Electrodiagnostic findings
suggested a severe, asymmetric process affecting anterior
horn cells or motor axons. Diffuse axonal polyneuropathy
was not evident, despite a slight sensory loss in the distal
extremities.
Case 4
On August 2, 2002, fever, headache, and neck stiffness
developed in a 69-year-old female Louisiana resident with
a history of diabetes and degenerative disc disease; the
next day acute weakness occurred in the right arm without
pain, numbness, or paresthesias. She was hospitalized on
August 4. On admission, physical examination document-
ed fever, vomiting, encephalopathy, nuchal rigidity, and a
bilateral rash on the lower extremities. Neurologic exam-
ination displayed a flaccid and areflexic right arm. Her
legs and left arm exhibited normal strength, reflexes, and
coordination, with normal sensation in all limbs. A coarse
tremor was noted in the chin, left arm, and legs (Table 2).
Laboratory findings included CSF pleocytosis (Table 3).
Differential diagnoses included meningoencephalitis with
associated motor polyradiculopathy and monoplegia sec-
ondary to stroke. The patient was treated with antibacter-
ial and antiviral medications. Results of CT and MRI of
the brain were normal. MRI of the cervical spine showed
multilevel degenerative disc disease. The patient
remained lethargic until day 13, when mental status
abruptly improved; right arm weakness persisted. On day
19, she was transferred to a rehabilitation facility.
Electrodiagnostic studies showed absent CMAPs and pro-
found denervation with no voluntary activation in muscles
of the right arm. Scattered denervation was also seen in
the other three limbs. SNAPs had borderline amplitudes
and conduction velocities bilaterally. The results were
most consistent with a severe, asymmetric process affect-
ing anterior horn cells or motor axons. The patient was
subsequently transferred back to intensive care because
her respiratory function was deteriorating, but she was not
intubated. After Guillain-Barré syndrome was diagnosed,
she was started on IVIG but had no improvement in
weakness.
790 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 3. Initial laboratory findings in patients with acute flaccid paralysis associated with acute West Nile virus infection
Case no.
Leukocytes
(x10
3
/mm
3
)
Hematocrit (%)
CSF
WBC (/mm
3
)
CSF
RBC (/mm
3
)
CSF
protein (mg/dL)
CSF
glucose (mg/dL)
1
17.6
38.0
3
1,778
89
54
2
3.6
38.2
2,600
87
204
99
3
11.8
44.4
140
40
234
74
4
9.5
37.8
143
4
116
119
5
7.9
45.6
Not performed
Not performed
Not performed
Not performed
6
13.0
45.4
329
7
75
66
7
10.3
Not performed
182
9
37
79
a
CSF, cerebrospinal fluid; WBC, leukocyte count; RBC, erythrocyte count.

Case 5
On August 11, 2002, severe nausea, vomiting,
headache, and diarrhea in the absence of fever developed
in a 50-year-old male Mississippi resident with a history of
alcohol abuse; the next day, progressive right arm weak-
ness developed. He was hospitalized on August 14.
Neurologic examination on admission showed flaccid
paralysis of the right arm and mild weakness of the right
leg, with normal sensation in all limbs (Table 2).
Laboratory values are shown in Table 3; a lumbar puncture
was not performed. Acute stroke was diagnosed, and the
patient was treated with heparin. Mental status changes,
dysarthria, and dysphagia subsequently developed but
resolved. Upon transfer to a rehabilitation center on day
12, the patient had paralysis and areflexia limited to the
right arm, with normal sensation and diffuse tremor in all
limbs. Brain MRI results were normal; cervical spine MRI
displayed mild multilevel foraminal stenosis on the left.
Electrodiagnostic studies showed markedly reduced motor
responses in the right arm with normal sensory responses,
consistent with a severe asymmetric process affecting
anterior horn cells.
Case 6
On August 16, 2002, a 46-year-old male Louisiana res-
ident with a history of coronary artery disease was hospi-
talized with fever, headache, fatigue, and leg weakness of
3 days’ duration. He reported no nuchal rigidity or mental
status changes, although family members described him as
intermittently confused. Neurologic examination showed a
plegic and areflexic right leg and mild left leg weakness;
sensation was intact throughout. A bilateral tremor of the
upper extremities and jaw was noted (Table 2). Laboratory
abnormalities included a CSF pleocytosis (Table 3). He
was diagnosed with Guillain-Barré syndrome and started
on IVIG. Brain CT and MRI results were normal. Results
of an enhanced MRI of the spine suggested meningitis
involving the conus medullaris and cauda equina.
Electrodiagnostic studies performed on day 4 demonstrat-
ed early denervation and absent activation in muscles of
the right leg and reduced activation of muscles in the right
arm. CMAPs and SNAPs in the right arm and leg were
normal. These findings were consistent with a severe,
asymmetric process affecting anterior horn cells or motor
axons. He was transferred to a rehabilitation facility on day
6 with no improvement of weakness.
Case 7
On September 1, 2002, a previously healthy, 39-year-
old male Louisiana resident had onset of fever, headache,
and nuchal rigidity followed the next day by dysphagia and
bilateral arm and leg weakness that was worse on the left.
He was hospitalized on September 6 for acute respiratory
failure and intubated. Neurologic examination showed
normal cognition, asymmetric flaccid paralysis of the left
arm and leg with absent reflexes, hyporeflexic weakness of
the right arm and leg, and weakness of bulbar muscles
(Table 2). A partial supranuclear gaze palsy, cogwheel
rigidity, and bilateral Babinski signs were also evident.
Admission laboratory findings showed peripheral leukocy-
tosis and CSF pleocytosis (Table 3). Brain MRI showed
increased T2 signal in the periaqueductal gray matter, sub-
stantia nigra, and trigeminal motor nuclei.
Electrodiagnostic studies performed on day 15 showed dif-
fuse denervation in all myotomes, reduced CMAPs (worse
on the left), and preserved SNAPs. On day 25, he was
transferred to a long-term care facility with no improve-
ment of limb weakness.
Discussion
The clinical and electrodiagnostic findings in these
patients with WNV infection suggest involvement of
spinal cord gray matter, specifically anterior horn cells,
and a resulting acute poliomyelitis-like syndrome. All
patients exhibited features typical for polio, including
acute flaccid paralysis without paresthesias or sensory
loss, marked asymmetric weakness, diminished or absent
deep tendon reflexes in the affected limbs, and weakness
that developed during an acute infectious process. Other
typical features of poliomyelitis included CSF pleocytosis
in five of six patients with CSF examination, acute respi-
ratory distress in four, and acute changes in bowel or blad-
der function in two. In addition, electrodiagnostic findings
showed asymmetric muscle denervation, reduced CMAPs,
and preserved SNAPs. No patients had evidence of
demyelinating polyneuropathy or myopathy. The absence
of new sensory abnormalities localizes the disease process
to the anterior horn cells or motor axons. Although muscle
denervation and reduced CMAP amplitudes do not distin-
guish loss of anterior horn cells from loss of motor axons
(15), these patients’ clinical features can be explained only
by anterior horn cell disease, since no known infectious
processes limited to motor axons produce widespread,
asymmetric paralysis without sensory involvement. While
MRI signal abnormalities in the anterior spinal cord have
been noted in patients with poliomyelitis (16,17), these
findings are inconsistent (18,19), and the absence of such
changes in our four patients in which imaging was per-
formed does not preclude a diagnosis of a poliomyelitis-
like syndrome.
Since immunization has eradicated wild-type
poliovirus from the developed world, most cases of para-
lytic polio-like conditions in the United States have been
linked to other RNA viruses, including echoviruses,
enteroviruses, and coxsackieviruses (20). Case reports
have documented a poliomyelitis-type syndrome associat-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 791
RESEARCH

ed with other flaviviruses (21–23), as well as anterior
myelitis associated with WNV infection (24).
The assertion that WNV infection involves anterior
horn cells and causes a polio-type syndrome has a patho-
logic basis. The neuropathology of experimental WNV
infection in monkeys was most pronounced in the cerebel-
lum, medulla, and the cervical and lumbar regions of the
spinal cord (25). Anterior horn cells showed degeneration
and neuronal cell death; conversely, no changes were seen
in the oligodendroglia or peripheral nerves. Similarly,
WNV-infected horses displayed multifocal polioen-
cephalomyelitis, with involvement of the ventral and later-
al horns of the thoracic and lumbar spinal cord (26,27).
WNV antigen was mainly localized within the gray matter
of the spinal cord, with no lesions apparent in peripheral
nerves or ganglia. In WNV-infected birds, lesions and viral
antigen were most prominent in the cerebellum and the
gray matter of the spinal cord (28).
Previous case studies have attributed WNV-associated
acute flaccid paralysis to Guillain-Barré syndrome, motor
axonopathy, or severe axonal polyneuropathy (4–6). The
clinical signs and symptoms and electrodiagnostic findings
reported in those cases, and those described here, are most
consistent with a polio-like condition, and would be atypi-
cal for Guillain-Barré syndrome or other peripheral nerve
disorders. Although acute poliomyelitis and polio-like con-
ditions may occasionally simulate Guillain-Barré syn-
drome (29), our cases had several clinical, laboratory, and
electrodiagnostic features that differed from typical
Guillain-Barré syndrome (30–32; Table 4).
In Guillain-Barré syndrome, electrodiagnostic findings
generally suggest peripheral nerve demyelination or, less
commonly, a combined demyelinating and axonal process
(30,31). The cases reported here displayed reduced or
absent CMAPs with preserved SNAPs, no evidence of
demyelination, a neurogenic pattern of recruitment, and
widespread denervation; combined with the clinical pic-
ture of an asymmetric paralysis, these findings are typical
for a polio-like condition and uncommon for Guillain-
Barré syndrome. A pure axonal variant of Guillain-Barré
syndrome has been described (33) and may be confused
with poliomyelitis and polio-like conditions; however,
such cases are generally characterized by distally promi-
nent weakness and show subclinical sensory nerve
involvement on electrodiagnostic testing. Thus, in the con-
text of WNV infection, electrodiagnostic studies previous-
ly interpreted as motor axonal polyneuropathy or motor
axonopathy without sensory nerve involvement (4–6) are
more suggestive of anterior horn cell loss than of Guillain-
Barré syndrome.
Three of the seven patients had acute flaccid paralysis
without other findings, suggestive of severe central nerv-
ous system involvement caused by WNV infection.
Physicians should suspect WNV infection in patients from
areas where WNV is being transmitted and who have
acute, painless, asymmetric weakness, even if unaccompa-
nied by fever or apparent meningoencephalitis. Diagnostic
studies should include testing for WNV-specific IgM anti-
body in CSF or acute- and convalescent-phase serum sam-
ples. In patients from such areas who have acute flaccid
paralysis, CSF analysis, thorough electrodiagnostic stud-
ies, and spinal imaging should be considered before initi-
ating diagnostic evaluations or therapies directed at
Guillain-Barré syndrome, stroke, inflammatory
myopathies, or other peripheral inflammatory processes.
These therapies are ineffective for polio-like syndromes
and can produce serious sequelae (34–37).
Continued surveillance and investigation of WNV-
infected patients are needed to fully define the scope of
clinical illness and determine the incidence of acute flaccid
paralysis. In addition to assessing clinical outcome, the
identification of risk factors and the pathologic confirma-
tion of anterior horn cell involvement in patients with
WNV-associated acute flaccid paralysis remain important
public health goals.
792 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 4. Clinical characteristics of patients with West Nile virus–associated acute flaccid paralysis compared with patients with typical
Guillain-Barré syndrome (25–27)
a
Characteristic
West Nile virus–associated flaccid paralysis
Guillain-Barré syndrome
Timing of onset
Acute phase of infection
1–8 weeks after acute infection
Fever and leukocytosis
Present
Absent
Weakness distribution
Asymmetric; occasional monoplegia
Generally symmetric; proximal and distal muscles
Sensory symptoms
Absence of numbness, paresthesias, or
sensory loss; occasional myalgias
Painful distal paresthesias and sensory loss
Bowel/bladder involvement
Often present
Rare
Concurrent encephalopathy
Often present
Absent
CSF profile
Pleocytosis and elevated protein
No pleocytosis; elevated protein
(albuminocytologic dissociation)
Electrodiagnostic features
Anterior horn cell/motor axon: reduced/absent
CMAPs, preserved SNAPs; asymmetric denervation
Demyelination: marked slowing of conduction velocity;
conduction block, temporal dispersion; reduced SNAPs
a
CSF, cerebrospinal fluid; CMAPs, compound muscle action potentials; SNAPSs, sensory nerve action potentials.

Acknowledgments
We are grateful to Stanley W. Chapman, the Wilson
Research Foundation, Raoult Ratard, Andrea Vicari, Grant L.
Campbell, Michael Bunning, and Susan P. Montgomery for their
important contributions to this investigation.
Dr. Sejvar is a neurologist and epidemiologist with the
Centers for Disease Control and Prevention’s Division of Viral and
Rickettsial Diseases, National Center for Infectious Diseases. His
current areas of research include the epidemiology of encephalitis,
prion diseases, and other infections of the nervous system.
References
1. Tsai T, Popovici F, Cernescu C, Campbell G, Nedelcu N. West Nile
encephalitis epidemic in southeastern Romania. Lancet 1998;
352:767–71.
2. Mostashari F, Bunning M, Kitsutani P, Singer D, Nash D, Cooper M,
et al. Epidemic West Nile encephalitis, New York, 1999: results of a
household-based seroepidemiological survey. Lancet 2001;
358:261–4.
3. Chowers M, Lang R, Nassar F, Giladi D, Rubinshtein E, Itzhaki A, et
al. Clinical characteristics of the West Nile fever outbreak, Israel,
2000. Emerg Infect Dis 2001;7:675–8.
4. Nash D, Mostashari F, Fine A, Miller J, O’Leary D, Murray K, et al.
The outbreak of West Nile virus infection in the New York City area
in 1999. N Engl J Med 2001;344:1807–14.
5. Sampson B, Ambrosi C, Charlot A, Reiber K, Veress J,
Armbrustmacher V, et al. The pathology of human West Nile virus
infection. Hum Pathol 2000;31:527–31.
6. Asnis D, Conetta R, Teixeira A, Waldman G, Sampson B. The West
Nile virus outbreak of 1999 in New York City: the Flushing Hospital
experience. Clin Infect Dis 2000;30:413–8.
7. Leis A, Stokic D, Polk J, Dostrow V, Winkelmann M. A
poliomyelitis-like syndrome from West Nile virus infection. N Engl J
Med 2002;347:1279–80
8. Glass J, Samuels O, Rich M. Poliomyelitis due to West Nile virus. N
Engl J Med 2002;347:1280–1.
9. Centers for Disease Control and Prevention. Acute flaccid paralysis
syndrome associated with West Nile virus infection—Mississippi and
Louisiana, July–August 2002. MMWR Morb Mortal Wkly Rep
2002;51:825–7.
10. Martin D, Muth D, Brown T, Johnson A, Karabatsos N, Roehrig J.
Standardization of immunoglobulin M capture enzyme-linked
immunosorbent assays for routine diagnosis of arboviral infections. J
Clin Microbiol 2000;38:1823–6.
11. Beaty B, Calisher C, Shope R. Arboviruses. In: Lennette E, Lennette
D, and Lennette E, editors. Diagnostic procedures for viral, rick-
ettsial, and chlamydial infections. Washington: American Public
Health Association; 1995. p. 89–212.
12. Prince H, Hogrefe W. Performance characteristics of an in-house
assay system used to detect West Nile virus (WNV)-specific
immunoglobulin M during the 2001 WNV season in the United
States. Clin Diagn Lab Immunol 2003;10:66–9.
13. Martin D, Biggerstaff B, Allen B, Johnson A, Lanciotti R, Roehrig J.
Use of immunoglobulin M cross-reactions in differential diagnosis of
human flaviviral encephalitis infections in the United States. Clin
Diagn Lab Immunol 2002;9:544–9.
14. Petersen L, Marfin A. West Nile virus: a primer for the clinician. Ann
Intern Med 2002;137:173–9.
15. Kimura J. Electrodiagnosis in diseases of nerve and muscle: princi-
ples and practice. 2nd ed. Philadelphia: F.A. Davis Co.; 1989. p.
249–74.
16. Kornreich L, Dagan O, Grunebaum M. MRI in acute poliomyelitis.
Neuroradiology 1996;38:371–2.
17. Rao D, Bateman D. Hyperintensities of the anterior horn cells on
MRI due to poliomyelitis. Journal of Neurology, Neurosurgery, and
Psychiatry 1997;63:720.
18. Huang C, Liu C, Chang Y, Chen C, Wang S, Yeh T, et al. Neurologic
complications in children with enterovirus 71 infection. N Engl J
Med 1999;341:936–42.
19. Ohry A, Karpin H, Yoeli D, Lazari A, Lerman Y. West Nile virus
myelitis. Spinal Cord 2001;39:662–3.
20. Rotbart H. Viral meningitis and the aseptic meningitis syndrome. In:
Scheld W, Whitley R, Durack D, editors. Infections of the central
nervous system. Philadelphia: Lippincot-Raven Publishers; 1997. p.
239–63.
21. Solomon T, Kneen R, Dung N, Khan V, Thuy T, Ha D, et al.
Poliomyelitis-like illness due to Japanese encephalitis virus. Lancet
1998;351:1094–7.
22. Kho L, Sumarmo W, Jahja E, Gubler D. Dengue hemorrhagic fever
accompanied by encephalopathy in Jakarta. Southeast Asian J Trop
Med Public Health 1981;12:83–6.
23. Sumarmo W, Wulur H, Jahja E, Gubler D, Suharyono W, Sorensen K.
Clinical observations on virologically confirmed fatal dengue infec-
tions in Jakarta, Indonesia. Bull World Health Organ
1983;61:693–701.
24. Gadoth N, Weitzman S, Lehmann E. Acute anterior myelitis compli-
cating West Nile fever. Arch Neurol 1979;36:172–3.
25. Manuelidis EE. Neuropathology of experimental West Nile virus
infection in monkeys. J Neuropathol Exp Neurol 1956;15:448–60.
26. Cantile C, Di Guardo G, Eleni C, Arispici M. Clinical and neu-
ropathological features of West Nile virus equine encephalomyelitis
in Italy. Equine Vet J 2000;32:31–5.
27. Cantile C, Del Piero F, Di Guardo G, Arispici M. Pathologic and
immunohistochemical findings in naturally occurring West Nile virus
infection in horses. Vet Pathol 2001;38:414–21.
28. Steele K, Linn M, Schoepp R, Komar N, Geisber T, Manduca R, et al.
Pathology of fatal West Nile virus infection in native and exotic birds
during the 1999 outbreak in New York City, New York. Vet Pathol
2000;37:208–24.
29. Gorson K, Ropper A. Nonpoliovirus poliomyelitis simulating
Guillain-Barré syndrome. Arch Neurol 2001;58:1460–4.
30. Asbury A, Arnason B, Karp H. Criteria for diagnosis of Guillain-
Barré syndrome. Ann Neurol 1978;3:565–6.
31. Weinberg D. AAEM case report #4: Guillain-Barré syndrome.
Muscle Nerve 1999;22:271–81.
32. Hurwitz E, Holman R, Nelson D, Schonberger L, Breman D, Kaslow
R, et al. National surveillance for Guillain-Barré syndrome. January
1978–March 1979. Neurology 1983;33:150–7.
33. Visser L, Van der Meche F, Van Doorn P, Meulstee J, Jacobs B,
Oomes P, et al. Guillain-Barré syndrome without sensory loss (acute
motor neuronopathy): a subgroup with specific clinical, electrodiag-
nostic, and laboratory features. Brain 1995;118:841–7.
34. Norda R, Berseus O, Stemayr B. Adverse events and problems in
therapeutic hemapheresis. A report from the Swedish registry.
Transfusion and Apheresis Sciences 2001;25:33–41.
35. Stangel M, Muller M, Marx P. Adverse events during treatment with
high-dose intravenous immunoglobulins for neurological disorders.
Eur Neurol 1998;40:173–4.
36. Gottlieb S. Intravenous immunoglobulin increases the risk of throm-
botic events. BMJ 2002;324:1056.
37. Struble E, Dice Y. Intravenous immune globulin (IVIG) precipitating
acute myocardial infarction. J Miss State Med Assoc 2002;43:115.
Address for correspondence: James J. Sejvar, Medical Epidemiologist,
Division of Viral and Rickettsial Diseases, National Center for Infectious
Diseases, Centers for Disease Control and Prevention, 1600 Clifton Road,
Mailstop A39, Atlanta, GA 30333, USA; fax: 404-639-3163; email:
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 793
RESEARCH

Seven alligators were submitted to the Tifton
Veterinary Diagnostic and Investigational Laboratory for
necropsy during two epizootics in the fall of 2001 and 2002.
The alligators were raised in temperature-controlled build-
ings and fed a diet of horsemeat supplemented with vita-
mins and minerals. Histologic findings in the juvenile alliga-
tors were multiorgan necrosis, heterophilic granulomas,
and heterophilic perivasculitis and were most indicative of
septicemia or bacteremia. Histologic findings in a hatchling
alligator were random foci of necrosis in multiple organs
and mononuclear perivascular encephalitis, indicative of a
viral cause. West Nile virus was isolated from submissions
in 2002. Reverse transcription-polymerase chain reaction
(RT-PCR) results on all submitted case samples were pos-
itive for West Nile virus for one of four cases associated
with the 2001 epizootic and three of three cases associat-
ed with the 2002 epizootic. RT-PCR analysis was positive
for West Nile virus in the horsemeat collected during the
2002 outbreak but negative in the horsemeat collected after
the outbreak.
W
est Nile virus (WNV) has been reported in a variety
of species but primarily endotherms. Arboviruses
have been reported to affect ectotherms, and in some cases
ectotherms are thought to serve as a reservoir (1–4). The
mode of transmission of the arbovirus to ectotherms has
often been presumed to be through ingestion or a bite from
the insect carrier (5).
During the fall of 2001 and 2002, two epizootics
occurred among captive alligators on a south Georgia alli-
gator farm that houses over 10,000 animals.
Approximately 250 alligators died between November and
December 2001, and >1,000 alligators died in 2002. These
epizootics tended to occur approximately 2 weeks after the
first abrupt drop in ambient temperature, which occurred
both years in mid-October and was characterized by mini-
mum temperatures between 0°C and 8°C and maximum
temperatures between 10°C and 18°C for a period of 1 to
3 days.
Methods
Animals and Housing
Animals were housed in six barns that were divided
into 10 pens; each pen contained approximately 100–200
alligators. The nursery animals are obtained either as eggs
from Florida or as hatchlings from onsite breeders.
All pens are cleaned in the morning starting at 6 a.m.
An automatic flushing system is used to drain the pens,
flush them, and fill them with clean water. Well water is
chlorinated with an automated system that injects chloride
gas into the water. The water is then piped into a central
collecting area and heated. The water temperature is main-
tained at 32.2°C year-round, and the buildings are kept
dark to reduce environmental stress on the animals. The
reduced stress and warm environment allow continued
growth (i.e., growth of >
1 m per year rather than 0.30 m
per year).
Alligators are fed in the mid- to late afternoon. The diet
consists of 95% ground raw horsemeat (obtained frozen
from a source in Pennsylvania) to which vitamins and min-
erals are added in a pelleted alligator diet carrier. The
ingredients are thoroughly mixed in a large commercial
mixer. The source of the horsemeat has remained constant
since 1985. The source of the vitamins and minerals has
varied, based upon availability.
The breeding population is maintained in a separate
fenced enclosure on the premises. This enclosure is a
native swampland and therefore subjected to ambient
weather conditions. A rookery was recently established in
the breeding area by native birds. Attempts to depopulate
the rookery (using U.S. Department of Agriculture–
approved methods) have been unsuccessful. The alligators
eat fledglings and older birds that fall from the nests and
branches or otherwise get within reach. Alligators do not
nest under the rookery. No mosquito control is practiced
on the farm.
Tissue Collection
Animals were seen moribund or dead upon arrival at
the laboratory. Blood was collected from the occipital
794 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
West Nile Virus in
Farmed Alligators
Debra L. Miller,* Michael J. Mauel,* Charles Baldwin,* Gary Burtle,* Dallas Ingram,*
Murray E. Hines II,* and Kendal S. Frazier*
*University of Georgia, Tifton, Georgia, USA

sinus or caudal vein of live animals. Gross observations
were made, and the animals were humanely euthanized.
Tissues were collected from the eye, thyroid gland, lymph
node, lung, heart, brain, spinal cord, kidney, liver, spleen,
pancreas, adrenal gland, gallbladder, tonsil, trachea, stom-
ach, intestines, and reproductive tract. Fresh tissue speci-
mens were submitted for virus isolation, reverse transcrip-
tion-polymerase chain reaction (RT-PCR), and bacterial
culture. Tissues were also collected in 10% buffered for-
malin, processed, and embedded in paraffin. Five-microm-
eter-thick sections were stained with hematoxylin and
eosin and viewed by light microscopy. Tissues opportunis-
tically collected from an adult clinically normal, free-rang-
ing alligator served as a control.
Multiple aliquots (totaling 1 g) of the ground raw horse-
meat (without additives) that was being fed during the
2002 epizootic (October and November) were collected
and processed for RT-PCR. Subsequent aliquots from
postepizootic horsemeat shipments (in December and
January) were similarly processed.
Virus Isolation
A 10% homogenate in Earle’s minimal essential media
(MEM) containing gentamicin was made of each speci-
men. The homogenate was centrifuged for 10 min at 2,000
RPM and 4°C. The supernatent was filtered and spread
onto a preformed monolayer of Vero cells. In 2001, fathead
minnow (FHM), white sturgeon skin (WSS), epithelioma
papillosum caprini (EPC), and channel catfish ovary
(CCO) cells were used instead of the Vero cells. Inoculated
cells were incubated in a 5% CO
2
atmosphere at 37°C.
Cells were examined each day for viral cytopathic effect
(CPE). If no CPE was observed, aliquots of the first pas-
sage were transferred to a second preformed monolayer of
Vero cells (FHM, WSS, EPC, and CCO cells in 2001) on
day 7. If no CPE was observed after a second 7 days of
passage, the culture was considered negative. Monolayers
demonstrating viral CPE were passaged to chambered
slides. The slides were fixed in cold methanol, and a West
Nile fluorescent-antibody test was conducted to confirm
the isolate.
Fluorescent-Antibody Testing
Mouse anti–WNV-specific polyclonal antibody
(Centers for Disease Control and Prevention [CDC],
Division of Vector-Borne Infectious Diseases, Fort
Collins, CO) was applied to the chamber and the slide
incubated in 5% CO
2
at 37°C for 30 min. The slide was
rinsed two times for 5 min in a sodium carbonate/bicarbon-
ate buffer (pH 9.3). The slide was then air-dried, followed
by an anti-mouse fluorescein-conjugated antibody, and
incubated as before for 30 min. The slide was washed
twice in carbonate buffer, followed by 5 min in 0.5%
Evans blue counter stain. Slides were dipped in distilled
water, and a glycerin/water mounting media and coverslip
was added. Slides were examined with a fluorescent
microscope. All isolates were tested for WNV. All isolates
were also tested for Eastern equine encephalomyelitis
virus (EEEV) by using a similar protocol. We tested for
EEEV because of its known prevalence within the geo-
graphic area. The EEEV-specific monoclonal antibody
(CDC, Atlanta, GA) was prepared against the New Jersey
1960 strain of EEEV.
RNA Extraction
RNA was extracted from various specimens (fresh tis-
sue, virus isolation homogenate or cell culture lysate, and
formalin-fixed paraffin-embedded tissue). For extraction
from fresh specimens, approximately 1 g of tissue was
placed in a whirlpack bag and homogenized by using a
Stomacher Lab Blender 80 (Tekmar Co., Cincinnati, OH)
with three times the tissue volume of phosphate-buffered
saline (PBS). Three milliliters of the tissue homogenate
was processed with a Rneasy Midi kit (QIAGEN, Inc.,
Valencia, CA) per manufacturer’s directions. If a virus iso-
lation homogenate or cell culture lysate in Earle’s MEM
was used, approximately 4 mL of the homogenate or lysate
was washed with 5 mL of PBS, the supernatant removed,
and the pellet processed with the Rneasy Midi kit. For
paraffin sections, several 5-µm sections from paraffin
blocks were cut and deparaffinized with xylene. The
xylene was removed, and samples were washed two times
with 100% ethanol for 10 min, once with 95% and once
with 70% ethanol. Samples were incubated overnight at
56°C in 80 µL of proteinase K with 5 mL of Buffer RLT
from the Rneasy Midi kit and then processed per manufac-
turer’s directions.
RT-PCR
RT-PCR for WNV was performed on the tissues
according to the procedure described by Kuno (6) and
using the RT-nested primer sets described by Johnson et al.
(7). In brief, a RT-PCR mixture was prepared by using the
outside primer set (P1401 – ACCAACTACTGTGGAGTC
and P1845 – TTCCATCTTCACTCTACACT) to amplify a
445-bp product. Forty microliters of the RT-PCR mixture
and 10 µL of sample were dispensed into a 0.2-mL thin
wall PCR tube, and 10 µL of Rnase-free water was added
for a final volume of 50 µL. With the use of a model PTC-
200 thermal cycler (MJ Research, Inc., Waltham,
Massachusetts), cycling conditions for the RT-PCR were
as follows: 53°C for 30 min, followed by 40 cycles of
94°C for 1 min, 53°C for 1 min, 72°C for 1 min, and then
held at 4°C. Ten microliters of RT-PCR first-round product
was used for the nested PCR (nPCR). The nPCR mixture
was prepared by using 40 µL of PCR mixture (now with
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 795
RESEARCH

the inside primer set [P1485 – GCCTTCATACACAC-
TAAAG and P1732 – CCAATGCTATCACAGACT]) to
amplify a 248-bp product. The cycling conditions for the
nPCR were as described above, but the first ramp was
omitted (53°C for 30 min). A 10-µL aliquot of each reac-
tion with 1 µL of loading buffer added was loaded onto a
1.5 % agarose gel in Tris-borate-EDTA (TBE) buffer and
run at 70 V for approximately 1.5 h.
This protocol was repeated on all samples with primer
sets for EEEV and St. Louis encephalitis virus (SLEV).
For the 262-bp EEEV genomic fragment, an outer set of
forward (P4 (EEE-4) - CTAGTTGAGCACAAACACCG-
CA) and reverse (P7 (cEEE-7) - CACTTGCAAGGT-
GTCGTCTGCCCTC) primers, followed by a nested set of
forward (P5 (EEE-5) - AAGTGATGCAAATCCAACTC-
GAC) and reverse (P6 (cEEE-6) - GGAGCCACACG-
GATGTGACACAA) primers, was used (8). The RT-PCR
mixture was similar to that described by Kuno (6). The
thermal cycling parameters varied from those of WNV as
follows: 94°C for 90 s followed by 30 cycles of 94°C for
20 s, 65°C for 35 s, 72°C for 17 s, and then a final elonga-
tion step of 72°C for 4 min. A single RT-PCR procedure
was used for SLEV. The 393-bp genomic fragment was
generated by using forward (SLE727 – GTAGCCGACG-
GTCAATCTCTGTGC) and reverse (SLE119c - ACTCG-
GTAGCCTCCATCTTCATCA) primers and using param-
eters as for WNV (9).
Bacterial Culture
Swabs of individual tissues were streaked onto 5%
bovine blood agar (BBA), Wilkins-Chalgren anaerobe
agar, mycoplasma agar, Lowenstein-Jensen agar slant, and
Hektoen Enteric agar (HE) agar (intestines only). Blood
was inoculated into thioglycolate broth and streaked onto
BBA. Inoculated media were incubated at 30°C with
duplicate blood agar plates incubated in the presence or
absence of 5% CO
2
, with the exception of the anaerobic
cultures, which were incubated at 37°C. The thioglycolate
broth was subcultured onto BBA after 24 h. Plates were
examined each day for growth and subcultured onto BBA
as needed. Bacterial colonies selected from pure cultures
were Gram stained. Cultures were injected into Sensititre
(Trek Diagnostic Systems, Westlake, OH) gram-negative
AP80 or gram-positive AP90 autoidentification plates and
the antibiotic sensitivity plate CMVIECOF and allowed to
incubate for 18 h at 37°C before automated reading of the
reactions per the manufacturer’s directions. Any isolates
that failed to be identified by the Sensititre system were
identified by using the RapID NF Plus System (Remel,
Norcross, GA) or the API20E system (API Analytab
Products, Plainview, NY).
Results
Clinical Findings
The affected alligators appeared to “star gaze” in the
water just before death, suggesting neurologic lesions (10).
Alligators sometimes became stranded in the dry part of
the pen with loss of leg control and neck spasms. No long-
term signs of stress were noted, and most animals were
eating well until a few days before death. The hatchlings
(approximately 30-cm long at the time of the epizootic)
and juveniles (1–2 m long) seemed to be more severely
affected.
A specific pattern of transmission was not noted in
2001. However, in 2002, the alligator deaths initially
occurred in one building and spread throughout the build-
ing in the opposite direction from that taken to feed and
clean the animals. At least one interruption of chlorine
addition to flush water occurred before the 2002 epizootic.
Deaths were not incurred in the breeding colony, and no
deaths were reported in birds that inhabited the rookery.
Gross Findings
2001
Both Florida and Georgia stock animals were affected,
but, in general, the Florida stock was affected first.
Initially, three juvenile alligators were sent for necropsy
during the 2001 epizootic. In general, the alligators were in
good to excellent body condition. One alligator had
approximately 25 mL of serosanguinous fluid in the peri-
cardial sac and 50 mL yellow serous fluid in the peritoneal
cavity. Two of the three had yellow-tan, caseous necrosis
of the palatine tonsils and multiple caseous yellow-tan
plaques, 2- to 10-mm in diameter, on the mucosal surfaces
of the esophagus, corpus, and pars pylorica. Only scant
ingesta were noted throughout the gastrointestinal (GI)
tract, and the intestinal mucosa was hemorrhagic in rare
instances. The liver and spleen of one alligator had multi-
ple 1- to 3-mm tan foci scattered throughout the parenchy-
ma. One alligator was in poor to moderate body condition
and had scattered bronchiectasis, no ingesta throughout the
GI tract, and mild multifocal serous atrophy of fat. No
other gross lesions were noted.
Approximately 2 months after the 2001 epizootic
began, another juvenile, live alligator was submitted to our
laboratory. The gross lesions were similar to those
described above but with numerous 1- to 3-mm tan foci in
the parenchyma of the liver, spleen, and kidneys.
2002
Three alligators were examined from the fall 2002 epi-
zootic, two juveniles and a hatchling. The two juveniles
had lesions similar to those described in the previous year.
796 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
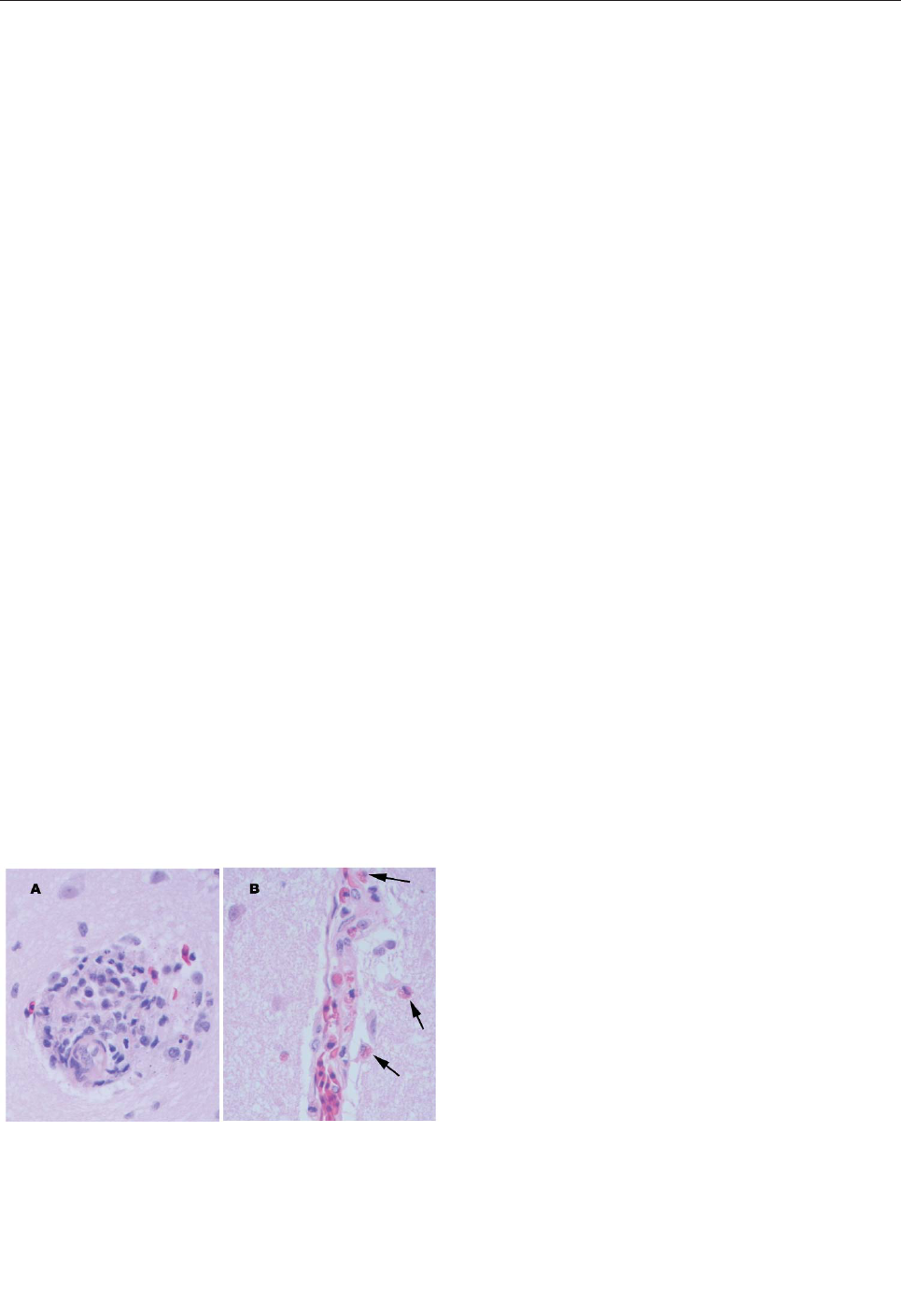
The liver and kidneys of the hatchling were pale and mot-
tled tan/brown. Ingesta were scant throughout the GI tract.
The free-ranging alligator was in excellent body condition.
No significant gross changes were noted in its tissues.
Light Microscopic Findings
2001
Tissues of the alligators from the 2001 epizootic were
examined and were similar in two of the three alligators. In
the brain, rare glial nodules that contained occasional het-
erophils were present (Figure 1). The spleen was congest-
ed with moderate diffuse reticuloendothelial hyperplasia
and moderate numbers of heterophils. The tonsil had
severe multifocal coalescing areas of caseous necrosis and
heterophilic inflammation with reactive lymphoid follicu-
lar hyperplasia. In the esophagus, a focally extensive,
mixed ulcerative, and proliferative lesion was present; it
had a marked mixed but predominantly mononuclear
inflammation, colonies of bacteria, and extensive fibrin
deposition. In the liver, multifocal lymphoplasmacytic
aggregates and heterophilic granulomas were present, con-
sisting of caseous necrotic foci with degenerate heterophils
surrounded by an outer layer of macrophages, lympho-
cytes, and heterophils. The lungs were congested with mild
diffuse or patchy lymphoplasmacytic and heterophilic
interstitial infiltrates. The kidney had multifocal het-
erophilic granulomas. The pars pylorica region of the
stomach had multifocal mucosal abscesses and moderate
diffuse lymphoplasmacytic and heterophilic infiltrates of
the lamina propria. The small intestine had moderate, dif-
fuse mucosal and submucosal infiltrates of lymphocytes,
heterophils, and plasma cells and multifocal areas of acute
necrosis associated with bacteria. The remaining tissues
appeared within normal limits. Special stains for fungi and
acid-fast bacteria were negative. A population of primarily
gram-negative and fewer gram-positive bacteria was
observed in the heterophilic granulomas.
The third alligator had primarily pulmonary changes.
The airways contained moderate numbers of heterophils,
occasional mucous plugs with degenerate inflammatory
cells, and scattered bacterial colonies. The remaining tis-
sues were as described for the first two alligators.
Tissues from the alligator seen 2 months after the epi-
zootic had similar findings to those of the first two alliga-
tors with the addition of rare, small caseating granulomas
within the lungs. The granulomas contained numerous
large macrophages and multinucleated cells. Acid-fast
stains demonstrated low numbers of slender, beaded, acid-
fast positive bacilli consistent with mycobacteria.
2002
Multiple tissues from the two juvenile alligators from
the 2002 epizootic were examined. The tissue changes
were similar to those described for the 2001 epizootic
except that the inflammatory component was primarily
heterophils. The meninges within the brain and all spinal
cord sections except those from the sacral spinal cord had
stasis of heterophils within the blood vessels and perivas-
cular infiltration of mild numbers of heterophils (Figure 1).
One alligator had a small focus of macrophages and het-
erophils noted within the endocardium.
Multiple tissues were examined from the hatchling alli-
gator, and lesions differed from the previous submissions
on the basis of cellular composition of the inflammatory
cell infiltrates. Lymphoplasmacytic perivascular cuffs were
present throughout the brain and meninges (Figure 1).
Rarely, heterophils were admixed within the cuffs. Similar
changes were not seen within the spinal cord. Random foci
of necrosis were seen within the liver, pancreas, and tonsil.
Mild to moderate perivascular infiltrates of lymphocytes,
plasma cells, and heterophils were seen within the kidney
and heart, and similar but fewer numbers of infiltrates were
seen within the pulmonary interstitium. The heart had mul-
tiple, random foci of patchy vacuolar degeneration of the
myocytes and random aggregates of lymphocytes, plasma
cells and heterophils. Mild numbers of mixed inflammato-
ry cells were seen within the intestinal lamina propria. The
remaining tissues were unremarkable. Major pathologic
changes were not observed by light microscopy in the tis-
sues from the free-ranging alligator.
Virus Isolation/RT-PCR
Virus isolation was negative for all animals from the
2001 epizootic. WNV was isolated from tissues from all
animals in the 2002 epizootic. Additionally, all animals
from the 2002 epizootic and one animal from the 2001 epi-
zootic were positive for WNV by RT-PCR from fresh or
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 797
RESEARCH
Figure 1. Perivascular changes observed within the brain of alliga-
tors infected with West Nile virus (400x). A. Perivascular infiltrates
were composed of primarily lymphocytes, plasma cells, and
macrophages in the hatchling alligator. B. Perivascular infiltrates
were composed of primarily heterophils (arrows) in juvenile alliga-
tors.

formalin-fixed, paraffin-embedded tissues (Figure 2). In
general, liver was the most likely tissue to yield positive
results. Positive results were not obtained from any of the
tissues from the free-ranging alligator. All tissues tested
negative by RT-PCR for EEEV and SLEV. Retrospective
attempts to culture WNV at both 37°C and room tempera-
ture on FHM, CCO, EPC, and WWS cells were negative.
Aliquots from the horsemeat that was being fed during
the 2002 epizootic tested positive for WNV by RT-PCR
(Figure 2). Aliquots of the horsemeat from two postepi-
zootic shipments were negative for WNV by RT-PCR.
Bacterial Culture
Aeromonas sobria and Edwardsiella tarda were consis-
tently cultured from the intestines. These organisms and
occasionally others (Escherichia coli, Pseudomonas fluo-
rescens, α- and β-hemolytic Streptococcus) were isolated
from various tissues (liver, lung, and kidney) from the alli-
gators dying during the 2001 epizootics and the juveniles
from the 2002 epizootics. Alcaligenes spp. were isolated
from a tonsil swab in one of the animals in 2001.
Salmonella Group D was isolated from the intestines of the
hatchling alligator submitted in 2002.
Discussion
The histologic findings from the hatchling alligator
were most suggestive of a viral etiology, whereas those of
the older alligators were most suggestive of a primary bac-
terial cause. Given that both the RT-PCR and virus isola-
tion were positive for WNV, that virus is suspected to be
the underlying cause of both epizootics. Contaminated
horsemeat is the presumed source of the outbreak. We
speculate that the WNV infection led to the alligators’
immune systems’ becoming immunocompromised, which
resulted in the animals being more susceptible to various
environmental stressors and subsequent invasion by
opportunistic pathogens. Failure to isolate virus from the
alligators in 2001 may have been due to the inability of the
virus to propagate in the four cell lines used (FHM, CCO,
EPC, and WWS cells), as determined by retrospective cul-
ture attempts, rather than absence of virus.
Two important points to examine further are time of
year and age of affected animals. Both epizootics occurred
in the late fall to early winter. Although the epizootics
appeared to be correlated with the first abrupt drop in envi-
ronmental temperature, this finding was likely coinciden-
tal, especially given that the animals were housed in envi-
ronmentally controlled barns. The most likely factor in the
time of year is correlation with the occurrence of WNV
infection in horses. Historically, horses become infected
with WNV during the mosquito season (summer through
early fall). Undiagnosed WNV-infected animals sold for
food would most likely end up in the food supply during
the late summer and early fall months. As was found in this
study, deaths traced to consumption of contaminated food
would taper off in late fall or early winter as the food sup-
ply was less likely to contain virus. Furthermore, all ani-
mals have equal potential for viral exposure through con-
sumption because individual packages of horsemeat are
combined before mixing with the vitamin supplements and
being divided between all barns. In general, reptiles
achieve immunocompetence at an early age (often in a
matter of days), but this immunocompetence may be tem-
perature dependent until the animals are several months of
age (11). This fact may partially explain why the hatchling
alligators tended to die from the viral infection, whereas
the juveniles tended to die from infections caused by sec-
ondary invaders.
Extrinsic stressors may have increased certain animals’
susceptibility to the virus or opportunistic pathogens. For
example, the pens where the epizootics originated tended
to be the first to be washed out at 6 a.m., the coolest time
of the day. During the first abrupt drop in environmental
temperature, the first wash water was possibly cooler
because of colder water in the line between the boiler and
the pens. This cold stressor would serve as a shock to the
animals’ systems. During the 2002 outbreak, an additional
stress was internal construction, undertaken 2 weeks before
the epizootic within the initially affected building. The
environmental (temperature and darkness) control of the
building was maintained during this time, but silence was
not maintained. Additionally, sanitation-related stress may
have occurred during periods of intermittent flushing, such
as over weekends and during pen renovation activities.
Whether brood stock source had an affect on the sus-
ceptibility of the animals is not clear. Although Florida
stock animals were those initially affected, this finding
was likely coincidental because of their location in the
pens. The pens that were more exposed to external stres-
798 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure 2. West Nile virus (WNV) reverse transcription-polymerase
chain reaction results from epizootic die-offs in farm-raised alliga-
tors. The expected amplicon is 248 bp. Lane 1, a 100-bp molecu-
lar weight ladder. Lane 2, the positive WNV control. Lane 3, fresh
tissue samples from a juvenile alligator in the 2002 epizootic. Lane
4, virus isolation cell homogenate from a juvenile alligator in the
2002 epizootic. Lane 5, horsemeat that was being fed to alligators
during the 2002 epizootic. Lane 6, initial postepizootic horsemeat
shipment. Lanes 7, 8, and 9, formalin-fixed, paraffin-embedded tis-
sues of juvenile alligators in 2001 and 2002. Lane 10, fresh tissue
from a wild alligator. Lane 11, negative WNV control.

sors contained Florida animals. Additionally, most animals
in the production unit are from Florida brood stock.
Several management recommendations were suggested
to the producer. The primary recommendation was to stop
feeding horsemeat and switch to another food source such
as beef or fish. We also recommended that the water tem-
perature be reduced to 29.4°C in an attempt to reduce the
stress of rapid growth and perhaps produce an environment
less conducive for viremia. To date, neither of these rec-
ommendations has been implemented, but subsequent
horsemeat shipments have tested negative. Future investi-
gation will include the testing of the eggs from the brood
stock, clinically healthy animals, rookery birds, and free-
ranging alligators to explore the epidemiology of this virus
in ectotherms.
Acknowledgments
We thank the staff of The University of Georgia Tifton
Veterinary Diagnostic and Investigational Laboratory for help in
processing the samples.
Dr. Miller is an assistant professor in the Department of
Pathology at the University of Georgia (UGA) College of
Veterinary Medicine. She works as a veterinary pathologist at the
UGA Tifton Veterinary Diagnostic and Investigational
Laboratory. Her research interests are in wildlife disease and
reproduction.
References
1. Doi R, Oya A, Shirasaka A, Yabe S, Sasa M. Studies on Japanese
encephalitis virus infection of reptiles. II. Role of lizards on hiberna-
tion of Japanese encephalitis virus. Jpn J Exp Med 1983;53:125–34.
2. McLean RG, Ubico SR, Bourne D, Komar N. West Nile virus in live-
stock and wildlife. Curr Top Microbiol Immunol 2002;267:271–308.
3. Nir Y, Lasowski Y, Avivi A, Cgoldwasser R. Survey for antibodies to
arboviruses in the serum of various animals in Israel during
1965–1966. Am J Trop Med Hyg 1969;18:416–22.
4. Thomas LA, Patzer ER, Cory JC, Coe JE. Antibody development in
garter snakes (Thamnophis spp.) experimentally infected with west-
ern equine encephalitis virus. Am J Trop Med Hyg 1980;29:112–7.
5. Oya A, Doi R, Shirasaka A, Yabe S, Sasa M. Studies on Japanese
encephalitis virus infection of reptiles. I. Experimental infection of
snakes and lizards. Jpn J Exp Med 1983;53:117–23.
6. Kuno G. Universal diagnostic RT-PCR protocal for arboviruses. J
Virol Methods 1998;72:27–41.
7. Johnson DJ, Ostlund EN, Pedersen DD, Schmitt BJ. Detection of
North American West Nile virus in animal tissue by a reverse tran-
scription-nested polymersase chain reaction assay. Emerg Infect Dis
2001;7:739–41.
8. Linssen B, Kinney RM, Aquilar P, Russell KL, Watts DM, Kaaden
OR, et al. Development of reverse transcription-PCR assays specific
for detection of equine encephalitis viruses. J Clin Microbiol
2000;38:1527–35.
9. Lanciotti RS, Kerst AJ. Nucleic acid sequence-based amplification
assays for rapid detection of West Nile and St. Louis encephalitis
viruses. J Clin Microbiol 2001;39:4506–13.
10. Done LB. Postural Abnormalities. In: Mader DR, editor. Reptile med-
icine and surgery. Philadelphia: W.B. Saunders Co; 1996. p. 406–11.
11. Frye FL. Biomedical and surgical aspects of captive reptile hus-
bandry, Vol 1. 2nd edition. Malabar (FL): Krieger Publishing Co;
1991.
Address for correspondence: Debra L. Miller, University of Georgia,
Veterinary Diagnostic and Investigational Laboratory, Tifton, GA 31793,
USA; fax: 229-386-7128; email: [email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 799
RESEARCH
The opinions expressed by authors contributing to this journal do
not necessarily reflect the opinions of the Centers for Disease
Control and Prevention or the institutions with which the authors
are affiliated.

Over the past two decades, dengue virus serotype 3
(DENV-3) has caused unexpected epidemics of dengue
hemorrhagic fever (DHF) in Sri Lanka, East Africa, and
Latin America. We used a phylogenetic approach to evalu-
ate the roles of virus evolution and transport in the emer-
gence of these outbreaks. Isolates from these geographi-
cally distant epidemics are closely related and belong to
DENV-3, subtype III, which originated in the Indian subcon-
tinent. The emergence of DHF in Sri Lanka in 1989 corre-
lated with the appearance there of a new DENV-3, subtype
III variant. This variant likely spread from the Indian sub-
continent into Africa in the 1980s and from Africa into Latin
America in the mid-1990s. DENV-3, subtype III isolates
from mild and severe disease outbreaks formed genetical-
ly distinct groups, which suggests a role for viral genetics in
DHF.
A
rthropod-borne viruses are responsible for the emer-
gence of unexpected diseases in humans, as illustrat-
ed by the identification of West Nile virus encephalitis in
the American hemisphere in 1999 (1). The emergence of a
new disease is often attributable to the transport of a
pathogen (as in the case of West Nile virus) or changes in
the evolution or ecology of a native pathogen that hitherto
caused mild or no disease in humans (2,3). We studied
unexpected outbreaks of dengue hemorrhagic fever (DHF)
in Sri Lanka, East Africa, and Latin America caused by
dengue serotype 3 (DENV-3) virus.
Most persons infected with dengue viruses are asymp-
tomatic or develop dengue fever (DF). DHF and dengue
shock syndrome (DSS), which can be fatal, develop in a
minority of infected persons. The pathogenesis of DHF is
poorly understood, although factors such as age and previ-
ous exposure to dengue infections increase the risk for
severe disease (4). Epidemiologic studies point to particu-
lar DENV strains being more virulent than others (5–8).
For example, the dengue genotypes endemic to Central
and South America have caused mild disease, while the
Asian genotypes introduced to the region have led to DHF
epidemics (9–16). Similarly, outbreaks of DHF in some
Pacific islands have been traced to the introduction of
Southeast Asian dengue strains (17). DENV-2 subtypes
associated with mild and severe disease epidemics have
distinct mutations in the E gene and 5′ and 3′ untranslated
segments of the viral genome, although whether these
mutations directly contribute to pathogenesis is unproven
(18).
The distribution of DHF and DSS in Asia has been par-
ticularly puzzling. Before 1989, DHF was common in
Southeast Asia but rare in the Indian subcontinent despite
the circulation of all four serotypes in both regions. After
1989, this pattern of disease changed and regular epi-
demics of DHF were reported from several countries in the
Indian subcontinent (19). Sri Lanka, in particular, experi-
enced a dramatic and persistent increase in DHF cases
(20). Epidemiologic studies of dengue in Sri Lanka have
demonstrated that the intensity of virus transmission, as
well as the relative abundance of each serotype, remained
constant before and after the emergence of DHF (21).
Thus, DHF did not emerge in Sri Lanka because of an
overall increase in virus transmission or shift in serotype.
Although all four serotypes of dengue circulate in Sri
Lanka, persons who have the severe form of the disease
are most frequently infected with DENV-3 (20,22).
Lanciotti et al. characterized the genetic relatedness of
DENV-3 isolates from regions throughout the tropics and
subtropics and identified four geographically distinct sub-
types (23). All Sri Lankan isolates were classified as sub-
type III, which also includes isolates from East Africa and
India, as well as recent isolates from Latin America.
Because DENV-3 isolates from Sri Lanka isolated before
and after 1989 (when DHF emerged) formed separate
groups within subtype III, Lanciotti and colleagues postu-
lated that a genetic shift in DENV-3 may have been
responsible for the emergence of DHF (23).
In the current study, using phylogenetic methods, we
analyzed DENV-3 viruses isolated from Sri Lanka for up
to 10 years after the emergence of DHF to confirm the
800 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Emergence and Global Spread
of a Dengue Serotype 3,
Subtype III Virus
William B. Messer,* Duane J. Gubler,† Eva Harris,‡ Kamalanayani Sivananthan,§
and Aravinda M. de Silva*
*University of North Carolina, Chapel Hill, North Carolina, USA;
†Centers for Disease Control and Prevention, Fort Collins,
Colorado, USA; ‡University of California, Berkeley, California,
USA; and §Medical Research Institute, Colombo, Sri Lanka

establishment of a new genotype and evaluate the roles of
virus evolution and transport in establishing a new geno-
type. DENV-3, subtype III was introduced into Latin
America in 1994 (11), and the virus has subsequently been
isolated from DF and DHF outbreaks throughout Central
and South America (12–16). We also examined the genet-
ic relationships between DENV-3, subtype III isolates
from Latin America, East Africa, and the Indian subconti-
nent. On the basis of our results, we describe the most
likely scenario of events that led to the emergence of
DENV-3 –associated DHF in the Indian subcontinent and
the Americas.
Materials and Methods
Virus Strains
The dengue virus strains sequenced for this study as
well as sequences obtained from GenBank for this study
are listed in Table 1. The virus isolates were obtained
from the Centers for Disease Control and Prevention,
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 801
RESEARCH
Table 1. Dengue virus type 3 sequences used
a
Strain
Y
Location
Name
Subtype
Sequence source
GenBank accession no.
D1266
1983
Sri Lanka
83SriLan1
III
This study
AF547225
D1306
1983
Sri Lanka
83SriLan2
III
This study
AF547226
D1307
1983
Sri Lanka
83SriLan3
III
This study
AF547227
D1336
1983
Sri Lanka
83SriLan4
III
This study
AF547228
D1440
1984
Sri Lanka
84SriLan1
III
This study
AF547229
073
1985
Sri Lanka
85SriLan
III
This study
AF547241
D2783
1989
Sri Lanka
89SriLan1
III
This study
AF547230
D2863
1989
Sri Lanka
89SriLan2
III
This study
AF547231
D2803
1989
Sri Lanka
89SriLan3
III
This study
AF547232
D3197
1990
Sri Lanka
90SriLan1
III
This study
AF547233
D5231
1993
Sri Lanka
93SriLan1
III
This study
AF547234
D9397
1994
Sri Lanka
94SriLan1
III
This study
AF547235
L57
1997
Sri Lanka
97SriLan1
III
This study
AF547242
K1
1998
Sri Lanka
98SriLan
III
This study
AF547243
1557
1985
Mozambique
85Mozamb1
III
This study
AF547236
1558
1985
Mozambique
85Mozamb2
III
This study
AF547237
1559
1985
Mozambique
85Mozamb3
III
This study
AF547238
251991
1991
Kenya
91Kenya
III
This study
AF547239
SOM079
1993
Somalia
93Somalia
III
This study
AF547240
32267
1994
Nicaragua
94Nicara1
III
This study
AF547244
6845
1998
Nicaragua
98Nicara1
III
This study
AF547245
7431
1998
Nicaragua
98Nicara2
III
This study
AF547246
7071
1998
Nicaragua
98Nicara3
III
This study
AF547262
BC 96/94
1994
Panama
94Panama1
III
This study
AF547247
032231
1994
Panama
94Panama2
III
This study
AF547248
BC 13/96
1994
Panama
94Panama3
III
This study
AF547249
BC 20/97
1996
Mexico
96Mexico1
III
This study
AF547250
BC 172/97
1996
Mexico
96Mexico2
III
This study
AF547251
BC 184/97
1996
Mexico
96Mexico3
III
This study
AF547252
BC173/97
1996
Mexico
96Mexico4
III
This study
AF547253
17605
1995
Costa Rica
95CostaR1
III
This study
AF547254
17608
1995
Costa Rica
95CostaR2
III
This study
AF547255
322473
1995
Costa Rica
95CostaR3
III
This study
AF547256
322488
1995
Costa Rica
95CostaR4
III
This study
AF547257
20/8
1997
Guatemala
97Guatem1
III
This study
AF547263
366-781
1998
Puerto Rico
98PuertoR1
III
This study
AF547258
400-996
2000
Puerto Rico
00PuertoR1
III
This study
AF547264
MK
1998
El Salvador
98ElSalv1
III
This study
AF547259
612210
2001
Venezuela
01Venezue1
III
This study
AF547260
VEN03
2001
Venezuela
01VEN03
III
This study
AF547261
Ref. 18
1981–91
Sri Lanka
81,85,89,91 SriLanA
III
GenBank
L11431,L11436–L11438
Ref. 18
1984
India
84IndiaA
III
GenBank
L11424
Ref. 18
1986
Samoa
86Samoa
III
GenBank
L11435
Ref. 18
1962–86
Thailand
62,73,86,86 Thailand
II
GenBank
L11440–L11442,L11620
Ref. 18
1983
Philippines
83Philipp
I
GenBank
L11432
Ref. 18
1989
Tahiti
89Tahiti
I
GenBank
L111619
Ref. 18
1992
Fiji
92Fiji
I
GenBank
L11422
Ref. 18
1973–85
Indonesia
73,78,85 Indones
I
GenBank
L11425,L11426,L11428
Ref. 18
1974–81
Malaysia
74,81 Malaysi
I
GenBank
L11429,L11427
Ref. 18
1956
Philippines
D3H-87
I
GenBank
L11423
Ref. 18
1963–77
Puerto Rico
63,77 PuertoR
IV
GenBank
L11433,L11434
Ref. 18
1965
Tahiti
65 Tahiti
IV
GenBank
L11439
a
Includes original identifier for strain, year of isolation, taxa name used in this paper, dengue virus 3 subtype, source of viral sequence, an
d GenBank accession numbers.

Dengue Branch, Puerto Rico, and Division of Vector-
Borne Infectious Diseases, Ft. Collins, Colorado;
Medical Research Institute, Colombo, Sri Lanka; School
of Public Health, Berkeley, California; Walter Reed
Army Institute for Research, Washington, D.C.; and
University of Massachusetts Medical Center, Worcester,
Massachusetts.
RNA Extraction
QiaAmp Viral RNA Mini Kit (QIAGEN, Valencia, CA)
was used to extract viral RNA from both the mosquito
grind supernatants and infected tissue culture media fol-
lowing the manufacturer’s protocol. Extracted RNA was
stored at –70°C or immediately subjected to reverse tran-
scription–polymerase chain reaction (RT-PCR).
RT-PCR
DENV-3 RT-PCR was carried out as described by
Lanciotti (23). Primers were designed to amplify and
sequence a 966-bp fragment from positions 179–1,144,
encompassing part of Capsid, all of PreM, and part of the
E gene sequences. The reverse primer (DEN3/735)
hybridized to positions 1,189–1,171 (5′-ctcctcaggcaaaac-
cgct-3′) and the forward primer (D1 consensus) hybridized
to positions 132–159 (5′-tcaatatgctgaaacgcgcgagaaaccg-
3′). The reverse primer DEN3/735 was added to extracted
RNA, incubated at 85°C for 90 s, and allowed to cool to
room temperature. RT was carried out for 45–60 min in 20
µL of reaction mix containing 25 U avian myeloblastosis
virus reverse transcriptase (Roche, Nutley, NJ), deoxynu-
cleoside tripophosphate, MgCl
2
, and RT buffer. PCR was
performed by adding a 30-µL cocktail containing D1 con-
sensus primer, PCR buffer, and EXPAND polymerase
(Roche) to the 20-µL RT reaction. PCR conditions were 4
min at 94°C, 30–35 cycles of 94°C for 30 s, 54°C for 30 s,
and 72°C for 90 s with 5 s/cycle added to elongation step
after the first 10 cycles. We separated 5 µL of the reaction
products on 2% agarose gels and visualized it by ethidium
bromide staining. When necessary, target bands were
excised and purified by using the Qiagen QIAquick Gel
Extraction kit (QIAGEN) following manufacturer’s
instructions. All remaining PCR reaction products were
purified by using the Qiagen PCR Purification kit follow-
ing the manufacturer’s protocol.
DNA Sequencing
Purified PCR products were sent to the automated DNA
sequencing facility at the University of North Carolina,
Chapel Hill, NC. The DENV-3 sequences used in this man-
uscript included 40 newly determined sequences, which
have been submitted to GenBank (accession nos.
AF547225–AF547264).
Viral Sequence Analysis
Overlapping individual nucleic acid sequences were
assembled with the aid of VECTOR NTI ContigExpress
(InforMax, Inc., Bethesda, MD). Sequences were aligned
and analyzed by using the following software: Clustal X
(available from: URL: http://inn-prot.weizmann.ac.il/soft-
ware/ClustalX.html), PAUP* (available from: URL:
http://www.sinauer.com), PHYLIP (available from: URL:
http://evolution.genetics.washington.edu/phylip.html), and
MEGA II (available from: URL: http://www.megasoft-
ware.net). Genetic distances were calculated by using
Tamura-Nei distance algorithm with 1,000 bootstrap repli-
cates; the trees were generated by using the Minimum
Evolution method. The phylogenetic tree in Figure 1 is
based on a 708-base segment, positions 437–1,145, span-
ning pre-M/M and a portion of the E gene. The phyloge-
netic tree presented in Figure 2 is based on 966-base region
spanning positions 179–1,145 on the viral genome, captur-
ing a portion of the C gene, all of pre-M/M gene, and a
portion of the E gene.
Results and Discussion
Many investigators have used viral nucleotide sequence
data and phylogenetic methods to understand genetic rela-
tionships between viruses, as well as the epidemiology of
viral disease. Phylogenetic studies have shown that dengue
viruses can move long distances between continents (24)
as well as short distances between neighboring countries
(25). Our goal was to use a phylogenetic approach to
understand recent DHF outbreaks caused by DENV-3
infections in the Indian subcontinent and Latin America.
Previous phylogenetic analysis of DENV-3 has princi-
pally relied on complete or partial sequences of the pre-
M/M and E genes (13,16,17,23). Our analysis used a 708-
base segment, positions 437–1,145, spanning pre-M/M
and a portion of the E gene, coding for 236 amino acids.
This region was selected because it both conserved the
original phylogenetic relationship identified by Lanciotti
et al. and, in preliminary analysis with previously estab-
lished sequences, captured 44% of the variable sites with-
in DENV-3, subtype III Sri Lankan sequences. No inser-
tion/deletion mutations and no hypervariable regions were
detected in this span.
A total of 40 DENV-3 sequences, including 21
sequences available from GenBank and 19 newly deter-
mined Indian subcontinent and African sequences (Table
1) were compared. Dates of isolation ranged from 1963 to
1998. With the exception of 63PuertoR, all sequences
were from low-passage (<4) virus cultures. Several
approaches to phylogenetic analysis, including maximum
likelihood, parsimony, and distance methods, were com-
pared. All approaches yielded identical or nearly identical
802 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
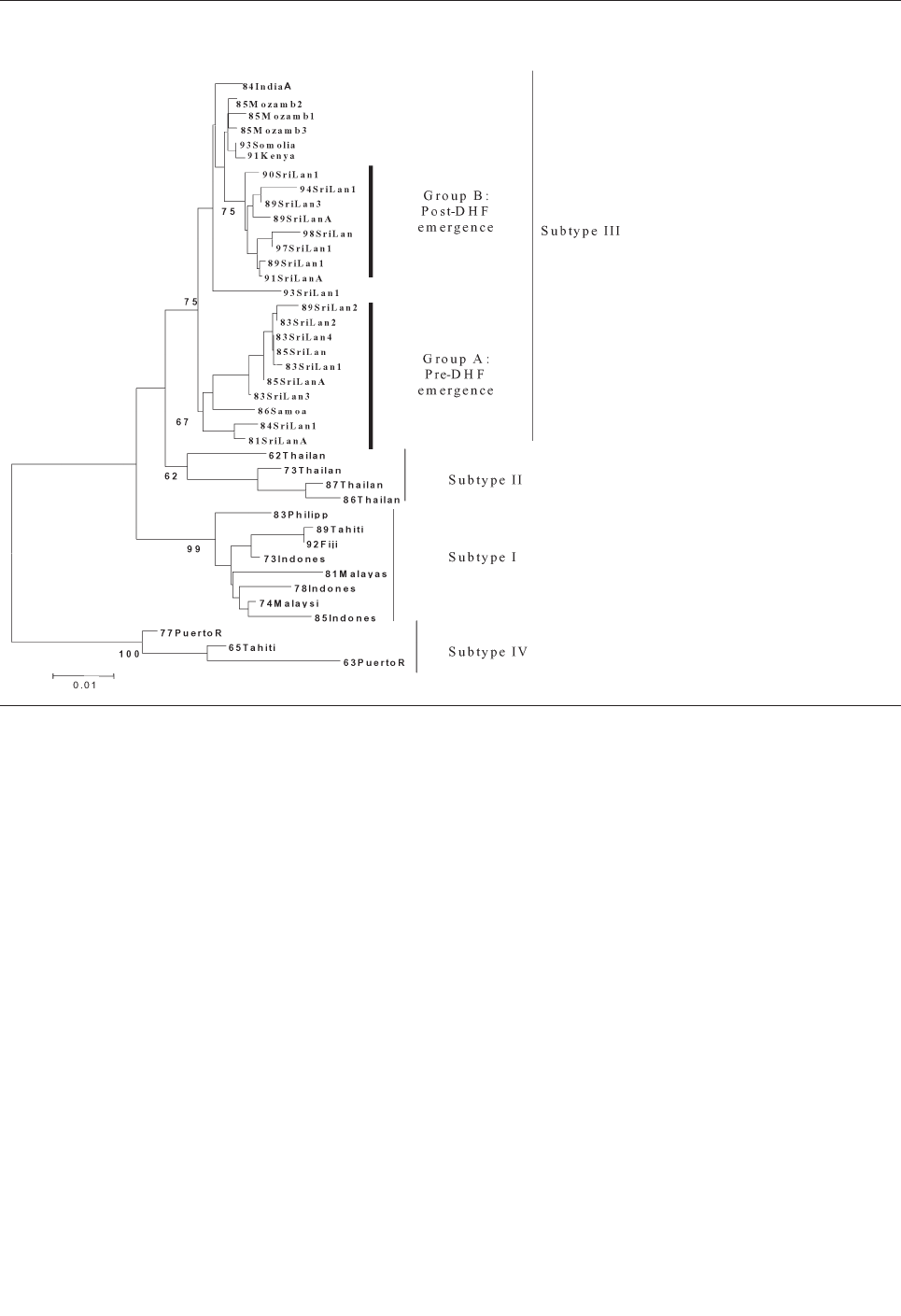
topologies. Results presented here used the Tamura-Nei
algorithm to calculate genetic distances and the minimum
evolution method to create the trees (Figure 1). The tree
identifies four distinct lineages that correspond to the
region of isolation, reproducing the same evolutionary
relationship first described by Lanciotti, et al. (23).
Subtype I includes isolates from Southeast Asia and the
South Pacific islands; subtype II consists of isolates from
Thailand; subtype III is comprised of isolates from the
Indian subcontinent, East Africa, and a single isolate from
Samoa; and subtype IV includes Puerto Rico and Tahiti.
Similarity within subtypes was high, with subtype III
showing the greatest mean similarity (98.4%), followed
by subtypes I, II, and IV (Table 4).
All 24 Sri Lankan, Indian, and East African strains fell
into subtype III (Figure 1). The circulating virus genotypes
within this region have remained closely related over the
relatively long period of 18 years (1981–1998), indicating
that countries bordering the western Indian Ocean form a
geographically distinct region with regard to DENV-3
viruses. DENV-2 viruses in the regions also form a subtype
with a similar geographic distribution (26,27). Frequent
trade between East Africa, Western Indian Ocean islands,
and the Indian subcontinent may have been responsible for
the movement of dengue viruses throughout the region
(26,28). Rico-Hesse, for example, demonstrated the intro-
duction of DENV-2 to Africa from islands in the Indian
Ocean (26). The earliest subtype III virus on record is an
isolate from India in 1966; this virus occupies a node that
is ancestral to all the subsequent Asian and African isolates
(R.S. Lanciotti, pers. comm.), suggesting that the DENV-
3, subtype III viruses have their origin in the Indian sub-
continent and have subsequently spread out of the region.
In Sri Lanka, regular epidemics of DHF have been
observed only after 1988. DENV-3 is responsible for many
of the infections that progress to DHF (20,22). DENV-3
isolates obtained before and after the emergence of DHF
are very closely related and belong to subtype III, indicat-
ing that the emergence of DHF on the island is not due to
the introduction of a new subtype from outside the region.
However, within subtype III, most Sri Lankan isolates
(except for 93SriLan1) from before and after the emer-
gence of DHF segregated into two distinct clades, desig-
nated groups A and B (Figure 1). Group A, with nine iso-
lates from 1981 to 1989, consists of viruses collected up to
the year epidemic DHF emerged in Sri Lanka but contains
no isolates from later than 1989. Group B includes eight
isolates from 1989 to 1998 but none from before 1989.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 803
RESEARCH
Figure 1. Phylogenetic tree of estab-
lished dengue virus 3 (DENV-3) sub-
types (23) and the relationship of Sri
Lanka pre– and post–dengue hemor-
rhagic fever DENV-3 isolates to the
established subtypes. This tree is based
on a 708-base segment, positions 437
to 1145, spanning pre-M/M and a por-
tion of the E gene. Scale bar shows
number of substitutions per bases
weighted by Tamura-Nei algorithm.
Horizontal distances are equivalent to
the distances between isolates.
Numbers at nodes indicate bootstrap
support values for the branch of the tree
inferred at that node. The origin of the
viruses and sequences used are listed
in Table 1. The amino acid substitutions
conserved within each DENV-3 subtype
are listed in Table 2. DHF, dengue hem-
orrhagic fever.

Temporally, the two groups are continuous, by virtue of
sharing isolates in 1989. Group A includes isolate
89SriLan2, while group B contains 89SriLan1, 89SriLan3,
and 89SriLanA. However, the groups do not form a contin-
uous lineage; they share a common ancestor only at the
node for subtype III (Figure 1). Group B shares ancestral
nodes with isolates from India and East Africa. Because
the Indian and East African isolates overlap temporally
with group A (all isolates are from the 1980s), group A and
group B lineages likely diverged sometime before 1981
and followed distinct evolutionary pathways.
We propose two likely scenarios that led to the emer-
gence of group B viruses in Sri Lanka. One possibility is
that the group B viruses were introduced from India or
East Africa into Sri Lanka because Indian and East African
isolates from the mid-1980s are closely related to Sri
Lankan group B viruses (Figure 1). Of the two regions,
India is the more likely source because of geographic prox-
imity to Sri Lanka, although the East African viruses could
be the direct ancestors of the group B viruses. Another pos-
sibility is that both groups co-circulated in Sri Lanka in the
early 1980s, with group B being a minor population. Some
804 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 2. Amino acid substitutions conserved within each dengue virus 3 subtype for the isolates used to create the phylogenetic tree in
Figure 1
a
Position
Subtype
Name
31
55
57
128
135
148
188
234
Outgroup
56Philipp
I
H
T
L
I
L
D
I
I
73Indones
–
L
–
F
–
–
–
V
I
74Malaysi
–
L
–
F
–
–
–
V
I
78Indones
–
L
–
F
–
–
–
V
I
81Malaysi
–
L
–
F
–
–
–
V
I
83Philipp
–
L
–
F
–
–
–
V
I
85Indones
–
L
–
F
–
–
–
V
I
89Tahiti
–
L
–
F
–
–
–
V
I
92Fiji
–
L
–
F
–
–
–
V
II
62Thailan
–
–
A
–
–
W
–
–
II
73Thailan
–
–
A
–
–
–
–
–
II
86Thailan
–
L
A
–
–
–
–
–
II
87Thailan
–
L
A
–
–
–
–
–
III
85Mozamb1
–
–
–
–
–
–
–
–
III
85Mozamb2
–
–
–
–
–
–
–
–
III
85Mozamb3
–
–
–
–
–
–
–
–
III
84IndiaA
–
–
–
–
–
–
–
–
III
91Kenya
–
–
–
–
–
–
–
–
III
93Somolio
–
–
–
–
–
–
–
–
III
81SriLanA
–
–
–
–
–
–
–
–
III
83SriLan1
–
–
–
–
–
–
–
–
III
83SriLan2
–
–
–
–
–
–
–
–
III
83SriLan3
–
–
–
–
–
–
–
–
III
83SriLan4
–
–
–
–
–
–
–
–
III
84SriLan1
–
–
–
–
–
–
–
–
III
85SriLanA
–
–
–
–
–
–
–
–
III
85SriLan
–
–
–
–
–
–
–
–
III
89SriLan2
–
–
–
–
–
–
–
–
III
89SriLanA
–
–
–
–
–
–
–
–
III
89SriLan1
–
–
–
–
–
–
–
–
III
89SriLan3
–
–
–
–
–
–
–
–
III
90SriLan1
–
–
–
–
–
–
–
–
III
91SriLanA
–
–
–
–
–
–
–
–
III
93SriLan1
–
–
–
–
–
–
–
–
III
94SriLan1
–
–
–
–
–
–
–
–
III
97SriLan1
–
–
–
–
–
–
–
–
III
98SriLan1
–
–
–
–
–
–
–
–
III
86Samoa
–
–
–
–
–
–
–
–
IV
63PuertoR
T
–
–
F
L
M
E
–
IV
65Tahiti
T
–
–
F
L
M
E
–
IV
77PuertoR
T
–
–
F
L
M
E
–
a
Reference strain 56Philipp is the highly passaged laboratory strain H87. Positions are numbered sequentially from the first position in the pre-M/M protein
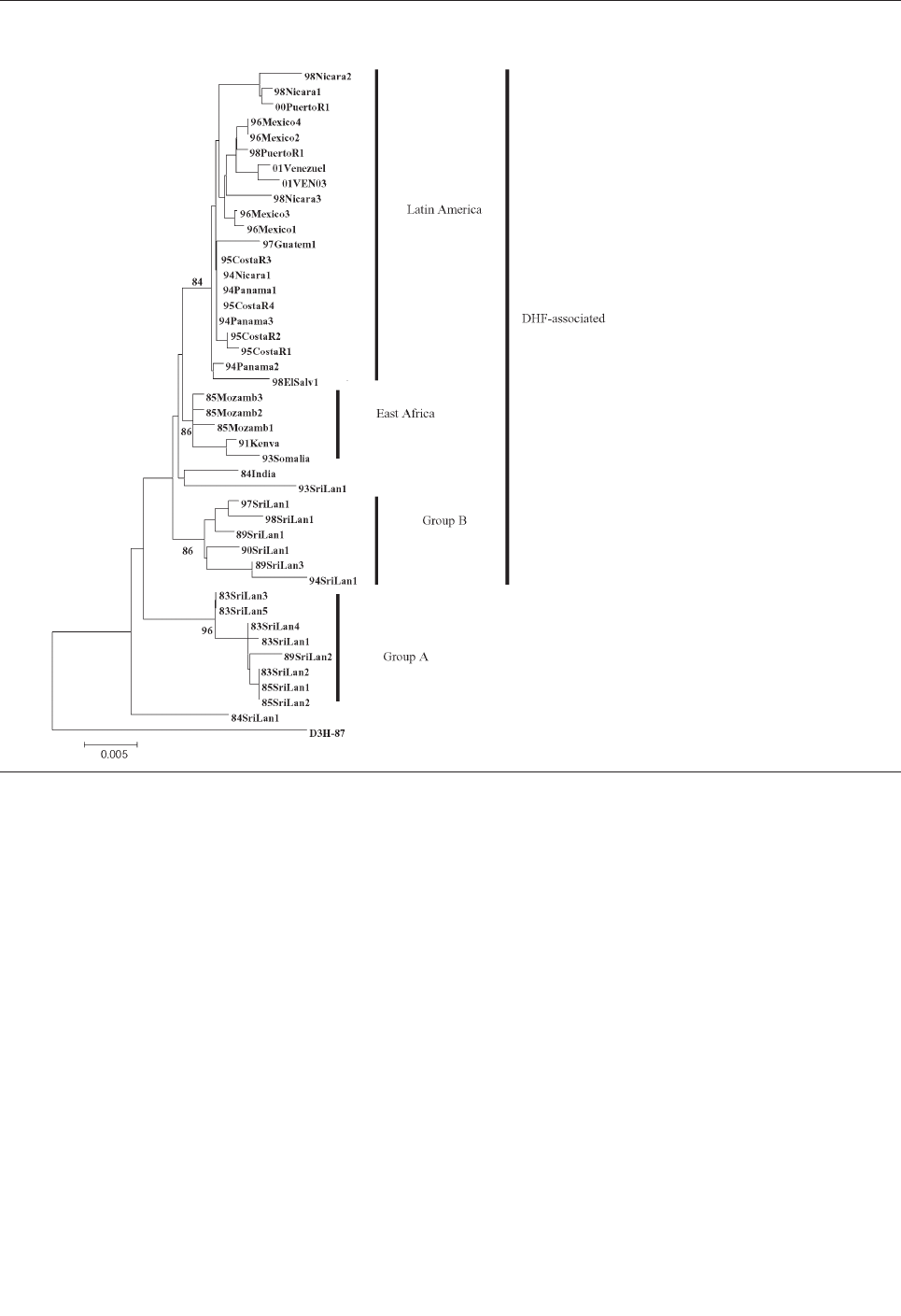
selective force operating in the late 1980s may have shift-
ed the balance in favor of group B viruses. In either case,
group B viruses emerged in Sri Lanka because a subtype
III variant already established in the greater region became
more common in Sri Lanka and not because a novel virus
evolved and emerged de novo on the island.
DENV-3, subtype III was detected in the Americas dur-
ing DF and DHF outbreaks in Nicaragua and Panama in
1994 (11). Subsequently, the virus has spread to many
countries in Latin America, and DENV-3–associated DHF
was confirmed in several countries (13,14,16,29–31)
(Figure 3). To establish the relationship of recent Latin
American DENV-3 isolates to each other and to the previ-
ously identified Indian subcontinent and East African sub-
type III isolates, we sequenced and analyzed a 966-base
region spanning positions 179–1,145 on the viral genome,
capturing a portion of the C gene, all of pre-M/M gene, and
a portion of the E gene. This region adds 288 positions to
the 5′ end of the sequences initially presented in this study.
Forty-three isolates were sequenced (21 from Mexico
and Central and South America, 16 from Sri Lanka, 1 from
India, and 5 from East Africa) (Table 1). The D3H-87
belonging to DENV-3, subtype 1 was used as an outgroup.
Years of isolation ranged from 1983 to 2001, an 18-year
span. Except for the D3H-87 outgroup, all other isolates
were low-passage clinical isolates. Most of the nucleotide
mutations were silent: only 12 amino acid positions
showed any variability and only 2 positions showed vari-
ability in more than one isolate. Consequently, the evolu-
tionary relationships observed in this analysis likely reflect
the results of genetic drift and are unlikely to have been
influenced by host-specific selection events on this portion
of the genome (26).
All sequences included in this analysis fell within sub-
type III (data not shown). Several approaches to phyloge-
netic analysis were compared, and all approaches yielded
identical or nearly identical topologies. We used the
Tamura-Nei algorithm to calculate genetic distances and
the minimum evolution method to create the trees (Figure
2). Bootstrap values are shown at critical nodes. Despite
the high overall similarity of the isolates in this analysis,
geographically and temporally distinct groups formed sep-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 805
RESEARCH
Figure 2. Phylogenetic tree of dengue
virus 3, subtype III group A, group B, East
Africa, and Latin America. Tree is based
on 966-base region spanning positions
179–1,145 on the viral genome, capturing
a portion of the C gene, all of pre-M/M
gene and a portion of the E gene.
Nucleotide substitutions conserved within
each dengue virus 3, subtype III group
(group A, B, East Africa, and Latin
America) are listed in Table 3. DHF,
dengue hemorrhagic fever.

arate lineages. Generally, two separate lineages formed
within subtype III. The first consists of group A viruses
isolated from 1981 to 1989 in Sri Lanka. These viruses
have been associated only with DF. The second is com-
posed of Sri Lankan group B, Indian, East African, and all
of the isolates from Mexico and Central and South
America.
Within group A, members are closely related, with a
nucleotide mean similarity of 99.4% (Table 5). Within the
expanded group B and related viruses, three distinct clades
exist: a group of closely related Sri Lankan isolates from
1989 to 1998, 5 East African isolates from 1985 to 1993,
and 21 isolates from 1994 to 2001 from Latin America.
Isolates 84India and 93SriLan1 are less closely related to
the other geographically distinct isolates in the larger sec-
ond lineage.
The isolates from Latin America all emerge from a
common node on the tree, suggesting a single introduction
806 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 3. Nucleotide substitutions conserved within dengue virus 3, subtype III groups
Position
Group
Strain
338
429
503
566
653
686
695
707
728
734
749
791
821
866
896
902
978
1010
1019
1056
Reference
H87
A
A
C
G
G
T
T
G
C
C
C
A
T
C
T
T
C
C
T
C
Group A
83SriLan1
-
G
T
A
-
-
C
-
-
T
T
-
-
T
C
C
T
T
-
-
Group A
83SriLan2
-
G
T
A
-
-
C
-
-
T
T
-
-
T
C
C
T
T
-
-
Group A
83SriLan3
-
G
T
A
-
-
C
-
-
-
T
-
-
T
-
C
-
T
-
-
Group A
83SriLan4
-
G
T
A
-
-
C
-
-
T
T
-
-
T
C
C
T
T
-
-
Group A
84SriLan1
-
G
-
-
-
-
-
-
-
-
T
-
-
-
C
-
-
-
-
-
Group A
85SriLan1
-
G
T
A
-
-
C
-
-
T
T
-
-
T
C
C
T
T
-
-
Group A
89SriLan2
-
G
T
A
-
-
C
-
-
T
T
-
-
T
C
C
T
T
-
-
unclassified
84India
G
G
-
-
A
-
-
-
-
-
T
-
-
-
-
-
-
-
G
-
unclassified
93SriLan1
G
G
-
-
A
-
-
-
-
-
-
-
-
-
-
-
-
-
G
-
East Africa
85Mozamb1
G
-
-
-
A
-
-
-
-
-
-
-
-
-
-
-
-
-
G
-
East Africa
85Mozamb2
G
-
-
-
A
-
-
-
-
-
-
-
-
-
-
-
-
-
G
-
East Africa
85Mozamb3
G
-
-
-
A
-
-
-
-
-
-
-
-
-
-
-
-
-
G
-
East Africa
91Kenya
G
-
-
-
A
-
-
-
-
-
-
-
-
-
-
-
-
-
G
-
East Africa
93Somalia
G
-
-
-
A
-
-
-
-
-
-
-
-
-
-
-
-
-
G
Group B
89SriLan1
G
G
-
-
A
-
-
A
T
-
-
-
C
-
-
-
-
-
G
-
Group B
89SriLan3
-
G
-
-
A
-
-
A
T
-
-
-
C
-
-
-
-
-
G
-
Group B
90SriLan1
G
G
-
-
A
-
-
A
T
-
-
-
C
-
-
C
-
-
G
-
Group B
94SriLan1
-
G
-
-
A
-
-
A
T
-
-
-
C
-
-
-
-
-
G
-
Group B
97SriLan1
G
G
-
-
A
-
-
A
T
T
-
-
C
-
C
-
-
-
G
-
Group B
98SriLan1
G
G
-
-
A
-
-
A
T
-
-
-
C
-
C
-
-
-
G
-
L. America
94Nicara1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
94Panama1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
94Panama2
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
94Panama3
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
CostaRica1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
CostaRica2
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
CostaRica3
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
CostaRica4
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
96Mexico1
G
G
-
-
A
C
C
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
96Mexico2
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
96Mexico3
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
96Mexico4
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
97Guatem1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
98Nicara1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
98Nicara2
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
98Nicara3
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
98ElSalv1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
01Venezue1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
01VEN03
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
98PuertoR1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
L. America
00PuertoR1
G
G
-
-
A
C
-
-
-
-
-
G
C
-
-
-
-
-
G
T
a
Reference strain is the highly passaged laboratory strain H87. Positions are numbered sequentially from the first nucleotide position at the 5′ end of the genome.
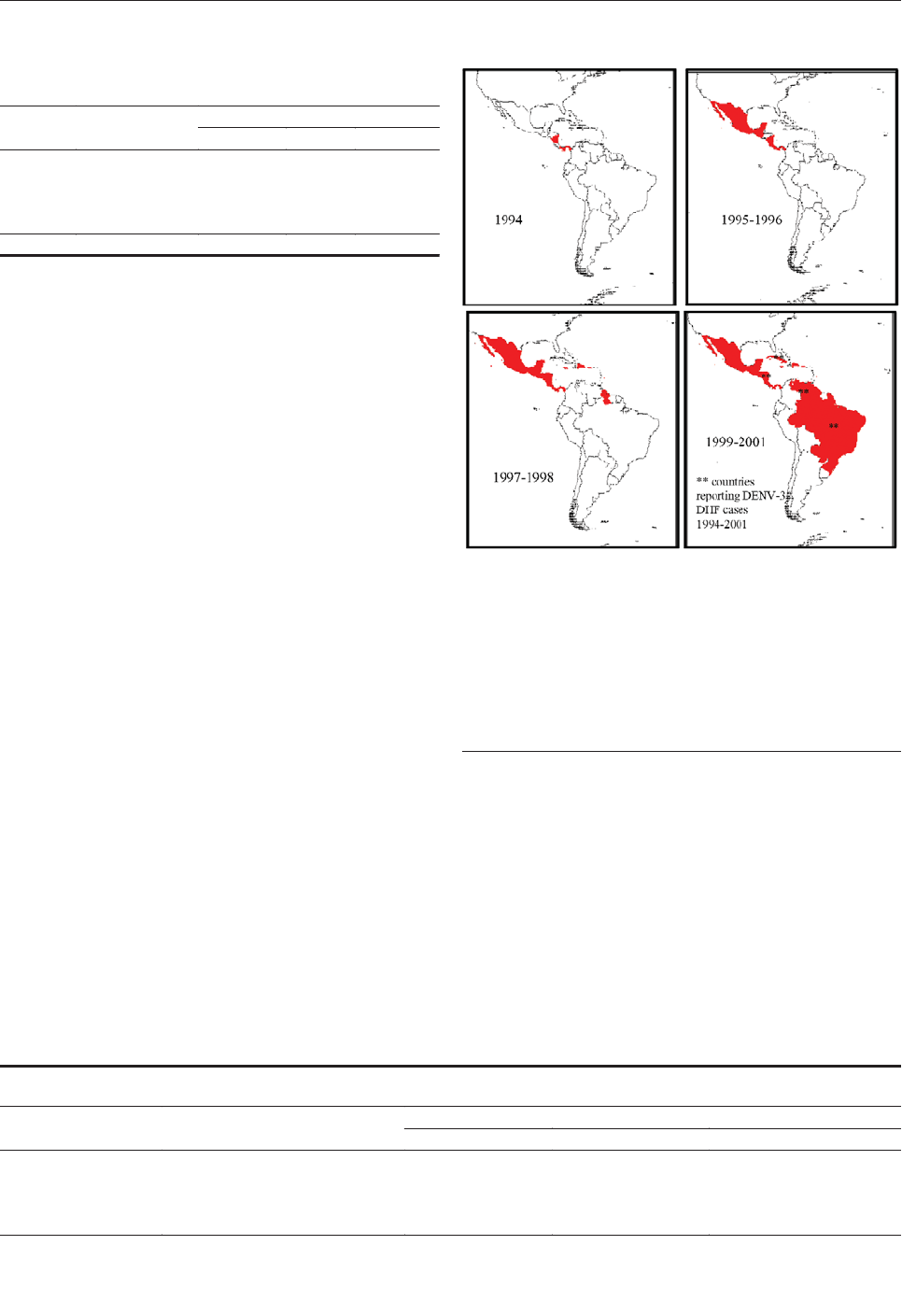
of a virus and the subsequent diversification of the virus
population from the founding strain. The DENV-3, sub-
type III isolates from Nicaragua, Panama, and Costa Rica
are closest to the Latin American group’s originating node,
with the more recent isolates found farther from that node,
reflecting the viral population’s ongoing evolution after
the point source introduction.
The internal branch from the Latin American group
shares a common node with the isolates from East Africa.
The common hypothetical ancestor for Latin America and
East Africa then shares a common node with the Sri
Lankan group B virus isolates. Both on the phylogenetic
tree and in pair-wise comparisons (Table 5), the Latin
American group was more closely related to the isolates
from East Africa than to the group B Sri Lankan isolates.
Furthermore, the East African isolates pre-date the earliest
Latin American isolates by 9 years, while the less closely
related Sri Lankan group B isolates are nearly contempo-
raneous with the Latin American isolates. Therefore, the
point source DENV-3 introduction into Latin America is
most likely to have its origins in East Africa and not the
Indian subcontinent (Figure 4).
Little is known about dengue activity in Africa, partic-
ularly DENV-3 (32). DENV-3 was first detected on the
African continent in 1984 to 1985 during an outbreak in
Mozambique (32). Later studies of U.S. troops in Africa
and the Persian Gulf suggested that DENV-3 is endemic in
those regions but largely undetected (33). Our results show
that all East African DENV-3 isolates belong to subtype
III. The fact that DENV-3 was only first isolated from East
Africa in 1985, whereas the viruses were present in the
Indian subcontinent at least as far back as 1966 (R.S.
Lanciotti, pers. comm.), suggests that DENV-3, subtype III
was introduced from the Indian subcontinent into East
Africa in or before 1984 (Figure 4). This introduction led
to the establishment of a stable East African group of
DENV-3, subtype III because all the isolates from
Mozambique, Kenya, and Somalia isolated from 1985 to
1993 form a distinct clade within subtype III (Figure 3).
The DENV-3, subtype III viruses introduced into Latin
America are most closely related to subtype III viruses in
East Africa (Figure 3). Although we can only speculate
about the exact mode of transport of DENV-3 into Latin
America, we propose that Panama, with its canal that
attracts goods as well as civilians and military personnel
from other parts of the world, may have been the point of
introduction of subtype III into the Americas. Similarly,
the introduction of DENV-2 in 1981 into Cuba may be
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 807
RESEARCH
Table 4. Summary of within- and between-subtype nucleotide
mean similarity for dengue virus 3 isolates shown in Figure 1
a
Between subtype similarity (%)
Subtype
Within subtype
similarity (%)
I
II
III
I
98.1
II
97.7
94.9
III
98.4
95.6
96.3
IV
97.6
92.3
92.5
92.7
a
Mean similarities were calculated with the Tamura-Nei distance algorithm.
Figure 3. Map of the spread of dengue virus 3 (DENV-3), subtype
III through Latin America and the Caribbean. The introduction of
DENV-3, subtype III was first reported in November 1994 in
Nicaragua and Panama. This virus strain has been isolated, iden-
tified, and reported in at least 16 other countries in the region.
*Represents countries with dengue hemorrhagic fever (DHF)
caused by DENV-3. These countries are Nicaragua in 1994 and
1998, Brazil and Venezuela in 2001 (Pan American Health
Organization, unpub. data).
Table 5. Summary of within- and between-group nucleotide mean similarity for the dengue virus 3, subtype III virus isolates shown in
Figure 2
a
Between group similarity
Subgroup
Within-group similarity
Subgroup A
East Africa
Subgroup B
Subgroup A
99.4%
East Africa
99.5%
98.2%
Subgroup B
98.8%
97.9%
98.7%
Latin America
99.5%
98.0%
99.0%
98.5%
a
Mean similarities were calculated with the Tamura-Nei distance algorithm.

attributable to Cuban military personnel traveling between
Southeast Asia and Cuba (24,34).
Epidemiologic and clinical studies on dengue in
Indonesia in the 1970s pointed to strain differences
between DENV-3 viruses contributing to transmission and
disease severity (35,36). Despite their overall similarity at
the nucleotide level, the DENV-3, subtype III isolates
examined in this study have been associated with severe or
mild disease outbreaks (Figure 3). Sri Lankan group A
viruses were isolated during a time of little to no DHF,
while group B viruses were isolated after the emergence of
DHF in Sri Lanka. The emergence of DHF in Sri Lanka
was not accompanied by a change in dengue transmission
or the abundance of any particular serotype (21).
Implicating DENV-3 directly as the cause of DHF in Sri
Lanka has been difficult because few virus isolates are
available from DHF patients in Sri Lanka. However, dur-
ing dengue surveillance studies in 1997, only DENV-3 was
isolated from hospitalized dengue cases, whereas DENV-
1, DENV-2, and DENV-3 were isolated from patients vis-
iting outpatient clinics (22). These observations suggest
that DENV-3 is responsible for severe dengue disease in
Sri Lanka. Further studies are required to better establish
the relative contribution of DENV-3 to severe disease in
Sri Lanka.
The current studies support a viral genetic basis for
severe and mild disease outbreaks. We found that the popu-
lation of DENV-3 viruses associated with DHF in Sri Lanka
did not appear to be direct descendants of the group A virus-
es that were circulating before DHF emerged in that coun-
try. The Sri Lankan 1989–1997 isolates are more closely
related to the isolates from East Africa and the isolates from
the Americas than they are to the isolates from 1981 to 1989
in Sri Lanka (Figure 3). All three groups of subtype III
viruses (Sri Lankan group B, East African group, and Latin
American group) associated with DHF are more closely
related to each other than they are to the pre–DHF group A
viruses from Sri Lanka (Figure 3). Thus, all the viruses
within subtype III are closely related (mean 98.4% identity
at the nucleotide level), yet they form distinct phylogenetic
groups associated with mild or severe disease.
The Sri Lankan group B viruses may be associated with
severe disease unlike group A viruses because the group B
viruses are inherently more virulent. Alternatively, the
ability of preexisting dengue antibody to neutralize group
A viruses and enhance group B viruses may account for the
observed associations with severe and mild disease. In a
recent study, antibodies against American DENV-1 viruses
neutralized the Native American DENV-2 genotype better
than the Southeast Asian DENV-2 genotype that is current-
ly circulating in the Americas and causing DHF (37). This
study lends support to the idea that Asian DENV-2 may
produce a more severe disease not because of inherent vir-
ulence properties but because persons with previous pri-
mary DENV-1 infections may enhance infection with this
genotype and neutralize infections with the Native
American DENV-2 genotype. Similarly, DENV-2 and -3
are the common serotypes in Sri Lanka, and persons with
previous primary DENV-2 infections could neutralize the
DENV-3 group A viruses better than the group B viruses.
This difference may explain the unexpected emergence of
DHF associated with group B. Further comparative studies
with group A and B viruses are needed to understand their
association with mild and severe disease, respectively.
Acknowledgments
We thank Irene Bosch, Vance Vorndam, Niranjan Kanesa-
thasan, and Eric Wagar for virus isolates and Tissa Vitarana, Gaya
Colombage, Nalini Withana, and staff members in the Virology
Department at the Medical Research Institute in Colombo for
their help.
This work was supported by a Junior Faculty Development
Award and other funds from the University of North Carolina at
Chapel Hill (A.M.S.)
Dr. Messer earned his Ph.D. in ecology from the University
of North Carolina at Chapel Hill. His broad research interests
include the emergence, movement, and evolution of human
pathogens. For his doctoral research, he evaluated host, viral, and
environmental factors that may have contributed to the sudden
emergence of dengue hemorrhagic fever in Sri Lanka. He attends
medical school at the University of North Carolina.
808 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure 4. Global spread of dengue virus 3 (DENV-3), subtype III,
which has been continuously circulating in the Indian subcontinent
from the 1960s to the present. The virus was first isolated from
East Africa in 1985 in Mozambique and subsequently from Kenya
(1991) and Somalia (1993) (32,33). DENV-3 subtype III was first
detected in the American continent in 1994 (Nicaragua and
Panama) and the virus has subsequently spread through most of
Latin America (13,14, 16,29,30). The arrows depict the most likely
directions of spread based on the phylogenetic relationships
between the viruses (see text for details). The map also displays
countries in which dengue is known to occur.

References
1. Briese T, Jia XY, Huang C, Grady LJ, Lipkin WI. Identification of a
Kunjin/West Nile–like flavivirus in brains of patients with New York
encephalitis. Lancet 1999;354:1261–2.
2. Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, et
al. Origin of the West Nile virus responsible for an outbreak of
encephalitis in the northeastern United States. Science
1999;286:2333–7.
3. Morse SS. Emerging viruses. New York: Oxford University Press;
1993.
4. Halstead SB. Epidemiology of dengue and dengue hemorrhagic
fever. In: Kuno DJGaG, editor. Dengue and dengue hemorrhagic
fever. New York: CAB International; 1997. p. 23–44.
5. Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol
Rev 1998;11:480–96.
6. Rosen L. The emperor’s new clothes revisited, or reflections on the
pathogenesis of dengue hemorrhagic fever. Am J Trop Med Hyg
1977;26:337–43.
7. Gubler DJ, Reed D, Rosen L, Hitchcock JR Jr. Epidemiologic, clini-
cal, and virologic observations on dengue in the Kingdom of Tonga.
Am J Trop Med Hyg 1978;27:581–9.
8. Gubler DJ, Suharyono W, Lubis I, Eram S, Gunarso S. Epidemic
dengue 3 in central Java, associated with low viremia in man. Am J
Trop Med Hyg 1981;30:1094–9.
9. Guzman MG, Kouri GP, Bravo J, Soler M, Velasquez S, Morier L.
Dengue haemorrhagic fever in Cuba. A retrospective seroepidemio-
logic study. Am J Trop Med Hyg 1990;42:179–84.
10. Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos
C, et al. Origins of dengue type 2 viruses associated with increased
pathogenicity in the Americas. Virology 1997;230:244–51.
11. Dengue type 3 infection. Nicaragua and Panama, October–November
1994. Wkly Epidemiol Rec 1995;70:41–3.
12. Gubler DJ. The global pandemic of dengue/dengue haemorrhagic
fever: current status and prospects for the future. Ann Acad Med
Singapore 1998;27:227–34.
13. Guzman M, Huelva G, Saenz E, Quiroz E, De los Reyes J, Balmaseda
A. Reintroduccion del dengue 3 en las Americas: 1994–1996.
Archivos Venezolanos de Medicina Tropial 1998;2:8–19.
14. Balmaseda A, Sandoval E, Perez L, Gutierrez CM, Harris E.
Application of molecular typing techniques in the 1998 dengue epi-
demic in Nicaragua. Am J Trop Med Hyg 1999;61:893–7.
15. Nogueira RM, Miagostovich MP, de Filippis AM, Pereira MA,
Schatzmayr HG. Dengue virus type 3 in Rio de Janeiro, Brazil. Mem
Inst Oswaldo Cruz 2001;96:925–6.
16. Usuku S, Castillo L, Sugimoto C, Noguchi Y, Yogo Y, Kobayashi N.
Phylogenetic analysis of dengue-3 viruses prevalent in Guatemala
during 1996–1998. Arch Virol 2001;146:1381–90.
17. Chungue E, Deubel V, Cassar O, Laille M, Martin PM. Molecular
epidemiology of dengue 3 viruses and genetic relatedness among
dengue 3 strains isolated from patients with mild or severe form of
dengue fever in French Polynesia. J Gen Virol 1993;74:2765–70.
18. Leitmeyer KC, Vaughn DW, Watts DM, Salas RA, de Chacon IV,
Ramos C, et al. Dengue virus structural differences that correlate with
pathogenesis. J Virol 1999;73:4738–47.
19. Srivastava VK, Suri S, Bhasin A, Srivastava L, Bharadwaj M. An epi-
demic of dengue haemorrhagic fever and dengue shock syndrome in
Delhi: a clinical study. Ann Trop Paediatr 1990;10:329–34.
20. Vitarana UT, Jayasekera N, Withane N, Gubler DJ. Finding the cause
of dengue hemorrhagic fever outbreaks in Sri Lanka. Arbovirus
Research in Australia 1993;6:125–9.
21. Messer WB, Sivananthan K, Elvitigala J, Preethimala LD, Ramesh R,
Withana N, et al. Epidemiology of dengue in Sri Lanka before and
after the emergence of epidemic dengue hemorrhagic fever. Am J
Trop Med Hyg 2002;66:765–73.
22. de Silva AM, Sivananthan K, Withana N, Vorndam V, Gubler DJ.
Dengue 3 virus is responsible for recent epidemics of dengue hemor-
rhagic fever in Sri Lanka. In: Annual Meeting of the American
Society for Tropical Medicine and Hygiene. San Juan, Puerto Rico;
1998.
23. Lanciotti RS, Lewis JG, Gubler DJ, Trent DW. Molecular evolution
and epidemiology of dengue-3 viruses. J Gen Virol 1994;75:65–75.
24. Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos
C, et al. Origins of dengue type 2 viruses associated with increased
pathogenicity in the Americas. Virology 1997;230:244–51.
25. Kobayashi N, Thayan R, Sugimoto C, Oda K, Saat Z, Vijayamalar B,
et al. Type-3 dengue viruses responsible for the dengue epidemic in
Malaysia during 1993–1994. Am J Trop Med Hyg 1999;60:904–9.
26. Rico-Hesse R. Molecular evolution and distribution of dengue virus-
es type 1 and 2 in nature. Virology 1990;174:479–93.
27. Lewis JA, Chang GJ, Lanciotti RS, Kinney RM, Mayer LW, Trent
DW. Phylogenetic relationships of dengue-2 viruses. Virology
1993;197:216–24.
28. Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, et al.
Evolutionary relationships of endemic/epidemic and sylvatic dengue
viruses. J Virol 2000;74:3227–34.
29. Gubler D. Dengue and dengue hemorrhagic fever: its history and
resurgence as a global public health problem. In: Kuno DGaG, editor.
Dengue and dengue hemorrhagic fever. New York: CAB
International; 1997.
30. Isturiz RE, Gubler DJ, Brea del Castillo J. Dengue and dengue hem-
orrhagic fever in Latin America and the Caribbean. Infect Dis Clin
North Am 2000;14:121–40.
31. Briseno-Garcia B, Gomez-Dantes H, Argott-Ramirez E, Montesano
R, Vazquez-Martinez AL, Ibanez-Bernal S, et al. Potential risk for
dengue hemorrhagic fever: the isolation of serotype dengue-3 in
Mexico. Emerg Infect Dis 1996;2:133–5.
32. Gubler DJ, Sather GE, Kuno G, Cabral JR. Dengue 3 virus transmis-
sion in Africa. Am J Trop Med Hyg 1986;35:1280–4.
33. Kanesa-thasan N, Chang GJ, Smoak BL, Magill A, Burrous MJ,
Hoke CH Jr. Molecular and epidemiologic analysis of dengue virus
isolates from Somalia. Emerg Infect Dis 1998;4:299–303.
34. Gubler DJ, Trent DW. Emergence of epidemic dengue/dengue hem-
orrhagic fever as a public health problem in the Americas. Infect
Agents Dis 1993;2:383–93.
35. Gubler DJ, Suharyono W, Lubis I, Eram S, Sulianti Saroso J.
Epidemic dengue hemorrhagic fever in rural Indonesia. I. Virological
and epidemiological studies. Am J Trop Med Hyg 1979;28:701–10.
36. Lee E, Gubler DJ, Weir RC, Dalgarno L. Genetic and biological dif-
ferentiation of dengue 3 isolates obtained from clinical cases in Java,
Indonesia, 1976–1978. Arch Virol 1993;133:113–25.
37. Kochel TJ, Watts DM, Halstead SB, Hayes CG, Espinoza A, Felices
V. Effect of dengue-1 antibodies on American dengue-2 viral infec-
tion and dengue haemorrhagic fever. Lancet 2002;360:310–2.
Address for correspondence: Aravinda de Silva, Department of
Microbiology and Immunology, CB#7290, University of North Carolina,
Chapel Hill, NC 27599, USA; fax: (919) 962-8103; email:
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 809
RESEARCH

Vibrio cholerae in O-group 139 was first isolated in
1992 and by 1993 had been found throughout the Indian
subcontinent. This epidemic expansion probably resulted
from a single source after a lateral gene transfer (LGT)
event that changed the serotype of an epidemic V. choler-
ae O1 El Tor strain to O139. However, some studies found
substantial genetic diversity, perhaps caused by multiple
origins. To further explore the relatedness of O139 strains,
we analyzed nine sequenced loci from 96 isolates from
patients at the Infectious Diseases Hospital, Calcutta, from
1992 to 2000. We found 64 novel alleles distributed among
51 sequence types. LGT events produced three times the
number of nucleotide changes compared to mutation. In
contrast to the traditional concept of epidemic spread of a
homogeneous clone, the establishment of variant alleles
generated by LGT during the rapid expansion of a clonal
bacterial population may be a paradigm in infections and
epidemics.
A
n epidemic of cholera began in Madras, India, in 1992
and within a year had spread across the Indian sub-
continent, with cases numbering in the millions (1,2).
Vibrio cholerae isolates from this epidemic had a previous-
ly unidentified serotype, subsequently designated as O139
Bengal (1,2). This new serotype appears to have resulted
when a lateral gene transfer (LGT) event occurred that
replaced the 22 kb of the wbf region (encoding the O1 anti-
gen) of a seventh pandemic V. cholerae O1 El Tor strain
with a 37-kb region encoding the O139 polysaccharide
(3–5). The epidemic spread rapidly through all age groups,
as persons with previous exposure to V. cholerae O1 were
not immune to O139 infection. Since 1992, O139 strains
have established endemicity in this geographic region and
account for a variable percentage of cholera cases every
year (6).
Genetic variation observed in O139 isolates has been
attributed to many causes. Variation in restriction fragment
length polymorphism (RFLP) analysis of rDNA genes (7)
and in recA sequence (8) has been interpreted as evidence
for multiple origins. Genetic variability in RFLP of the
CTX element (6) has been attributed to phage-mediated
recombination. Variation in antimicrobial susceptibility (9)
has been attributed to plasmid exchange in response to
selective pressure from drug use. The variation in pulsed-
field gel electrophoresis (PFGE) analysis of genomic
restriction fragments (6,10) has been attributed to point
mutations. Multilocus sequence typing (MLST), which has
been used in the evaluation of a number of other bacterial
species (11–14), provides an alternative method for meas-
uring genetic relatedness and has provided data for identi-
fying both point mutations and LGT events (14). MLST
has improved discriminatory power over PFGE in some
cases, e.g., Enterococcus (15) and Salmonella (16); how-
ever, in the case of Escherichia coli O157, it does not
because of an absence of sequence variation in the clonal-
ly derived isolates (17). A small MLST study of O139 iso-
lates of V. cholerae did not identify any LGT events (18).
To understand the evolutionary dynamics of V. choler-
ae O139, we sequenced segments from nine loci, including
seven that may be classified as traditional housekeeping
genes, one that carries the genes for cholera toxin, and
another that is next to the insertion sequence within the
O139 wbf region (3–5). Thus, the last two loci might be
expected to show LGT, because they are associated with
known mobile elements, but the other seven loci would not
be expected to show LGT. However, we found putative
LGT alleles at all nine loci in the 96 clonally related O139
isolates.
Materials and Methods
We evaluated nine loci—dnaE, lap, recA, pgm, gyrB,
cat, chi, rstR, and gmd—from 96 V. cholerae O139 isolat-
ed from patients seen at the Infectious Diseases Hospital,
Calcutta, from 1992 to 2000 (see Appendix, online only).
DNA was prepared from overnight cultures by using
PrepMan Ultra (Applied Biosystems Inc., Foster City, CA)
at the University of Maryland School of Medicine. Each
810 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Molecular Epidemiology of O139
Vibrio cholerae: Mutation, Lateral
Gene Transfer, and Founder Flush
Pallavi Garg,*† Antonia Aydanian,‡ David Smith,‡ J. Glenn Morris, Jr.,‡ G. Balakrish Nair,*†
and O. Colin Stine‡
*National Institute of Cholera and Enteric Diseases, Calcutta,
India; †International Centre for Diarrheal Diseases Research,
Dacca, Bangladesh; and ‡University of Maryland School of
Medicine, Baltimore, Maryland, USA

locus was amplified by using polymerase chain reaction
(PCR) with primers (Table 1) selected from a conserved
region of the locus, as determined by aligning sequences
from GenBank. Our primers selectively amplified the orig-
inal O139 rstR gene found in all isolates and not the addi-
tional one found in some recently inserted CTX elements
(19). The presence of amplified products was confirmed on
agarose gels. Purification of the products was performed by
using Millipore filters. The purified PCR products were
sequenced in both directions by using the same primers
used for amplification and Big Dye cycle sequencing kit
(ABI) in accordance with manufacturer’s instructions. The
fluorescently labeled products were separated and detected
by using either an ABI 377 or 3700 Automatic Sequencer
(ABI). The trace files were read by using Phred (20,21) and
Phrap (22). Low-quality sequence at the ends was trimmed,
and the contigs from each individual isolate were aligned
by using Clustal X (23). Variable nucleotides were identi-
fied manually. Isolates with identical alleles were identified
from a distance matrix obtained from PAUP (24). The alle-
les have been assigned GenBank accession numbers
AY297845 to AY297921.
The expected number of alleles that were a result of
point mutations was calculated. All point mutations were
assumed to occur independently; thus, the expected num-
ber of alleles with >2 nucleotides (nt) can be calculated,
and the excess number of observed alleles was attributed to
conspecific LGT of homologous genes. If one assumes
that p is the probability of seeing a single mutation in an
allele, the chance of seeing two mutations on the same
allele is p
2
; the probability of seeing three or more muta-
tions is p
3
. Probability can be calculated from the data by
dividing the number of alleles with a single nucleotide dif-
ference, 34, by 785, the number of alleles in which observ-
ing a point mutation is possible (the 6-bp deletion; the
recombinant gmd, recA alleles; and all duplicate novel
alleles were excluded). Thus, p equals 0.043, p
2
equals
0.0018, and p
3
equals 8 x 10
-5
. When these probabilities are
multiplied by the total number of alleles, 785, the expect-
ed number of alleles containing two independent point
mutations is 1.45, and the expected number containing
three or more is 0.06 (Figure 1).
Results
Each of the loci examined had a variable number of
observed alleles: 9 for dnaE, 20 for lap, 11 for rstR, 11 for
gmd, 2 for recA, 8 for pgm, 4 for gyrB, 7 for cat, and 5 for
chi. The most variable, lap with 20 alleles, was expected
because it is a highly variable locus when analyzed with
multilocus enzyme electrophoresis (25). The most com-
mon allele was present in 91% of isolates (n=87) for dnaE,
77% (n=86) for lap, 79% (n=90) for rstR, 82% (n=87) for
gmd, 99% (n=96) for recA, 90% (n=94) for pgm, 97%
(n=92) for gyrB, 93% (n=88) for cat, and 94% (n=89) for
chi. Thus, the pattern for each locus consists of a common
or ancestral allele and a series of rare alleles, as expected
for the expansion of a clone.
Of the 64 less frequent alleles, some result from LGT
and others from mutation. The three alleles with the largest
changes are unlikely to be due to point mutations. First, a
gmd allele that differed by 113 of the 360 bp sequenced,
when compared with sequences in GenBank using BLAST
(available from: URL: www.ncbi.nlm.nih.gov/BLAST/)
showed greater similarity to gmd from E. coli (AF061251)
than gmd from V. cholerae, consistent with LGT of a
homologous gene into the V. cholerae genome. Second, an
alternative recA allele that differs by 24 nt is likely to be
the result of LGT of a homologous gene. Although sub-
stantial, the number of nucleotide differences is not large
enough for the allele to be clustered with sequences from
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 811
RESEARCH
Table 1. Primers used for multilocus sequence typing
Locus
Primer 1
Primer 2
dnaE
CGRATMACCGCTTTCGCCG
GAKATGTGTGAGCTGTTTGC
lap
GAAGAGGTCGGTTTGCGAGG
GTTTGAATGGTGAGCGGTTTGCT
rstR
CGTGTTAGAGCACAC
GAGTGAATCGTCGTG
gmd
CCTTATGCKGTGGCRAA
CTWGGATCACCTAACA
recA
GAAACCATTTCGACCGGTTC
CCGTTATAGCTGTACCAAGCGCCC
pgm
CCKTCSCAYAACCCGCC
TCRACRAACCATTTGAADCC
gyrB
GAAGGBGGTATTCAAGC
GAGTCACCCTCCACWATGTA
cat
ATGGCTTATGAATCGATGGG
TCCCATTGCCATGCACC
chi
CAYGAYCCRTGGGCWGC
ACRTCTTCAATCTTGTC
Figure 1. Bar graph of the number of novel alleles (y-axis) with a
specific number of nucleotide differences from the ancestral allele.
Two alleles with 24-bp and 113-bp differences are excluded from
the graph.
V. mimicus, the closest sibling species to V. cholerae (8), a
finding that suggests that recombination occurred within V.
cholerae. Third, a lap allele had a 6-bp deletion and a sin-
gle nucleotide difference that may be the result of a dou-
ble-strand break repair.
We calculated that at least 26 putative conspecific LGT
events occurred in the 96 isolates studied. Figure 1 shows
the number of nucleotide differences between each novel
allele and the ancestral allele. If all point mutations are
assumed to occur independently, the expected number of
alleles with two or more variable nucleotides can be calcu-
lated and the excess number of observed alleles attributed
to conspecific LGT of homologous genes. The expected
number of alleles containing two independent point muta-
tions is 1.45, and the expected number containing three or
more is 0.06. Since 11 alleles were observed with 2 nt dif-
ferences, 9 more than expected, and 16 were observed with
>
3 differences, 16 more than expected, all of these alleles
probably did not occur through mutation; more likely,
these alleles are the result of LGT. Thus, we would esti-
mate that 26 alleles (9 + 16 + recA allele above) are puta-
tively due to conspecific LGT of homologous genes.
The putative conspecific LGT alleles, although fewer in
number (26 alleles) than the assumed number of mutation-
derived alleles (34 alleles), provide most of the nucleotide
differences between alleles. The 120 nt changes introduced
by conspecific LGT events are approximately three times
the 38 (34 single mutations + 4 2x2 double mutations)
introduced by mutation. This calculation is conservative:
The 26 conspecific LGT events may represent an underes-
timate of the number because some of the alleles differing
by <
1 nt may have resulted from LGT.
The analysis of all nine loci from each isolate was
based on the sequence type (ST). Each isolate was defined
by a 9-digit number composed of the assigned allele num-
ber at each of the nine loci in the following order: dnaE,
lap, rstR, gmd, recA, pgm, gyrB, cat, and chi. The most
common allele was arbitrarily assigned as number 1. Thus,
the ST of all the most common alleles is ST
1,1,1,1,1,1,1,1,1. Missing data were assigned the most
common allele. This assumption is conservative, mini-
mizes the observed amount of variation, and is consistent
with the preponderance of common alleles found at each
locus.
Fifty-one unique STs were found in the 96 isolates test-
ed, reflecting relatively extensive genetic diversity. The
overall average of 0.53 unique STs per isolate examined is
similar to that seen in every year including 1992 (Table 2).
Six STs occur more than once. As expected, the ancestral
ST:1,1,1,1,1,1,1,1,1, found in 40 isolates, occurred in all
years. Among the others, ST:1,1,2,1,1,1,1,1,1 was found
three times, once each in 1995, 1996, and 1997.
ST:1,2,1,1,1,1,1,1,1 and ST:1,1,7,1,1,1,1,1,1 were found
twice in 1992 and 1994, respectively. ST 1,1,1,6,1,1,1,1,1
was found once in 1998 and again in 1999.
ST:1,1,1,1,1,4,1,1,1 was found in 1995 and 1998. Since the
number of STs is large (51 types), and number of samples
in a collection period is small (8–13 samples; Table 2), STs
seen in multiple years must not only persist but also repre-
sent a substantial portion of the epidemic O139 V. choler-
ae population.
Five of the novel STs are related to other novel STs by
allelic change at another second or third locus. One
sequence type evolved into three related types found in
subsequent years (Figure 2a). The starred gmd allele is one
related to the E. coli sequence, and its presence in two dis-
tinct related STs in two different years demonstrates its
establishment in the population. That the pattern seen in
Figure 2b of ancestral alleles rstR 1 and chi 1 and two vari-
ant alleles, rstR 7 and chi 5, was found in all combinations
is indicative of an LGT event. Figure 2c-e shows three
additional groups of related sequences. In Figure 2a, b, and
d, the ST with the larger number of novel alleles occurred
in later years. In contrast, in Figure 2c and e, the ST with
the larger number of novel alleles occurred in the earlier
years. The lack of an overall temporal relationship may
result from the small sample size (8–13 isolates) in any
year.
One isolate, CRC5, is unusual because it has no
sequenced alleles of the ancestral type. Nevertheless, the
alleles from this isolate are closely related to those of the
ancestral type. Each CRC5 allele differs from the ancestral
allele by 7 nt for dnaE, 3 nt for lap, 4 nt for rstR, 24 nt for
recA, 6 nt for pgm, 10 nt for gyrB, and 4nt for chi. A com-
prehensive survey of the genetic distances for these loci
could determine the average distance between alleles for
each of these loci. The data would provide insight into
whether this isolate represents a second derivation of the
O139 clinical type from an environmental strain or if it is
a genetic outlier within the clonally related, but diversified,
O139 epidemic type.
Discussion
The emergence and pandemic spread of V. cholerae
O139 Bengal represented a chance to examine evolution of
a bacterial strain in the midst of a clonal expansion. Our
results are consistent with clonal expansion and subse-
quent divergence as described by Spratt and Maiden (26).
Putative recombinant alleles were found at all nine loci
among the 96 clonally related O139 isolates. One gmd
allele from V. cholerae was most similar to a gmd allele
from E. coli. The number of base-pair differences among
other alleles was higher than expected on the basis of a
simple computation for the accumulation of independent
mutations. This finding suggests that many of these events
were due to LGT. When we applied our criteria to the
812 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
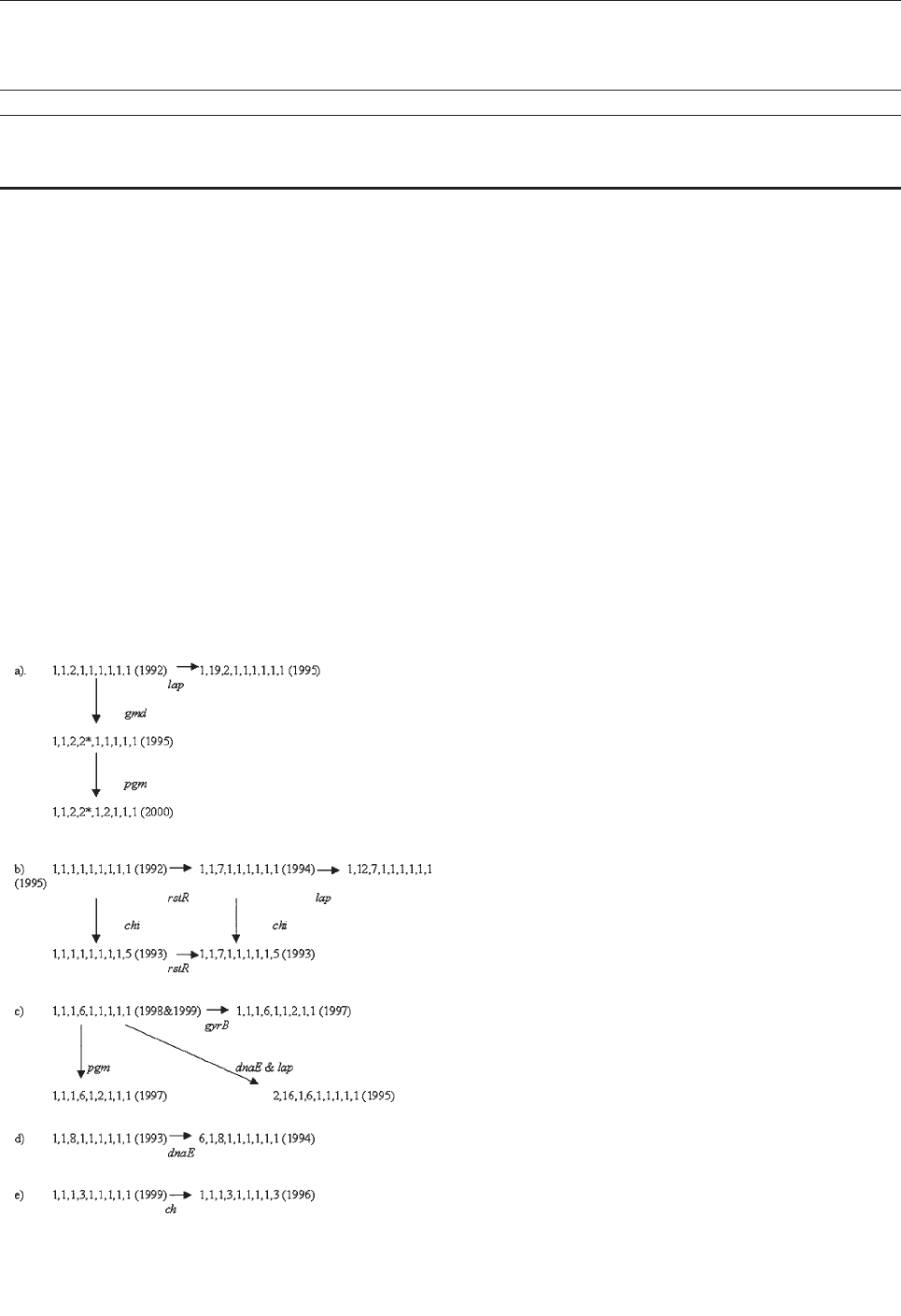
novel alleles identified in a previous study (18), 11 of the
13 would be considered to have resulted from LGT, since
the number of nucleotide differences to the ancestral allele
varied from 4 to 19. Thus, for V. cholerae, like Neisseria,
Streptococcus, and other bacterial species (11–14), conspe-
cific recombination of homologous genes appears to be
common and responsible for most of the alleles with mul-
tiple nucleotide differences and the majority of the
nucleotide differences. The genetic variability at the nine
loci alters our understanding of evolution in bacteria,
showing that recombination in V. cholerae occurs fre-
quently and most nucleotide changes occur by means of a
recombination that can alter any gene.
The proportion of recombinants from conspecific
recombination, 3.5% (28/785) is greater than that from
transgeneric recombination (0.01% from the acquisition of
E. coli gmd by one isolate). One potential implication of a
greater rate of conspecific recombination may be that, over
time, it will maintain the species identity of each individ-
ual bacterium, despite the constant bombardment of
homologous genes from other genera. Although at first
glance the frequency of the novel sequence types appears
to conflict between our study and an earlier study (18), the
observations may be reconciled on the basis of both the
observed frequencies and the timing of the observations.
Both studies identified a common ancestral allele in from
77% to 99% of isolates in our study and a series of rare
alleles with 1–19 variant alleles for each locus. These stud-
ies reported 10% novel sequence types in 29 isolates that
were collected from “the first epidemic period,” from 1992
to 1993 (18). Our data from 1992 showed 33% novel
sequence types from a sample of nine. These data are not
statistically different (chi-square test=2.4, p=0.12).
However, the researchers’ estimate of frequency (18) is
more likely to be correct because of the larger sample size.
The dates of collection may also be important because our
collection of isolates from 1993 began in March, when the
number of O139 cases at the Infectious Diseases Hospital
rose from <10 to >80 per month, corresponding to a rapid
population expansion or flush. Thus, we can predict that
we would see substantial variation in our sample.
The genetic diversity was greater in the V. cholerae
O139 isolates than in other clinically associated clones. In
V. parahaemolyticus O3:K6, a pandemic strain, 94% of
strains were identical at four loci (N. Chowdhury et al.,
unpub. data). In E. coli O157, all 77 isolates were identical
at seven loci in spite of variation between isolates on
PFGE (17). Although V. parahaemolyticus and E. coli are
widespread pathogens, they differ from V. cholerae O139
because their population size has expanded much more
slowly.
Among O139 isolates, the substantial genetic diversity
found in the first year of the epidemic may reflect a
“founder flush” phenomenon. During times of population
expansion, i.e., a flush, any novel genotype with similar or
even slightly deleterious fitness compared to the founder
genotype will produce sufficient offspring to become
established in the population (27). A founder flush appears
to have occurred in the establishment of Helicobacter
pylori in a single person (28). Although other previous
descriptions of this phenomenon have been limited to
insects, specifically butterflies (29) and drosophilids (30),
we believe that the founder flush phenomenon may
become the paradigm for epidemic bacterial expansion in
individual patients and populations. This founder flush
phenomenon, in turn, has implications for our interpreta-
tion of “clonality” among epidemic isolates and for our
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 813
RESEARCH
Table 2. Number of isolates tested and distinct sequence types, by year
No.
1992
1993
1994
1995
1996
1997
1998
1999
2000
Total
Isolates examined
9
9
12
10
13
11
12
12
8
96
Novel sequence types
3
6
6
7
8
6
7
4
7
51
Novel sequence types per isolate examined
0.33
0.66
0.5
0.7
0.62
0.55
0.58
0.33
0.88
0.53
Figure 2. Five groups of related sequence types of Vibrio cholerae
O139.

understanding of factors that contribute to the emergence
of new pathogenic strains.
This work was supported in part by an American Society for
Microbiology travel fellowship to Dr. Garg.
Dr. Garg completed this work as part of her Ph.D. disserta-
tion at University of Calcutta. She is a postdoctoral fellow at the
University of Maryland School of Medicine, pursuing studies of
thrombospondin in human microvascular endothelial cells.
References
1. Bhattacharya SK, Bhattacharya MK, Nair GB, Dutta D, Deb A,
Ramamurthy T, et al. Clinical profile of acute diarrhoea cases infect-
ed with the new epidemic strain of Vibrio cholerae O139: designation
of the disease as cholera. J Infect 1993;27:11–8.
2. Nair GB, Ramamurthy SK, Mukhopadhyay T, Garg S, Bhattacharya
MK, Takeda T, et al. Spread of Vibrio cholerae O139 Bengal in India.
J Infect Dis 1994;169:1029–34.
3. Comstock LE, Johnson JA, Michalski JM, Morris JG, Kaper JB.
Cloning and sequence of a region encoding surface polysaccharide of
Vibrio cholerae O139 and characterization of the insertion site in the
chromosome of Vibrio cholerae O1. Mol Microbiol 1966;19:815–26.
4. Stroeher UH, Parasivam G, Dredge BK, Manning PA. Novel Vibrio
cholerae O139 genes involved in lipopolysaccharide biosynthesis. J
Bacteriol 1997;179:2740–7.
5. Bik EM, Bunschoten AE, Willems RJL, Chang ACY, Mooi FR.
Genetic organization and functional analysis of the otn DNA essen-
tial for cell-wall polysaccharide synthesis in Vibrio cholerae O139.
Mol Microbiol 1996;20:799–811.
6. Basu A, Garg P, Datta S, Chakraborty S, Bhattacharya T, Yamasaki S,
et al. Vibrio cholerae O139 in Calcutta, 1992–1998: incidence, antibi-
ograms, and genotypes. Emerg Infect Dis 2000;6:139–47.
7. Faruque SM, Saha MN, Asadulghani, Bag PK, Bhadra RK,
Bhattacharya SK, et al. Genomic diversity among Vibrio cholerae
O139 strains isolated in Bangladesh and India between 1992 and
1998. FEMS Microbiol Lett 2000;184:279–84.
8. Stine OC, Sozhamannan S, Gou Q, Zheng S, Morris JG, Johnson JA.
Phylogeny of Vibrio cholerae based on recA sequence. Infect Immun
2000;68:7180–5.
9. Garg P, Chakraborty S, Basu I, Datta S, Rajendran K, Bhattacharya T,
et al. Expanding multiple antibiotic resistance among clinical strains
of Vibrio cholerae isolated from 1992–7 in Calcutta, India. Epidemiol
Infect 2000;124:393–9.
10. Kurazono H, Yamasaki S, Ratchtrachenchai O, Nair GB, Takeda Y.
Analysis of Vibrio cholerae O139 Bengal isolated from different geo-
graphical areas using macrorestriction DNA analysis. Microbiol
Immunol 1996;40:303–5.
11. Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R,
et al. Multilocus sequence typing: a portable approach to the identifi-
cation of clones within populations of pathogenic microorganisms.
Proc Natl Acad Sci U S A 1998;95:3140–5.
12. Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, et al.
Multilocus sequence typing system for Campylobacter jejuni. J Clin
Microbiol 2001;39:14–23.
13. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus
sequence typing for characterization of methicillin-resistant and
methicillin-susceptible clones of Staphylococcus aureus. J Clin
Microbiol 2000;38:1008–15.
14. Feil EJ, Smith JM, Enright MC, Spratt BG. Estimating recombina-
tional parameters in Streptococcus pneumoniae from multilocus
sequence typing data. Genetics 2000;154:1439–50.
15. Nallapareddy SR, Duh RW, Singh KV, Murray BE. Molecular typing
of selected Enterococcus faecalis isolates: pilot study using multilo-
cus sequence typing and pulsed-field gel electrophoresis. J Clin
Microbiol 2002;40:868–76.
16. Kotetishvili M, Stine OC, Kreger A, Morris JG Jr., Sulakvelidze A.
Multilocus sequence typing for characterization of clinical and envi-
ronmental Salmonella strains. J Clin Microbiol 2002;40:1626–35.
17. Noller AC, McEllistrem MC, Stine OC, Morris JG, Boxrud DJ, Dixon
B, et al. Lack of DNA sequence diversity among Escherichia coli
O157:H7 isolates that are distinct by pulsed-field gel electrophoresis.
J Clin Microbiol 2003;41:675–9.
18. Farfan M, Minana-Galbis D, Fuste MC, Loren JG. Allelic diversity
and population structure in Vibrio cholerae O139 Bengal based on
nucleotide sequence diversity. J Bacteriol 2002;184:1304–13.
19. Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE.
Characterization of VPI pathogenicity island and CTXΦ prophage in
environmental strains of Vibrio cholerae. J Bacteriol
2001;183:4737–46.
20. Ewing B, Greene P. Base-calling of automated sequencer traces using
Phred. II. Error probabilities. Genome Res 1998;8:186–94.
21. Ewing B, Hillier L, Wendl M, Greene P. Base-calling of automated
sequencer traces using Phred. I. Accuracy assessment. Genome Res
1998;8:175–85.
22. Green P. 1998. Phrap, SWAT and CrossMatch. Available from: URL:
http://www.washington.edu
23. Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ.
Multiple sequence alignment with Clustal X. Trends Biochem Sci
1998;23:403–5.
24. Swofford D. PAUP* beta version. Sunderland (MA): Sinauer Assoc;
2000.
25. Beltran P, Delgrado G, Navarro A, Trujillo F, Selander RK, Cravioto
A. Genetic diversity and population structure of Vibrio cholerae. J
Clin Microbiol 1999;37:581–90.
26. Spratt BG, Maiden MCJ. Bacterial population genetics, evolution and
epidemiology. Philos Trans R Soc Lond Biol Sci 1999;354:701–10.
27. Wallace B. Basic populations genetics. New York: Columbia
University Press; 1981. p. 429–32.
28. Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S,
et al. Helicobacter pylori genetic diversity within the gastric niche of
a single host. Proc Natl Acad Sci U S A 2001;98:14625–30.
29. Ford HD, Ford EB. Fluctuation in numbers, and its influence on vari-
ation, in Melitaea aurina, Rott. (Lepidoptera). Transactions of the
Royal Entomological Society London 1930;78:345–51.
30. Carson HL. The population flush and its genetic consequences. In:
Lewontin RC, editor. Population biology and evolution. Syracuse
(NY): Syracuse University Press;1968. p. 123–37.
Address for correspondence: O. Colin Stine, Department of
Epidemiology and Preventive Medicine, University of Maryland School
of Medicine, Baltimore, MD 21201, USA; fax: 410-706-1644; email:
814 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH

To evaluate the role of amoeba-associated bacteria as
agents of ventilator-associated pneumonia (VAP), we test-
ed the water from an intensive care unit (ICU) every week
for 6 months for such bacteria isolates; serum samples and
bronchoalveolar lavage samples (BAL) were also obtained
from 30 ICU patients. BAL samples were examined for
amoeba-associated bacteria DNA by suicide-polymerase
chain reaction, and serum samples were tested against
ICU amoeba-associated bacteria. A total of 310 amoeba-
associated bacteria from 10 species were isolated. Twelve
of 30 serum samples seroconverted to one amoeba-asso-
ciated bacterium isolated in the ICU, mainly Legionella
anisa and Bosea massiliensis, the most common isolates
from water (p=0.021). Amoeba-associated bacteria DNA
was detected in BAL samples from two patients whose
samples later seroconverted. Seroconversion was signifi-
cantly associated with VAP and systemic inflammatory
response syndrome, especially in patients for whom no eti-
ologic agent was found by usual microbiologic investiga-
tions. Amoeba-associated bacteria might be a cause of
VAP in ICUs, especially when microbiologic investigations
are negative.
H
ospital-acquired pneumonia occurs in 0.5% to 1% of
admitted patients admitted, representing 10% to 15%
of all nosocomial infections; pneumonia is the most com-
mon cause of nosocomial infection in intensive-care units
(ICUs) (1). This pneumonia is associated with high death
rates. As the etiologic agent of pneumonia remains
unknown in 20% to 50% of cases (2), identifying new lung
pathogens is a major public health goal. Aquatic bacteria
such as Legionella spp., Pseudomonas spp., Stenotro-
phomonas spp., Burkholderia spp., or Acinetobacter spp.
may colonize in hospital water supplies and have previous-
ly been shown to be causally associated with cases of noso-
comial infections (3). Free-living amoebae have been
shown to be a reservoir of pathogens such as Legionella sp.,
Burkholderia picketti, and Cryptococcus neoformans (4–7).
The most studied amoebae-associated bacterium is
Legionella pneumophila, the agent of Legionnaires’ disease
(8), which frequently results from exposure to contaminat-
ed aerosols. Additional amoeba-associated bacteria might
be implicated in community-acquired pneumonia, includ-
ing Legionella-like amoebal pathogens (9) and members of
the genus Parachlamydia (10). As part of research into the
diversity of bacterial agents associated with amoebae in
hospital water supplies, we have identified a new α-
Proteobacteria belonging to the Bradyrhizobiaceae family
(11–13). We demonstrated that patients with nosocomial
pneumonia hospitalized in the vicinity of the contaminated
water in a public hospital of our city have elevated antibody
titers to these bacteria (14). In this study, we performed the
same kind of analysis but focused our work on a single ICU
during a 6-month period. Amoeba-associated bacteria were
periodically evaluated in all ICU water taps. To evaluate
contact of patients hospitalized in this ICU and amoeba-
associated bacteria in the water, serum and bronchoalveolar
lavages (BAL) samples were periodically sampled. Serum
samples were tested in an immunofluorescence assay
against the isolated bacteria to detect seroconversions, and
DNA of these bacteria were detected in BAL samples by
suicide-polymerase chain reaction (PCR) (15,16), a PCR
technique without positive controls that incorporates “dis-
posable” primers to avoid false-positive results. The second
part of this work was to evaluate if exposure to the amoe-
ba-associated bacteria in the ICU could be associated with
disease. Thus, we specifically studied some clinical mark-
ers of infection, including fever, systemic inflammatory
response syndrome (SIRS), and pneumonia for patients
admitted to the ICU. As a definition of pneumonia based
only on clinical and roentgenographic criteria has been crit-
icized for low specificity (17–19), we used strict criteria in
the definition. These criteria were applied to cases in which
bacterial documentation was negative to determine if dis-
ease observed in patients hospitalized in an ICU may be
attributed to amoeba-associated bacteria.
Materials and Methods
All patients admitted to the ICU during a 26-week
period who needed intubation and mechanical ventilation
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 815
RESEARCH
Amoeba-Resisting Bacteria and
Ventilator-Associated Pneumonia
Bernard La Scola,* Ioanna Boyadjiev,*† Gilbert Greub,* Atieh Khamis,*
Claude Martin,† and Didier Raoult*†
*Unité des Rickettsies, Marseille, France; and †Hôpital Nord,
Marseille, France

were included. Patients were evaluated at admission by the
Acute Physiology and Chronic Health Evaluation II score
(20). At admission and every week thereafter, temperature,
leukocytes and platelet counts, hepatic enzymes, and pres-
ence of SIRS and ventilator-associated pneumonia (VAP)
were recorded. Serum samples were obtained from
patients at admission, every 7 days afterwards, and at dis-
charge. BAL was obtained at admission immediately after
intubation by using protected mini-bronchoalveolar lavage
(Combicath, Plastimed, Le-Plessy-Bouchard, France),
then performed when a lung infiltrate suggestive of pneu-
monia appeared, and repeated every week until pneumonia
resolved. BALs are part of the routine diagnosis and fol-
low-up of pneumonia in the ICU and were not performed
specifically for the study. Informed consent was obtained
from the patient’s family, according to French legislation.
Data about bacteria isolated from blood cultures, lung
secretions, and urine by conventional procedures were
recorded. When isolated from urine and lung secretions,
bacteria were only considered as pathogenic when concen-
tration in the specimen was >
10
5
and >
10
6
CFU/mL,
respectively (21).
Definitions of VAP and SIRS were based on previously
published criteria (22–25), but to increase specificity, we
limited our study to severe cases of VAP by adding strict
criteria (Table 1). Taps and ice machine water were sam-
pled every other week, as previously reported (11). The
procedure for isolating bacteria from water and lung secre-
tions by using cocultivation with Acanthamoeba polypha-
ga followed by subculture onto BCYE agar plates has been
detailed elsewhere (11,26). Bacteria were identified by
using 16S rRNA gene sequence comparisons as previous-
ly described (27). Legionella species were identified by
using mip gene amplification and sequencing (28).
Twenty bacterial antigens were tested by microim-
munofluorescence as previously reported (14,29).
Bacterial species isolated from the ICU water during the
studied period and 10 other species previously isolated in
the same conditions from other sites (Bosea eneae, B.
vestrisii, B. thiooxydans, Mesorhizobium amorphae,
Azorhizobium caulinodans, Afipia felis, A. felis
genospecies A, A. clevelandensis, A. birgiae, and A. mas-
siliae [11–14]) were tested. The serologic tests were per-
formed on the first serum samples from all patients admit-
ted to the ICU; these patients were available for a second
sample (30 patients). The control group comprised 10
patients in the same ICU. These patients had shorter stays
and had samples taken at admission but was not available
(100 patients) and 114 patients with other diseases, includ-
ing Q fever (10 samples), trench fever (5 samples),
tularemia (8 samples), Mediterranean spotted fever (10
samples), epidemic typhus (5 samples), syphilis (10 sam-
ples), cat scratch disease (5 samples), pneumonia caused
by Chlamydia pneumoniae (5 samples), C. psittaci (5 sam-
ples), Mycoplasma pneumoniae (10 samples), L. pneu-
mophila (10 samples), hepatitis C virus (5 samples), infec-
tions caused by cytomegalovirus (5 samples), Epstein Barr
virus (11 samples), and HIV (10 samples). Serum samples
were diluted at 1:25, 1:50, and 1:100 for immunoglobulin
(Ig) G and IgM determination. The cutoff titer for a posi-
tive detection was determined as the lowest titer for which
all first serum samples and control serum samples were
negative. Then, all patients’ serum samples were tested and
serial twofold dilutions from 1:50 to 1:1600 were made on
these samples with titers at least equal to the cutoff titer.
Tests by Western blot were performed as previously
described (29) for any patient with a seroconversion.
DNA was extracted from BAL samples by using
QIAMP Tissue kit (QIAGEN, Hilden, Germany), accord-
ing to the manufacturer’s instructions. PCR detection was
attempted on all BAL samples for bacteria against which at
least one seroconversion was generated. DNA extraction
from serum samples was performed in a laboratory other
than the one in which the isolates were identified to avoid
vertical contamination from previous amplifications, and
no positive control was used to avoid horizontal contamina-
tion from the same experiment (16). We used a nested PCR
that incorporated two primer pairs used only once (Table 2)
followed by sequencing and comparison to the targeted
sequence, as previously described for suicide-PCR (15,16).
All samples were tested the same day in the same assay.
Sample testing was blinded, and positive amplicons were
sequenced. Negative controls consisted of BAL samples
from 10 patients of the same ICU obtained at admission,
BAL samples from 200 patients with nosocomial pneumo-
nia hospitalized in other medical centers of the city, water
samples, and a suspension of A. polyphaga. At least one
negative control was used for every two serum samples.
816 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 1. Definition criteria for ventilator-associated pneumonia (VAP) and systemic inflammatory response syndrome (SIRS)
VAP
SIRS
Unexplained VAP
Unexplained SIRS
New and persistent roentgenographic lung
infiltrate and new onset of:
a) Increase in white blood cells >10 g/L
b) Fever or hypothermia (>38°C or <36°C)
c) Purulent sputum
d) Duration of at least 2 weeks
e) PaO
2
/FiO
2
ratio <100
At least two of:
a) T° >38°C or <36°C
b) Heart rate >90/min
c) Respiratory rate >20/min or PaCO
2
<32 mmHg
d) Leukocytes >12 or <4 g/L or immature
(band) forms
Lack of recovery of
bacteria from:
a) Lung secretions
b) Blood cultures
Lack of recovery of bacteria from:
a) Lung secretions
b) Blood cultures
c) Urine

Comparisons of demographic, clinical, and laboratory
data between patients with evidence of amoeba-associated
bacteria contact (seroconversion or positive detection in
BAL samples) were performed by using chi square and
Mann-Whitney tests, respectively. The tested variables
were age; sex; an underlying disease; severity score; intu-
bation duration; hospitalization duration; SIRS; death;
increase of hepatic enzymes, platelet count, and leuko-
cytes; VAP; and fever. We also compared demographic,
clinical, and laboratory data between patients with and
without unexplained VAP, fever, and SIRS. Multivariate
analysis adjusted for age, sex, prolonged intubation, and an
underlying disease was performed to confirm observed
associations. STATA software (v. 7.0, Stata Corporation,
College Station, TX) was used for analysis.
Results
Ten species (310 isolates) were identified from 864
water samples (Table 3). B. massiliensis and L. anisa were
the two most commonly isolated species (62.3% of the 310
isolates). In the ICU admission rooms , isolation of L.
anisa ranged from 75% to 100% of the tested taps during
the first 20 weeks; whereas all were negative during the
last 6 weeks, after taps were changed. No amoeba-associ-
ated bacteria were isolated from BAL.
Ninety serum samples from the 30 patients and the 214
control serum samples were tested by an immunofluores-
cence assay for IgG and IgM on the 20 amoeba-associated
bacteria antigens (12,160 tests). The 30 first serum sam-
ples and the samples from blood donors did not have an
IgG titer of >1:50 or IgM titer >1:25. Cutoff titers for pos-
itive serologic tests with 100% specificity are shown in
Table 4. Twelve (40%) patients seroconverted from 10 to
35 days after admission to at least one antigen; 10 showed
IgM antibodies (Table 5). Five patients seroconverted to L.
anisa, six to B. massiliensis, including one to both, one to
L. quinlivanii, and one to A. broomeae. Western blots con-
firmed the seroconversions, with the appearance of sever-
al reacting bands on convalescent-phase serum samples
(Figure). Patients also seroconverted to amoeba-associated
bacteria not detected in the water in this study: two to
A. clevelandensis (patient 2, IgG=1:100 and IgM=1:100;
patient 8, IgG=1:50 and IgM=1:200;) and two to A. felis
(patient 2, IgG=1:100 and IgM=1:100; patient 9,
IgG=1:1600 and IgM=1:25). Seroconversions were signif-
icantly more frequent against amoeba-associated bacteria
obtained in this ICU than against amoeba-associated bac-
teria isolated in previous studies: 13 of 300 tests versus 4
of 300 tests and 12 of 30 patients versus 4 of 30 patients
(p=0.046 and p=0.039, respectively). Patients also sero-
converted more frequently to the most commonly isolated
bacteria, L. anisa and B. massiliensis (>50 isolates,
p=0.021).
Table 6 shows the clinical characteristics of patients
with serologic evidence of exposure to amoeba-associated
bacteria isolated in the ICU. Analysis for risk and potential
confounding factors did not indicate differences between
patients with or without seroconversion. However, sero-
conversion was statistically associated with VAP
(p=0.026), unexplained VAP (p=0.034), SIRS (p=0.024),
and unexplained SIRS (p=0.045). Multivariable logistic
regression demonstrated that seroconversion was inde-
pendently associated with VAP even after we adjusted for
intubation duration, hospitalization duration, number of
serum samples, and underlying disease (p=0.014 to 0.030).
The DNA of L. anisa and B. massiliensis were each
detected once in the 66 BAL samples from 30 patients. For
two samples, seroconversion to the identified bacteria, L.
anisa and B. massiliensis, respectively, was observed 4
and 2 weeks after the PCR was positive in BAL. None of
the 210 control patients was positive for these bacteria in
BAL samples compared to 2 of 30 patients in the ICU
(p<0.01).
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 817
RESEARCH
Table 2. Polymerase chain reaction primers used for amplification and sequencing of bacterial DNA within human samples
Primers
Bosea-related strain
Legionella anisa
Afipia broomeae
L. quinlivanii
External forward
5′-TGCGAGTGTA
GAGGTGAAATT-3′
5′-TATTGGTGC
TGATTTAGGAA-3′
5′-TCTTTTGTGCG
GGAAGATAATG-3′
5′-TTGTTGATGTTT
GTTTTGAGACC-3′
External reverse
5′CGCTCGTTG
CGGGACTTAA-3′
5′-GCTAAGTCTGAA
GGTACA-3′
5′-TAAACTTTCCAA
CGGCTGGCAT-3′
5′-TTCAACACTTCT
TTCATCTGATC-3′
Internal forward
5′-GAGGTGAAA
TTCGTAGATATT-3′
5′-GCCCAATTG
ATTTTGACAG-3′
5′-GCTAACTTCGT
GCCAGCAG-3′
5′-TCCAAGAATAA
AAGGGGATTG-3′
Internal reverse
5′-GAGCTGACG
ACAGCCAT-3′
5′-GCATTAATTGT
AATGCTTCA-3′
5′-GTTTGCTCC
CCACGCTTTC-3′
5′-CCATACCAT
CCTGTAAGCCTT-3′
Table 3. Identification of the 310 bacterial strains isolated by
using amoebal co-culture procedure
Species
No. of isolates
Legionella anisa
126
Bosea massiliensis
67
Rasbo bacterium
45
Bradyrhizobium liaoningense
29
L. quinlivanii
12
L. pneumophila
11
L. rubrilucens
7
L. worsleiensis
6
B. japonicum
5
Afipia broomeae
2

The death rate was 33.3%; disease was mainly associ-
ated with fever (96.6%), VAP (56.6%), and SIRS (76.6%).
No microbial etiologic agent was found in 18 (62%) of 29
patients with fever, 10 (43%) of 23 patients with SIRS, and
in 8 (47%) of 17 patients with VAP. VAP was significantly
associated with duration of hospitalization (median hospi-
talization days/interquartile range of 23/18–41 with VAP
versus 14/10–21 without VAP, p=0.04), SIRS (16/17 with
VAP versus 7/13 without VAP, p=0.025) and seroconver-
sion to amoeba-associated bacteria (10/17 with VAP versus
2/13 without VAP, p=0.026). No other statistical difference
was observed between patients with VAP and without VAP
in terms of demographic, clinical, and paraclinical data,
risks, and potential confounding factors. Most patients
received cephalothin for the first 3 days of hospitalization
as an antibiotic prophylaxis. The most commonly used
antibiotics in patients with VAP were third-generation
cephalosporins. Patients with VAP received antibiotics
more frequently and for more than 1 week (16/17 with
VAP versus 7/13 without VAP, p=0.025).
Discussion
We confirmed that several amoeba-associated bacteria
are common in the water in ICU; we recovered 310 amoe-
ba-associated bacteria isolates from 10 species from the
water of the ICU. As exposure to a microorganism is a pre-
requisite to infectious disease, we first evaluated contact of
patients hospitalized in the ICU with amoeba-associated
bacteria in water. We found that 12 (40%) of 30 of patients
seroconverted to amoeba-associated bacteria and that these
seroconversions were significantly more common against
local isolates than to amoeba-associated bacteria from
other ICUs. Moreover, antibody response parallels that of
water contamination with B. massiliensis and L. anisa,
which cause 83% of seroconversions identified in 62% of
isolates (p=0.021). The cutoff titers, chosen to have 100%
specificity, and the detection of several reactive antigens in
the Western blots (Figure) suggest that these seroconver-
sions represent specific serologic reactions. Detection of
antibodies reacting to amoeba antigens would also be
important because infections could occur through inhala-
tion (perhaps after colonization) of infected amoebae act-
ing as a “Trojan horse” (6). In future studies, isolation of
amoebae in the aquatic environment as in the BAL sam-
ples of patients will be performed for further use as anti-
gens.
Patient exposure to amoeba-associated bacteria from
the ICU was also evident in results of amoeba-associated
bacteria DNA detection in BAL samples from 2 of 30
patients as compared with 0 of 210 control patients
(p<0.01). Moreover, for these patients, bacterial DNA was
detected in the BAL sample before the seroconversion to
the same bacteria 2 to 4 weeks after admission. This rate is
compatible with an acute infection occurring during hospi-
talization rather than colonization of the respiratory tracts
of patients. Contamination is unlikely when using a sui-
cide-PCR procedure (15,16). Isolation of these agents was
probably hampered by the antibiotic prophylaxis instituted
at admission in this ICU for trauma patients and likely
explains the lack of amoeba-associated bacteria isolation
in BAL samples.
Among the 30 patients, VAP occurred frequently
(56.6%) and was associated with hospitalization duration,
as previously reported (30,31). Of the patients with VAP,
58.8% seroconverted to amoeba-associated bacteria, as
compared to 15.4% of the remaining 13 patients
(p=0.026). In patients with seroconversion, SIRS was also
more prevalent (p=0.024). The percentages of unexplained
VAP and unexplained SIRS were four times more common
in amoeba-associated bacteria seroconverters than in non-
seroconverters and remained statistically significant in
spite of the small population studied (Table 6). Thus,
amoeba-associated bacteria may be a common cause of
unexplained VAP and SIRS.
Finally, we identified a cryptic outbreak in the ICU
caused by L. anisa, a pathogen commonly encountered in
the environment (32,33), only implicated in a few epi-
demics of Pontiac fever (34,35) and four cases of legionel-
losis (26,36–38). Five (16.7%) of 30 patients were infect-
ed with this bacterium, considered a relatively rare
pathogen. Serologic tests are not currently used for L.
anisa, and no urinary antigen test is available. Therefore,
818 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 4. Definition of cutoff titers for positive serologic tests and
100% specificity according to antigen tested by using the 224
control serum samples
a
Antigen
IgG
IgM
Legionella anisa
>1:50
>1:25
Bosea massiliensis
>1:50
>1:25
Rasbo bacterium
>1:100
>1:25
Bradyrhizobium liaoningense
>1:50
>1:25
L. quinlivanii
>1:100
>1:25
L. pneumophila
>1:50
>1:25
L. rubrilucens
>1:200
>1:25
L. worsleiensis
>1:200
>1:25
B. japonicum
>1:100
>1:25
Afipia broomeae
>1:200
>1:25
Bosea eneae
>1:800
>1:25
B. vestrisii
>1:100
>1:25
B. thiooxydans
>1:50
>1:25
Mesorhizobium amorphae
>1:100
>1:25
Azorhizobium caulinodans
>1:100
>1:25
Afipia felis
>1:100
>1:25
A. felis genospecies A
>1:100
>1:25
A. clevelandensis
>1:100
>1:25
A. birgiae
>1:50
>1:25
A. massiliae
>1:50
>1:25
a
Ig, immunoglobulin.

diagnosis based only on isolation may explain why L.
anisa is so rarely reported. However, its absence in the
BAL samples from the 200 patients from other ICUs
shows that local epidemiology plays a major role. This
study confirms that L. anisa is common in the environment
(32,33).
Members of the Bosea genus are gram-negative, oxi-
dase-positive, catalase-positive rods belonging to the α-2
subgroup of Proteobacteria. All are motile. They grow well
on BCYE agar from 25°C to 37°C but do not grow or grow
weakly on Columbia agar with 5% sheep blood. Colonies
are smooth, mucoid, round, and cream colored and are ure-
ase positive and α-hemolytic on Columbia agar with 5%
sheep blood and 0.2% yeast extract. B. massilensis are
negative in assays for arginine dihydrolase activity, esculin
and gelatin hydrolysis, β-galactosidase activity, maltose
assimilation, and acid production by fermentation or oxi-
dation of substrates tested in API 50 CH (Biomérieux,
Marcy l’étoile, France), especially D-glucose, D-fructose,
D-mannose, and sucrose. The species of the Bosea genus
have high MICs to penicillin and amoxicillin and low
MICs to doxycycline (13). In co-culture with A. polypha-
ga, B. vestrisii, B. eneae, and B. massiliensis are phagocyt-
ed and form progressively large vacuoles that lead to
amoebal lysis; however, they have never been reported as
pathogenic agents before. As B. massiliensis has not been
tentatively isolated elsewhere, whether our findings reflect
a local phenomenon or whether the bacterium is widely
encountered is not known. We think that our data support
the role of B. massiliensis in severe VAP, but confirmation
is needed to definitely establish a role.
Our study indicates that most patients with VAP
received β-lactam agents, mainly amoxicillin-clavulanic
acid and third-generation cephalosporin. These antibiotics
may have inhibited bacterial culture, which explains why
no amoeba-associated bacteria were isolated from BAL
samples. B. massiliensis has low MICs (<0.5 mg/L) to cef-
triaxone, doxycycline, rifampin, and erythromycin (13).
However, β-lactam agents are also active in vitro on
Legionella spp., but animal models and clinical studies
have demonstrated their inefficacy in the treatment of
legionellosis (39).
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 819
RESEARCH
Table 5. Antibody titers of 12 serum samples with seroconversion to at least one of the bacteria isolated in the intensive care unit
Bosea massiliensis
Legionella anisa
L. quinlivanii
Afipia broomeae
Case
Wk of sampling
IgG
IgM
IgG
IgM
IgG
IgM
IgG
IgM
1
<1:50
<1:25
4
1:50
1:100
1
a
7
1:800
1:50
2
a
1
<1:50
<1:25
3
1:400
1:100
7
1
<1:50
<1:25
3
<1:50
1:200
8
1
<1:50
<1:25
3
1:200
1:800
9
1
<1:50
<1:25
<1:50
<1:25
3
1:400
1:50
1:50
1:400
5
1:50
1:25
1:800
1:50
11
1
<1:50
<1:25
5
1:50
1:50
12
1
<1:50
<1:25
3
1:400
1:50
13
1
<1:50
<1:25
3
1:50
<1:25
5
1:100
<1:25
7
1:200
<1:25
19
1
<1:50
<1:25
3
1:400
<1:25
22
1
<1:50
<1:25
3
1:200
1:50
23
1
<1:50
<1:25
5
1:400
1:25
28
1
<1:50
<1:25
3
1:400
1:25
5
1:200
1:25
a
Patients with positive PCR results in bronchoalveolar lavage samples (patient 1, L. anisa; Patient 2, B. massiliensis); Ig, immunoglobulin. All patients were sampled at
admission (wk 1); titers in bold are those greater than or equal to defined cutoff titers (Table 4).

The results of our study confirm that the bacteriologic
tests of hospital water supplies is largely ignored. Our work
demonstrates that patients are exposed specifically to the
most common water amoeba–associated bacteria in their
environment, as evidenced by seroconversion against these
bacteria and DNA of these bacteria in BAL samples. The
route of infection, even if caused by aerosols generated in
the ICU, remains unclear. Patients of this ICU sometime
receive water through nasogastric tubes but only bottled,
sterile water. However, we cannot exclude mistakes caused
by not following recommended procedures. Patients for
whom exposure to these bacteria is supported by serocon-
version or DNA detection in BAL samples have unex-
plained VAP and SIRS more commonly. We speculate that
amoeba-associated bacteria in the environment of intubat-
ed patients may concurrently cause unexplained infections
and cryptic outbreaks. Research of new etiologic agents of
pneumonia in ICUs should be based on environmental
study of each ICU since ecologic findings of amoeba-asso-
ciated bacteria in water points in hospital vary.
Acknowledgments
The authors are indebted to Lina Barrassi for technical help
and Kelly Johnston and John Dumler for reviewing the manu-
script.
This work was supported by the Programme Hospitalier de
Recherche Clinique.
Dr. La Scola is associate professor at Marseilles Medicine
Faculty. He is a member of the Unité des Rickettsies (CNRS UMR
6020, World Health Organization reference center for rickettsiae
and rickettsial diseases). His fields of interest are isolation and
description of fastidious bacteria, including Coxiella, Rickettsia,
Bartonella, Tropheryma, and amoeba-associated bacteria.
820 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure. Western blot showing seroconversions in immunoglobulin
(Ig) G (Lanes 1, 3, 5, 7) and IgM (Lanes 2, 4, 6, 8) of patient 2
against Bosea massiliensis (Lanes 1 to 4) and patient 9 against
Legionella anisa (Lanes 5 to 8). Lanes 1, 2, 5, 6: acute-phase
sera; Lanes 3, 4, 7, 8: convalescent-phase sera.
Table 6. Clinical characteristics of patients with or without seroconversion to one of the amoeba-associated bacteria
a
Clinical characteristics
Seroconversion (N=12)
No seroconversion (N=18)
p value
Demographic data
Median age in y (IQR)
35 (25–43)
24 (21–52)
0.85
Male (%)
10 (83.3)
15 (83.3)
1
Risk and potential confounding factors
Underlying disease (%)
2 (16.6)
3 (16.6)
1
Circulation injury (%)
9 (75)
14 (77.8)
1
Median APACHE II
a
score (IQR)
21 (14–4)
23 (16–34)
0.12
Intubation in ICU (%)
8 (66.6)
7 (38.9)
0.26
Median hospitalization days (IQR)
25 (19–41)
17 (10–23)
0.094
Median intubation duration in days (IQR)
11 (7–20)
11 (7–20)
0.8
Median number of serum samples (IQR)
3 (3–5)
3 (2–4)
0.12
Clinical data
VAP (%)
10 (83.3)
7 (38.9)
0.026
Unexplained VAP (%)
6 (50)
2 (11.1)
0.034
Fever > 38.5 °C (%)
12 (100)
17 (84.4)
1
Unexplained fever (%)
7 (58.3)
11 (39.3)
0.27
SIRS (%)
12 (100)
11 (61.1)
0.024
Unexplained SIRS (%)
7 (58.3)
3 (14.3)
0.045
Death (%)
2 (16.7)
8 (44.4)
0.23
Paraclinical data
Leukocytes > 12 g/L (%)
12 (100)
14 (77.8)
0.13
Platelets > 500 g/L (%)
5 (41.7)
6 (33.3)
0.7
PCR detection of ARB in BAL samples(%)
2 (17)
0
0.15
a
IQR, interquartile range; VAP, ventilator-associated pneumonia; SIRS, Systemic Inflammatory Response Syndrome; APACHE II, Acute Physiology and Chronic Health
Evaluation II; PCR, polymerase chain reaction; ARB, angiotensin receptor blockers; BAL, bronchoalveolar lavage samples; bold p values are those that are significant
(<0.05).

References
1. Wenzel RP. Hospital-acquired pneumonia: overview of the current
state of the art for prevention and control. Eur J Clin Microbiol Infect
Dis 1989;8:56–60.
2. Marrie TJ, Durant H, Yates L. Community-acquired pneumonia
requiring hospitalization: 5-year prospective study. Rev Infect Dis
1989;11:586–99.
3. Rutala WA, Weber DJ. Water as a reservoir of nosocomial pathogens.
Infect Control Hosp Epidemiol 1997;18:609–16.
4. Rowbotham TJ. Preliminary report on the pathogenicity of
Legionella pneumophila for freshwater and soil amoebae. J Clin
Pathol 1980;33:1179–83.
5. Michel R, Hauröder B. Isolation of an Acanthamoeba strain with
intracellular Burkholderia pickettii infection. Zentralbl Bakteriol
1997;285:541–57.
6. Barker J, Brown MRW. Trojan horses of the microbial world: proto-
zoa and the survival of bacterial pathogens in the environment.
Microbiology 1994;140:1253–9.
7. Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neofor-
mans interactions with amoebae suggest an explanation for its viru-
lence and intracellular pathogenic strategy in macrophages. Proc Natl
Acad Sci U S A 2001;98:15245–50.
8. Stout JE, Yu VL. Legionellosis. N Engl J Med 1997;337:682–7.
9. Marrie TJ, Raoult D, La Scola B, Birtles RJ, de Carolis E. Legionella-
like and other amoebal pathogens as agents of community-acquired
pneumonia. Emerg Infect Dis 2001;7:1026–9.
10. Greub G, Raoult D. Parachlamydiaceae potential emerging
pathogens. Emerg Infect Dis 2002;8:625–30.
11. La Scola B, Barrassi L, Raoult D. Isolation of new fastidious α
Proteobacteria and Afipia felis from hospital water supplies by direct
plating and amoebal co-culture procedures. FEMS Microbiol Ecol
2000;34:129–37.
12. La Scola B, Mallet MN, Grimont PAD, Raoult D. Description of
Afipia birgiae sp. nov., Afipia massiliae sp. nov. and recognition of
Afipia felis genospecies A. Int J Syst Evol Microbiol
2002;52:1773–82.
13. La Scola B, Mallet MN, Grimont PAD, Raoult D. Bosea eneae sp.
nov., Bosea massiliensis sp. nov. and Bosea vestrisii sp. nov., isolat-
ed from water supplies, and emendation of the genus Bosea (Das
1996). Int J Syst Evol Microbiol. In press 2003.
14. La Scola B, Mezi L, Auffray JP, Berland Y, Raoult D. Intensive care
unit patients are exposed to amoeba associated pathogens. Infect
Control Hosp Epidemiol 2002;23:462–5.
15. Raoult D, Aboudharam G, Crubezy E, Larrouy G, Ludes B, Drancourt
M. Molecular identification by “suicide PCR” of Yersinia pestis as
the agent of Medieval Black Death. Proc Natl Acad Sci U S A
2000;97:12800–3.
16. Raoult D, Fournier PE, Fenollar F, Jensenius M, Prioe T, de Pina JJ,
et al. Rickettsia africae, a tick-borne pathogen in travelers to sub-
Saharan Africa. N Engl J Med 2001;344:1504–10.
17. A’Court CH, Garrard CS, Crook D, Bowler L, Conlon C, Peto T, et
al. Microbiologic lung surveillance in mechanically ventilated
patients using non-directed bronchial lavage and quantitative culture.
Q J Med 1993;86:635–48.
18. Kollef MH, Bock KR, Richards RD, Hearns ML. The safety and
diagnostic accuracy of minibronchoalveolar lavage in patients with
suspected ventilator-associated pneumonia. Ann Intern Med
1995;122:743–8.
19. Sanchez-Nieto JM, Torres A, Garcia-Cordoba F, El Ebiary M, Carillo
A, Ruiz J, et al. Impact of invasive and non-invasive quantitative cul-
ture sampling on outcome of ventilator-associated pneumonia: a pilot
study. Am J Respir Crit Care Med 1998;157:371–6.
20. Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II : a
severity of disease classification system. Crit Care Med
1985;13:818–29.
21. Reisner BS, Woods GL, Thomson RB Jr, Larone DH, Garcia LS, Shimizu
RY. Specimen processing. In: Murray PR, Baron EJ, Pfaller MA, Tenover
FC, Yolken RH, editors. Manual of clinical microbiology. 7th ed.
Washington: American Society for Microbiology; 1999. p. 64–104.
22. Kollef MH. Ventilator-associated pneumonia, a multivariate analysis.
JAMA 1993;270:1965–70.
23. Salata RA, Lederman MM, Shlaes DM, Jacobs MR, Eckstein E,
Tweardy D, et al. Am Rev Respir Dis 1987;135:426–32.
24. American Thoracic Society. Hospital-acquired pneumonia in adults:
diagnosis, assessment of severity, initial antimicrobial therapy, and
preventive strategies. Am J Respir Crit Care Med 1996;153:1711–25.
25. Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et
al. Definitions for sepsis and organ failure and guidelines for the use
of innovative therapies in sepsis. Chest 1992;101:1644–55.
26. La Scola B, Mezi L, Weiller PJ, Raoult D. Isolation of Legionella
anisa using an amoebal coculture procedure. J Clin Microbiol
2001;39:365–6.
27. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D.
16S ribosomal DNA sequence analysis of a large collection of envi-
ronmental and clinical unidentifiable bacterial isolates. J Clin
Microbiol 2000;38:3623–30.
28. Ratcliff RM, Lanser JA, Manning PA, Heuzenroeder MW. Sequence-
based classification scheme for the genus Legionella targeting the
mip gene. J Clin Microbiol 1998;36:1560–7.
29. La Scola B, Rydkina L, Ndihokubwayo JB, Vene S, Raoult D.
Serological differentiation of murine typhus and epidemic typhus
using cross-adsorption and Western blotting. Clin Diagn Lab
Immunol 2000;7:612–6.
30. George DL, Falk PS, Wunderink RG, Leeper KVJ, Meduri GU,
Steere EL, et al. Epidemiology of ventilator-acquired pneumonia
based on protected bronchosopic sampling. Am J Respir Crit Care
Med 1998;158:1839–47.
31. Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-
Chanoin MH, et al. The prevalence of nosocomial infection in inten-
sive care units in Europe. JAMA 1995;274:639–44.
32. Gorman GW, Feeley JC, Steigerwalt A, Edelstein PH, Moss CW,
Brenner DJ. Legionella anisa: a new species of Legionella isolated
from potable waters and a cooling tower. Appl Environ Microbiol
1985;49:305–9.
33. Bornstein N, Vieilly C, Marmet D, Surgot M, Fleurette J. Isolation of
Legionella anisa from a hospital hot water system. Eur J Clin
Microbiol 1985;4:327–30.
34. Fenstersheib MD, Miller M, Diggins C, Liska S, Detwiler L, Werner
SB, et al. Outbreak of Pontiac fever due to Legionella anisa. Lancet
1990;336:35–7.
35. Fields BS, Barbaree JM, Sanden GN, Morrill WE. Virulence of
Legionella anisa strain associated with Pontiac fever: an evaluation
using protozoan, cell culture, and guinea pig models. Infect Immun
1990;58:3139–42.
36. Bornstein N, Mercatello A, Marmet D, Surgot M, Deveaux Y,
Fleurette J. Pleural infection caused by Legionella anisa. J Clin
Microbiol 1989;27:2100–1.
37. Thacker WL, Benson RF, Hawes L, Mayberry WR, Brenner DJ.
Characterization of a Legionella anisa strain isolated from a patient
with pneumonia. J Clin Microbiol 1990;28:122–3.
38. Fallon RJ, Stack BH. Legionnaires disease due to Legionella anisa. J
Infect 1990;20:227–9.
39. Vergis EN, Yu VL. Legionella species. In: Yu VL, Merigan TC Jr,
Barriere SL, editors. Antimicrobial therapy and vaccines. Baltimore
(MD): Williams & Wilkins; 1999. p. 257–72.
Address for correspondence: Didier Raoult, Unité des Rickettsies, CNRS
UMR 6020, Faculté de Médecine, 27 Boulevard Jean Moulin, 13385
Marseille Cedex 05, France; fax: 33(0)4.91.83.03.90; email:
Didier[email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 821
RESEARCH

Municipal and agricultural pollution affects the Rio
Grande, a river that separates the United States from
Mexico. Three hundred and twenty-two Escherichia coli
isolates were examined for multiple antibiotic resistance
phenotypes and the prevalence of class 1 and class 2 inte-
gron sequences. Thirty-two (10%) of the isolates were
resistant to multiple antibiotics. Four (13%) of these iso-
lates contained class 1–specific integron sequences; one
isolate contained class 2 integron–specific sequences.
Sequencing showed that the class 1 integron–bearing
strain contained two distinct gene cassettes, sat-1 and
aadA. Although three of the four class 1 integron–bearing
strains harbored the aadA sequence, none of the strains
was phenotypically resistant to streptomycin. These results
suggest that integron-bearing E. coli strains can be present
in contaminated irrigation canals and that these isolates
may not express these resistance markers.
I
ntegron gene sequences contribute to the spread of
antimicrobial resistance alleles by lateral gene transfer
of gene cassettes in a variety of enteric bacteria, including
Campylobacter spp., Escherichia coli, and Salmonella
enterica serotype Typhimurium (1–4). The gastrointestinal
environment is suspected of serving as a reservoir for inte-
gron-bearing strains; when antimicrobial exposure occurs,
gene transfer events—which spread cassettes between
commensal organisms that are expelled into the environ-
ment (2)—would also occur.
The Rio Grande, the river separating the United States
from Mexico along the Texas-Mexico region, serves as a
source for irrigation water in Texas and Mexico. Previous
studies in our laboratory and others have shown that the
transboundary region is subject to extensive microbial and
chemical contamination. This contamination has been
associated with agricultural, municipal, and industrial
wastes originating from both sides of the border (5,6).
Leaking septic tanks and wastewater effluent discharges
result in fecal contamination levels as high as 2,000
CFU/mL of fecal coliforms (7,8).
Because of the strategic importance of the Rio Grande
for U.S. agriculture and the potential transmission of
antimicrobial resistance determinants by means of food
crops, we investigated the prevalence and characteristics
of class 1 and class 2 integron–bearing E. coli strains.
These strains were previously isolated from a study inves-
tigating fecal contaminants in irrigation water and associ-
ated sediments at specific locations along the river (9).
Methods
Three hundred and twenty-two E. coli isolates were
previously isolated from irrigation water and associated
sediments at the El Paso, Presidio, and Weslaco regions of
the river (9). After being confirmed as E. coli by MUG (4-
methyl umbelliferyl-β-D-glucoronide)–based fluores-
cence, these isolates were screened for antimicrobial sus-
ceptibility by using the agar dilution method (10,11). The
isolates were tested against ampicillin, tetracycline, ceftri-
axone, cephalothin, gentamicin, kanamycin, streptomycin,
chloramphenicol, ciprofloxacin, and trimethoprim/sul-
famethoxazole. The antibiotics were tested at concentra-
tions established by the National Antimicrobial Resistance
System (12).
Isolates that were multidrug resistant (resistant to two
or more antimicrobial agents) were grown overnight in 5
mL of Mueller-Hinton broth (Accumedia, Baltimore, MD)
with the appropriate concentration of antimicrobial com-
pound. A 1-mL aliquot of the culture was centrifuged at
10,000 rpm for 2 min. The cell pellet was resuspended in
500 µL of sterile water and boiled for 10 min. The result-
ing DNA suspension was used as template DNA in poly-
merase chain reaction (PCR) amplification for the class 1
and class 2 integrase gene and variable regions using the
primer sequences shown in the Table (13–15).
The PCR reactions used 10 µL of template DNA, 5 µM
of primers, 25 mM MgCl, 10 mM deoxynucleotide
triphosphate, and 23 ng bovine serum albumin. Nuclease-
free water (Ambion, Austin, TX) was added to achieve a
volume of 50 µL. A “hot start” method was used, and 1.25
U of Taq DNA polymerase (Sigma, St. Louis, MO) was
822 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Antimicrobial Resistance Markers
of Class 1 and Class 2 Integron-
bearing Escherichia coli from
Irrigation Water and Sediments
Matthew T. Roe,* Everardo Vega,* and Suresh D. Pillai*
*Texas A&M University, College Station, Texas, USA

added after initial template denaturation. The PCR cycle
was as follows: initial denaturation for 12 min at 94°C, hot
start pause at 80°C followed by 35 cycles of denaturation
at 94°C for 1 min, primer annealing at 60°C for 1 min, and
extension at 72°C for 5 min at first cycle. An additional 5
s was progressively added to each cycle to reach a final of
7 min, 55 s. PCR products were analyzed on 1% agarose
gel.
Amplification products were extracted from the gels
with the QIAGEN QIAquick gel extraction kit (Valencia,
CA). The amplified products were sequenced at a commer-
cial facility (MWG Biotech Inc., High Point, NC) with the
class 1 and class 2 integron variable region primers (integ
and hep) (Table). Contiguous sequences were created from
single sequence reads by using the CAP3 sequence assem-
bly program (16). Contiguous sequences were analyzed by
using the GenBank database of the National Center for
Biotechnology Information and the BLASTX search
engine (17). Putative gene relationships and sequence data
were analyzed by using a multiple sequence alignment cre-
ated by using Clustal W version 1.82 (18).
Results
Of the 322 E. coli isolates from sediment and irrigation
water samples analyzed for antimicrobial resistance, 104
(32%) isolates showed resistance to at least one of the
antimicrobial compounds (Figure 1). Approximately 10%
(32/322) of all the isolates showed a multidrug resistance
phenotype. Eighteen percent of the isolates were resistant
to cephalothin; however, only 5 (2%) of 322 were resistant
to ceftriaxone, which also belongs to the cephalosporin
family. Resistance to ampicillin was prevalent in approxi-
mately 35 (11%) of the isolates. Resistance to tetracycline
(9%), kanamycin (2%), gentamicin (0.3%), and strepto-
mycin (4%) was also observed. Resistance to the fluoro-
quinolone ciprofloxacin was seen in one isolate. Three
(<1%) of the 322 isolates were resistant to sulfonamide
sulfamethoxazole. On the basis of analysis of variance,
antimicrobial resistance and the sampling location were
correlated. Isolates from the El Paso sampling region had
significantly higher (p<0.05) antimicrobial resistance as
compared with the Presidio and Weslaco sampling regions
(data not shown).
The 32 isolates identified as multiple antimicrobial
resistant were assayed by PCR amplification for class 1
and class 2 integrase genes. Four isolates (approximately
13%) had the class 1 integrase gene intI1 (Figure 2A), and
one isolate had the class 2 integrase gene intI2 (Figure 2B).
Isolates identified as having the class 1 or class 2 integrase
genes were further characterized through PCR amplifica-
tion of the class 1 and class 2 variable regions. Of the four
amplified class 1 integron variable regions, three isolates
(isolate 16, isolate 19, and isolate 21) were approximately
1 kb in size, but the fourth isolate (isolate I-6) harbored a
2-kb fragment (Figure 3A). The 1-kb amplification prod-
ucts were observed in isolates from the El Paso area.
Nucleotide sequencing showed that all of the 1-kb
sequences contained a conserved configuration of a 780-
bp gene cassette identified as the aadA gene (Figure 4).
The 2-kb amplification product was seen in an isolate from
the Presidio sampling region. Nucleotide sequencing
showed that the variable region contained a 498-bp gene
cassette, identified as the dhfrXII gene, which encodes
trimethoprim resistance. The gene cassette did not exhibit
perfect homology with the dhfrXII gene (Figure 4). Within
the identified 498-bp gene cassette, a 323-bp stretch
showed 97% sequence homology; in addition, 59-bp and
56-bp fragments showed 88% and 89% homology, respec-
tively. “Islands” of sequence within the variable region
showed no sequence homology to any known genes.
When the 32 multiply antimicrobial-resistant isolates
were screened for class 2 integrons, only 1 isolate was pos-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 823
RESEARCH
Table. Oligonucleotide primer sequences used for amplification of class 1 and class 2 integrase and variable regions
Primer
Primer sequence
Target
Reference
integ-1
5´-GGCATCCAAGCAAG-3´
5′-Class 1 integron variable region
Levesque et al. 1995 (13)
integ-2
5´-AAGCAGACTTGACCTGA-3´
3´-Class 1 integron variable region
Levesque et al. 1995 (13)
hep51
5´-GATGCCATCGCAAGTACGAG-3´
5´-Class 2 integron variable region
White et al. 2001 (14)
hep74
5´-CGGGATCCCGGACGGCATGCACGATTTGTA-3′
3´-Class 2 integron variable region
White et al. 2001 (14)
intI1.F
5´-GGGTCAAGGATCTGGATTTCG-3´
5´-intI1 gene
Mazel et al. 2000 (15)
intI1.R
5´-ACATGGGTGTAAATCATCGTC-3´
3´-intI1 gene
Mazel et.al. 2000 (15)
intI2.F
5´-CACGGATATGCGACAAAAAGGT-3´
5´-intI2 gene
Mazel et al. 2000 (15)
intI2.R
5´-GTAGCAAACGAGTGACGAAATG-3´
5´-intI2 gene
Mazel et al. 2000 (15)
Figure 1. Frequency of antimicrobial resistance observed in 322
Escherichia coli isolates from irrigation water along the Texas-
Mexico border.
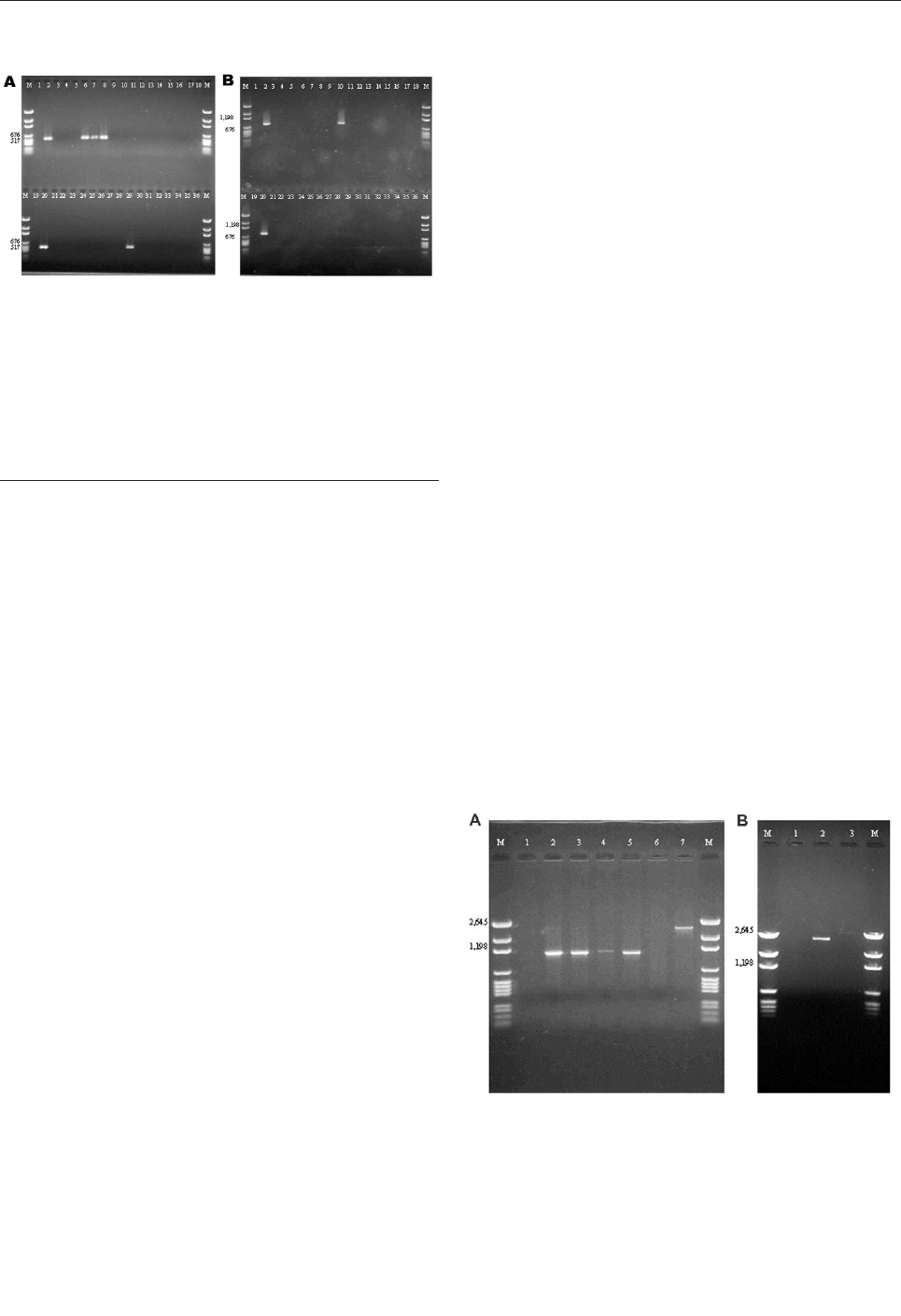
itive (Figure 2B). This particular isolate (isolate 29) was
obtained in the Presidio region and had a 2,600-bp variable
region (Figure 3B). Nucleotide sequencing identified two
distinct gene cassettes, namely, the sat-1 and aadA genes,
which code for streptothricin acetyl transferase, and amino-
glycosede adenyltransferase, respectively (Figure 5).
Discussion
Antimicrobial resistance in human pathogens has
become a major public health issue. Resistant organisms
have been isolated from a number of natural and man-
made environments (6,19,20). In natural environments,
resistant organisms can be indigenous or introduced
through natural or anthropogenic causes (21,22). Integron
gene sequences have been identified as a primary source of
resistance genes and are suspected to serve as reservoirs of
antimicrobial resistance genes within microbial popula-
tions (1,2,23,24). Previous studies along the Texas-Mexico
border have shown that fecal contamination of the Rio
Grande does occur (7,25). The isolation of 322 E. coli iso-
lates from irrigation water and associated sediments fur-
ther confirms that fecal wastes are affecting this body of
water. Previous studies have reported that municipal and
animal wastes regularly harbor multidrug-resistant E. coli
strains (6,26,27). In this study, 18% of the isolates were
resistant to cephalothin. These results are similar to those
from a recent survey of U.S. rivers, which found cefo-
taxime (a third-generation, cephalosporin-resistant, gram-
negative bacterium) to range from 16% to 96% across 22
rivers (19). The higher frequency of isolation of resistant
strains from the El Paso region compared with the other,
less urbanized sampling locations is not surprising since
the effluent from a number of wastewater treatment plants
enters the river at that region (W. McElroy, unpub. data;
28). Previous studies with sludge and septic tank wastes
showed relatively high levels of antimicrobial resistance in
E. coli (6). The precise sources of the E. coli isolates used
in this study could not be identified because of technical
limitations in source tracking (29).
Class 1 and class 2 integron gene sequences were found
within these E. coli isolates. Together, they accounted for
5 (16%) of 32 multidrug-resistant isolates characterized in
this study. This prevalence was higher than that reported
by Rosser et al. (30), who showed that 3.6% of gram-neg-
ative bacteria in an estuarine environment contained the
class 1 integron. Three of the four class 1 integron-bearing
E. coli in this study contained the nucleotide sequence of
the spectinomycin-streptomycin resistance gene aadA1
(31). Resistance to streptomycin was not observed in these
isolates, but resistance to the closely related kanamycin
was seen. These results are similar to those reported by
Zhao et al. (3), who identified that the aadA gene trans-
ferred to a strain of Hafnia alvei but did not report resist-
ance to streptomycin or spectinomycin. These researchers
attributed their findings to the inefficient expression of the
inserted gene cassette by the integron promoter. Previous
studies have also shown that the antimicrobial resistance
phenotype can be modulated once these strains are
exposed to specific environmental conditions (32).
The aadA gene cassette is not novel in class 1 inte-
grons. Earlier work by Zhao et al. (3) and Bass et al. (24)
has shown that the aadA gene is highly conserved among
Shiga toxin–producing and avian clinical E. coli isolates,
respectively. The only class 2 integron-bearing strain iso-
lated in this study also contained the aadA gene in addition
to the sat-1 gene, which codes for resistance to kanamycin,
a finding in agreement with the phenotypic expression.
824 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure 2. Agarose gel electrophoresis of integrase gene poly-
merase chain reaction (PCR) amplification products. A: PCR prod-
ucts of class 1 integrase gene intI1. Lane M; molecular marker;
lanes 1 and 19: no template (negative) control; lanes 2 and 20:
positive control (In2); lanes 3–36: multiple drug–resistant isolates.
B: PCR products of class 2 integrase gene intI2. Lane M: molecu-
lar marker; lanes 1 and 19: no template control (negative) control;
lanes 2 and 20: positive control (Tn7); lanes 3–36: multidrug-
resistant isolates.
Figure 3. A: Polymerase chain reaction (PCR) amplification prod-
ucts with primers targeted against the class 1–specific conserved
sequences. Lane 1: no template control; lane 2: positive control
(In2); lane 3: Escherichia coli isolate 16; lane 4: E . coli isolate 19;
lane 5: E. coli isolate 21; lane 6: blank; lane 7: E. coli isolate I-6.
B: PCR amplification products with primers targeted against the
class 2–specific conserved sequences. Lane 1: no template con-
trol; lane 2: positive control (In2); lane 3: E. coli isolate 29.
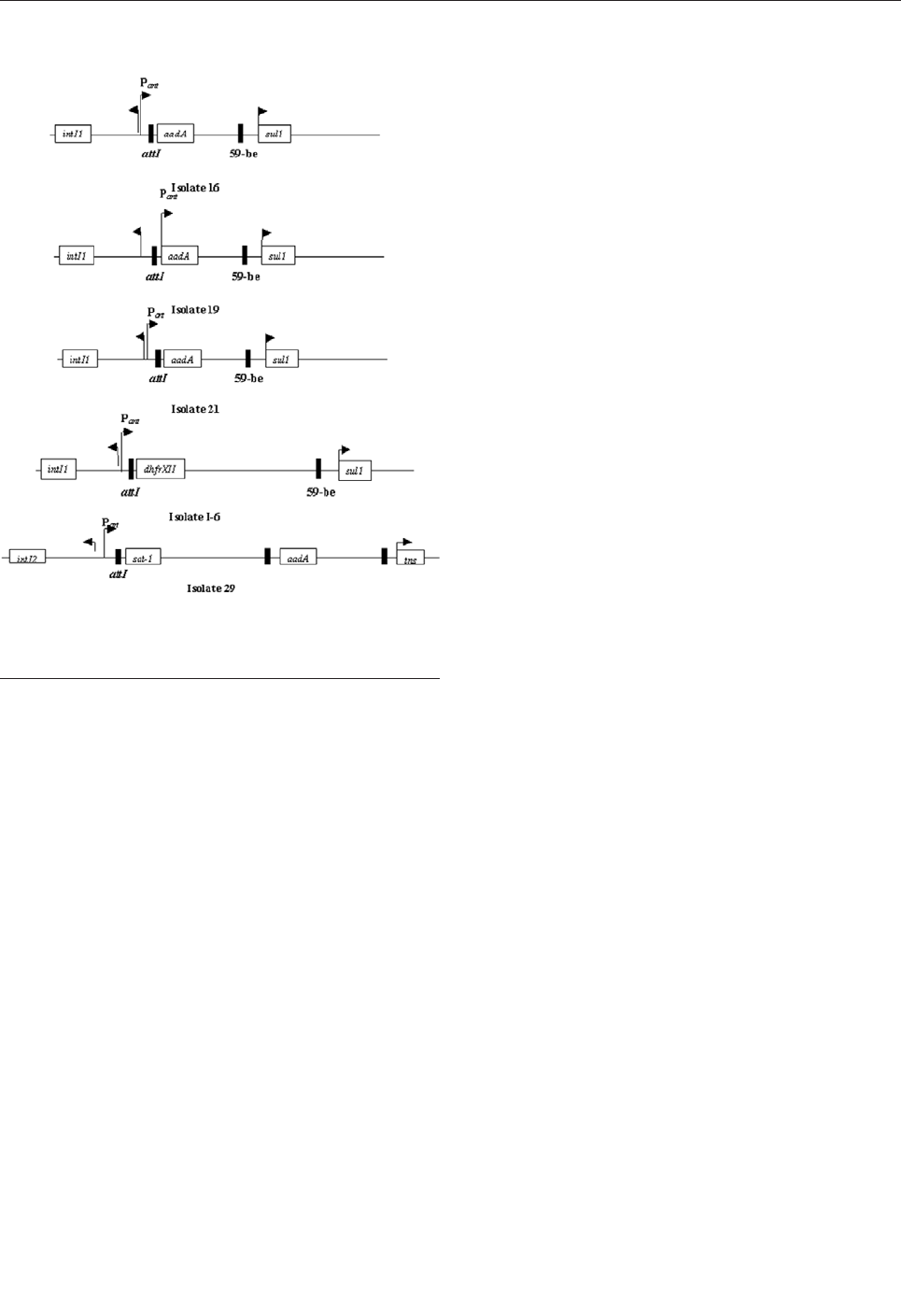
The sat-1 gene, which codes for the streptothricin acetyl
transferase, was not detected in any other E. coli isolate.
The presence of the sat-1 gene cassette, in combination
with the aadA gene, suggests that this class 2 integron is
likely a derivative of the class 2 integron found on transpo-
son Tn7 (33,34).
The aadA gene was conserved among the class 1 and
class 2 integrons, which suggests a possible selective
mechanism for this cassette in enteric bacteria from natu-
ral waters. The 2-kb integron-specific variable region–con-
taining strain, which was isolated from the Presidio area,
harbored the dihydrofolate reductase gene (dhfrXII)
instead of the aadA gene (35).
Overall, these results suggest that the irrigation canals
and sediments associated with the Rio Grande are contam-
inated by bacteria of fecal origin that contain antimicrobial
resistance genes. Of 322 E. coli isolates, 32 (approximate-
ly 10%) were resistant to multiple antimicrobial drugs.
Five of these 32 E. coli isolates harbored class 1 and class
2 integron sequences. This study did not investigate the
possibility that other integron-bearing nonfecal bacteria
were present. The occurrence of integron-bearing E. coli in
irrigation water is important since these organisms are
known fecal contaminants, and the potential for lateral
gene transfer exists. The results also indicate that integron-
bearing strains may not always express the antimicrobial
phenotype; thus, phenotype-based isolation of resistant
organisms can underestimate the levels of resistant organ-
isms. Studies are needed to identify whether integron-
mediated antimicrobial resistance transfer does indeed
occur within the irrigation canal sediments and on veg-
etable surfaces, when they are irrigated with contaminated
irrigation water.
Acknowledgments
We thank Anne Summers and Cynthia Liebert for providing
the integron-positive strains and James Zhu for helpful discus-
sions concerning the sequencing data.
This work was supported by funds from the State of Texas
ATP project 00517-0361-1999, the U.S. Department of Agriculture
(USDA)/CSREES-IFAFS grant 00-52102-9637, the USDA
CSREES grant 2001-34461-10405 and Hatch grant H8708.
Mr. Roe conducted this study while an M.S. student in the
Food Safety and Environmental Microbiology Laboratory in the
Poultry Science Department at Texas A&M University. His
research interests are in environmental microbiology and food
safety.
References
1. Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the
nature of bacterial innovation. Nature 2000;405:299–304.
2. Lucey B, Crowley D, Moloney P, Cryan B, Daly M, O’Halloran F, et
al. Integronlike structures in Campylobacter spp. of human and ani-
mal origin. Emerg Infect Dis 2000;6:50–5.
3. Zhao S, White DG, Ge B, Ayers S, Friedman S, English L, et al.
Identification and characterization of integron-mediated antibiotic
resistance among Shiga toxin–producing Escherichia coli isolates.
Appl Environ Microbiol 2001;67:1558–64.
4. Briggs CE, Fratamico PM. Molecular characterization of an antibiot-
ic resistance gene cluster of Salmonella typhimurium DT104.
Antimicrob Agents Chemother 1999;43:846–9.
5. Mroz RC Jr, Pillai SD. Bacterial populations in the groundwater on
the US-Mexico border in El Paso County, Texas. South Med J
1994;87:214–7.
6. Pillai SD, Widmer KW, Maciorowski KG, Ricke SC. Antibiotic
resistance profiles of Escherichia coli isolated from rural and urban
environments. J Environ Sci Health 1997;32:1665–75.
7. International Boundary and Water Commission. Water Bulletin #63:
Flow of the Rio Grande and related data. El Paso (TX): The
Commission; 1993. p. 132.
8. Scandura JE, Sobsey MD. Viral and bacterial contamination of
groundwater from on-site sewage treatment systems. Water Sci
Technol 1997;35:141–6.
9. Rayburn EL, Sternes KL, Weidenfeld RP, Pillai SD. Microbial
pathogens in irrigation canals and associated sediments. In:
Proceedings of the 101st General Meeting of the American Society
for Microbiology. 2001. Orlando (FL): American Society for
Microbiology; 2001.
10. Food and Drug Administration. Bacteriological analytical manual
online. 8th ed. 2001. Available from: URL: http://www.cfsan.fda.gov/
~ebam/bam-toc.html
11. National Committee for Clinical Laboratory Studies. Methods for
dilution antimicrobial susceptibility tests for bacteria that grow aero-
bically. Vol. 10. Villanova (PA): The Committee; 1999. p. 33.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 825
RESEARCH
Figure 4. Schematic representations of the four class 1 integrons
and one class 2 integron sequenced from multiple
antibiotic–resistant Escherichia coli isolates.

12. Centers for Disease Control and Prevention/Food and Drug
Administration; U.S. Department of Agriculture. National Antibiotic
Resistance Monitoring System. Atlanta: The Centers; 1999.
13. Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons
reveals several novel combinations of resistance genes. Antimicrob
Agents Chemother 1995;39:185–91.
14. White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes
in the enterobacteriaceae. Antimicrob Agents Chemother
2001;45:2658–61.
15. Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in
the ECOR collection: integrons and identification of a novel aad
gene. Antimicrob Agents Chemother 2000;44:1568–74.
16. Huang X, Madan A. CAP3: a DNA sequence assembly program.
Genome Res 1999;9:868–77.
17. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local
alignment search tool. J Mol Biol 1990;215:403–10.
18. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the
sensitivity of progressive multiple sequence alignment through
sequence weighting, position-specific gap penalties and weight
matrix choice. Nucleic Acids Res 1994;22:4673–80.
19. Ash RJ. Antibiotic resistance of gram-negative bacteria in rivers,
United States. Emerg Infect Dis 2002;8:713–6.
20. Goni-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P,
Quentin C, et al. Impact of an urban effluent on antibiotic resistance
of riverine Enterobacteriaceae and Aeromonas spp. Appl Environ
Microbiol 2000;66:125–32.
21. American Academy of Microbiology. Antimicrobial resistance: an
ecological perspective. Washington: American Society for
Microbiology; 1999. p. 1–14.
22. Wegener HC, Aarestrup FM, Jensen LB, Hammerum AM, Bager F.
Use of antimicrobial growth promoters in food animals and
Enterococcus faecium resistance to therapeutic antimicrobial drugs in
Europe. Emerg Infect Dis 1999;5:329–35.
23. Hall RM, Collis CM. Antibiotic resistance in gram-negative bacteria:
the role of gene cassettes and integrons. Drug Resist Updat
1998;1:109–19.
24. Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG,
et al. Incidence and characterization of integrons, genetic elements
mediating multiple-drug resistance, in avian Escherichia coli.
Antimicrob Agents Chemother 1999;43:2925–9.
25. Cech I, Essman A. Water sanitation practices on the Texas-Mexico
border: implications for physicians on both sides. South Med J
1992;85:1053–64.
26. McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in
animals. Clin Infect Dis 2002;34(Suppl 3):S93–106.
27. Schwarz S, Chaslus-Dancla E. Use of antimicrobials in veterinary
medicine and mechanisms of resistance. Vet Res 2001;32:201–25.
28. Texas Water Development Board, New Mexico Water Resources
Research Institute. Transboundary aquifers of the El Paso/Ciudad
Juarez/Las Cruces region. El Paso (TX): The Institute; 1997. p. 152.
29. Simpson JM, Santo Domingo JW, Reasoner DJ. Microbial source
tracking: state of the science. Environ Sci Technol 2002;36:5279–88.
30. Rosser SJ, Young HK. Identification and characterization of class 1
integrons in bacteria from an aquatic environment. J Antimicrob
Chemother 1999;44:11–8.
31. Sundstrom L, Radstrom P, Swedberg G, Skold O. Site-specific recom-
bination promotes linkage between trimethoprim- and sulfonamide-
resistance genes. Sequence characterization of dhfrV and sulI and a
recombination active locus of Tn21. Mol Gen Genet
1988;213:191–201.
32. Pillai SD, Pepper IL. Transposon Tn5 as an identifiable marker in
Rhizobia: survival and genetic stability of Tn5 mutant bean Rhizobia
under temperature stressed conditions in desert soils. Microb Ecol
1990;21:21–33.
33. Sundstrom L, Roy PH, Skold O. Site-specific insertion of three struc-
tural gene cassettes in transposon Tn7. J Bacteriol 1991;173:3025–8.
34. Hansson K, Sundstrom L, Pelletier A, Roy PH. IntI2 integron inte-
grase in Tn7. J Bacteriol 2002;184:1712–21.
35. Huovinen P, Sundstrom L, Swedberg G, Skold O. Trimethoprim and
sulfonamide resistance. Antimicrob Agents Chemother
1995;39:279–89.
Address for correspondence: Suresh D. Pillai, Department of Poultry
Science, 418D Kleberg Center, MS 2472 TAMUS, Texas A&M
University, College Station, Texas 77843-2472, USA; fax: 979-845-1921;
email: spillai@poultry.tamu.edu
826 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
OPPORTUNITIES FOR PEER REVIEWERS
The editors of Emerging Infectious Diseases seek to increase
the roster of reviewers for manuscripts submitted by authors all
over the world for publication in the journal. If you are interested
in reviewing articles on emerging infectious disease topics, please
e-mail your name, address, curriculum vitae, and areas of expert-
ise to [email protected]
At Emerging Infectious Diseases, we always request reviewers’
consent before sending manuscripts, limit review requests to three
or four per year, and allow 2-4 weeks for completion of reviews.
We consider reviewers invaluable in the process of selecting and
publishing high-quality scientific articles and acknowledge their
contributions in the journal once a year.
Even though it brings no financial compensation, participation
in the peer-review process is not without rewards. Manuscript
review provides scientists at all stages of their career opportunities
for professional growth by familiarizing them with research trends
and the latest work in the field of infectious diseases and by
improving their own skills for presenting scientific information
through constructive criticism of those of their peers. To view the
spectrum of articles we publish, information for authors, and our
extensive style guide, visit the journal web site at
www.cdc.gov/eid.
For more information on participating in the peer-review
process of Emerging Infectious Diseases, e-mail
[email protected] or call the journal office at 404-371-5329.

An epidemiologic and seroprevalence survey was con-
ducted (n=830) to assess the proportion of persons
exposed to hantavirus in IX Region Chile, which accounts
for 25% of reported cases of hantavirus cardiopulmonary
syndrome. This region has three geographic areas with dif-
ferent disease incidences and a high proportion of aborigi-
nals. Serum samples were tested for immunoglobulin (Ig) G
antibodies by enzyme-linked immunosorbent assay against
Sin Nombre virus N antigen by strip immunoblot assay
against Sin Nombre, Puumala, Río Mamoré, and Seoul N
antigens. Samples from six patients were positive for IgG
antibodies reactive with Andes virus; all patients lived in the
Andes Mountains. Foresting was also associated with
seropositivity; but not sex, age, race, rodent exposure, or
farming activities. Exposure to hantavirus varies in different
communities of IX Region. Absence of history of pneumo-
nia or hospital admission in persons with specific IgG anti-
bodies suggests that infection is clinically inapparent.
H
antaviruses, RNA-containing viruses, compose a
genus within the family Bunyaviridae. The natural
reservoirs of the pathogenic New World hantaviruses are
rodents of the family Muridae, subfamily Sigmodontinae,
in which a chronic and asymptomatic infection develops
(1). Hantavirus is a zoonosis transmitted from rodents to
humans by inhaling contaminated aerosols from feces,
urine, and saliva of infected mice (2).
Human infection with hantaviruses have been associat-
ed with two diseases. One is the hemorrhagic fever with a
renal syndrome (HFRS) caused by Hantaan, Puumala,
Seoul, and Dobrava/Belgrade viruses, first recognized dur-
ing the Korean War in 1950 to 1953. HFRS occurs mainly
in Asia and Europe; death rates range from 0.1% to 15%
(3,4). The other disease, a severe respiratory illness known
as hantavirus cardiopulmonary syndrome (HCPS), occurs
in the Americas and has a death rate of 40% (1,5–7). We
prefer the term hantavirus cardiopulmonary syndrome to
an alternative term, hantavirus pulmonary syndrome,
because most deaths associated with HCPS are related to
cardiac failure rather than pulmonary failure, and this
aspect of the syndrome remains underappreciated by prac-
titioners and others.
HCPS has been identified in several countries in North
and South America and is caused by different hantavirus-
es: Sin Nombre in North America, Juquitiba virus in Brazil
(8), Laguna Negra virus in Paraguay and Bolivia (9,10),
Andes in Argentina and Chile (11,12), Choclo virus in
Panama (13), and several subspecies or viral genotypes of
Andes virus in Argentina (e.g., Oran, Lechiguanas, et al.)
(14–16).
In 1995, Andes virus was first identified in the
Argentinean Patagonia and was recognized in central and
southern regions of Chile. The reservoir is the long-tailed
pygmy rice rat (Oligoryzomys longicaudatus), a species
that occurs primarily in temperate forest (17).
Since HCPS emerged in Chile, 287 cases have been
confirmed as of March 2003, causing a substantial impact
on the public health system; the death rate for HCPS in
Chile has exceeded 40%. In addition to the 287 cases of
HCPS, 17 cases of mild infection with no cardiopulmonary
involvement have been reported, demonstrating that han-
tavirus infection may have variability in its expression.
Serologic studies have established both clinically asymp-
tomatic infections and symptomatic infections not recog-
nized at the time as hantavirus infection (18–23).
The phenomenon of clinically nonapparent infections
varies in different areas and populations of the Americas.
In the United States, the proportion of infection versus dis-
ease is thought to be close to 1% (i.e., disease develops in
most of the infected patients, and human population sero-
prevalence varies between 0.2% to 1.7%) (4,24,25).
Seroprevalence studies in South America, however, have
shown that some populations have had much more fre-
quent exposure to hantaviruses in the absence of known
clinical manifestations, as seen in some populations native
to Paraguay and Northern Argentina (40% and 17%,
respectively) (26), the general population in Aysen,
Southern Chile (2.0% and 13.1% in urban and rural areas,
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 827
RESEARCH
Hantavirus Prevalence in the
IX Region of Chile
Marlis Täger Frey,*† Pablo C. Vial,*‡ Constanza H. Castillo,§ Paula M. Godoy,*
Brian Hjelle,¶ and Marcela G. Ferrés*
*Universidad Católica de Chile, Santiago, Chile; †Universidad
Austral de Chile, Valdivia, Chile; ‡Universidad del Desarrollo,
Santiago, Chile; §Universidad de La Frontera, Temuco, Chile; and
¶University of New Mexico School of Medicine, Albuquerque, New
Mexico, USA
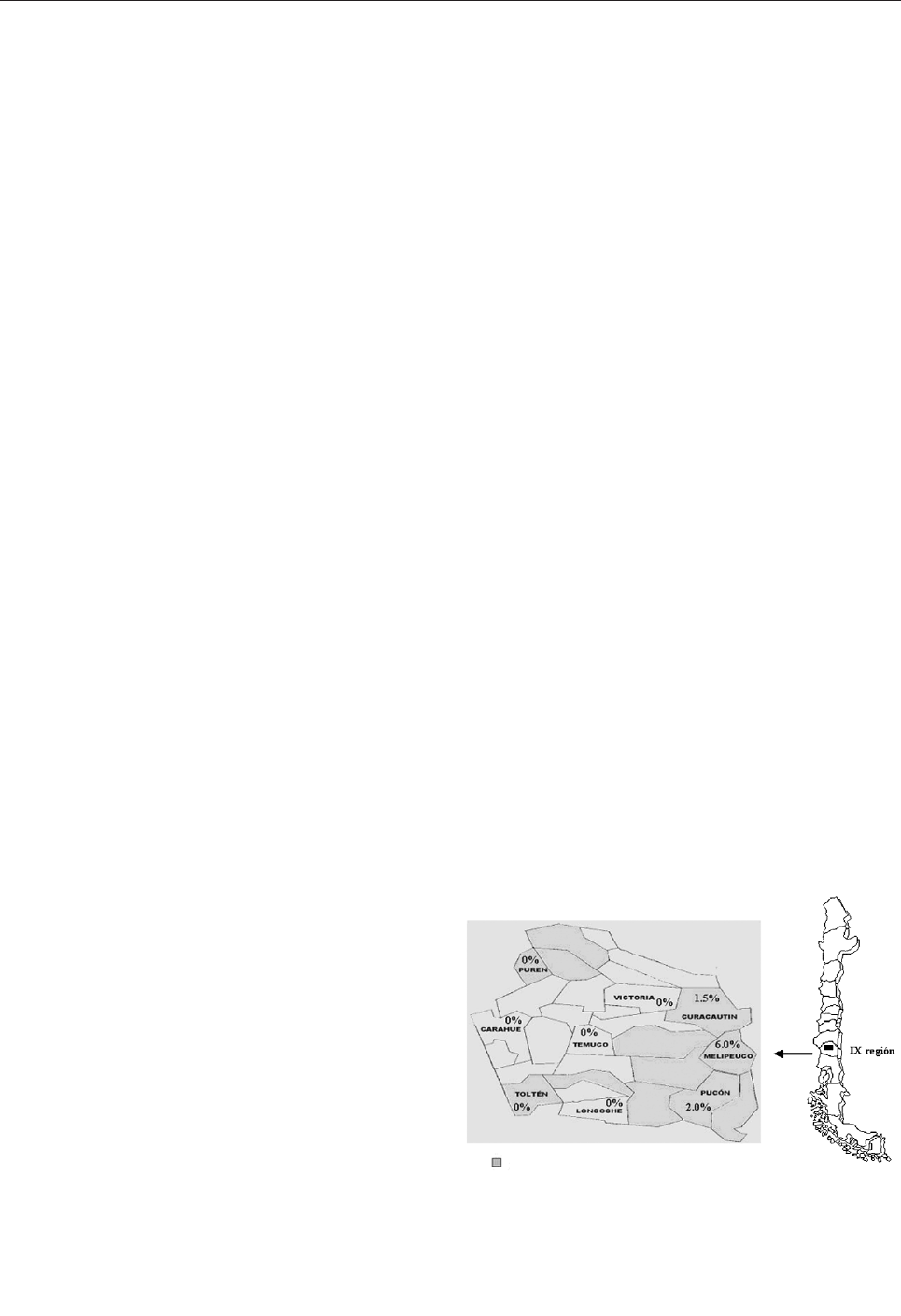
respectively) (27), São Paulo and Bahia, Brazil (1.23 %
and 13.1%, respectively) (28,29), and recently in Panama
(30%) (13). The populations with the highest seropreva-
lence were indigenous persons rather than those of
European ancestry.
Two hypotheses have been proposed to explain these
phenomena. The first one presumes that strains in South
America are of lesser pathogenicity, though in many cases
nonpathogenic viruses have yet to be detected in local
rodent populations. The second hypothesis involves the
existence of at least two variables: the nature of the expo-
sure and genetic constitution of the host population. The
high seroprevalence seen in Paraguay (26) could be caused
by a higher resistance in the aboriginal population and
greater exposure to the virus in some regions. The preva-
lence and high case-fatality ratios seen in North America
might be the result of a lower exposure, a lesser genetic
resistance to disease, or both.
More than 20 strains of hantaviruses are known world-
wide; not all of them are associated with human disease.
The observations discussed in this article suggest the need
to study whether hantaviruses of lesser pathogenicity and
other clinical entities until now unrecognized may occur in
areas where the seroprevalence is high and disease inci-
dence is low (11,14–16,18). For example, hantaviruses
previously thought to be endemic only in Europe and Asia
have also been recognized in the North American continent
(30–32).
To study the seroprevalence of hantavirus in Chile, we
studied the general population in the IX Region for the rea-
sons that follow. This area is ranked third among those
affected by HCPS, accounting for 25% of the known cases
in Chile. Three different areas can be distinguished on the
basis of geographic features in relation to the two moun-
tain ranges that traverse the region (the Pacific Coast and
the coastal range [Coastal], the Central Valley [Central],
and the pre-Andean Region [Andean]). The Mapuche, the
main native ethnic population of Chile, account for 26.3%
of the total regional population. Members of the tribe can
readily be identified from surnames, which are derived
from both parents. Therefore, we were able to study the
seroprevalence of hantavirus in the same region but in
areas having different incidence rates of the disease and
evaluate the asymptomatic infection in the Mapuche abo-
riginal population.
Methods
Study Design and Area
We conducted a cross-sectional epidemiologic and
serologic survey to determine the prevalence of IgG anti-
bodies against hantavirus N antigen in the adults living in
nine communities of the IX Region of Chile. The IX
Region of Chile is located in the southern part of the coun-
try, between parallel 37° and 40° South and the meridians
70° and 74° East (Figure). The region has a surface area of
31,842.3 km
2
and 781,242 inhabitants in 31 communities;
of the total population, 38.71% is rural. The characteristics
of these populations can be distinguished according to geo-
graphic features. The Mountainside Andes range (Andean)
is rural area with small towns; the population there is pri-
marily sustained with farming, woodcutting, and tourist
services. The population composes 13.8% of the region’s
total population, yet accounts for 82% of HCPS cases. The
Central Valley intermediate depression (Central) is a pre-
dominantly urban area in which the main sources of
income are industry, farming, and ranching. Temuco, the
capital and main urban center, is located in this area. The
Central Valley contains 67.3% of the total population and
accounts for 5.9% of HCPS cases. Coastal inhabitants
account for 19% of the region’s population; the communi-
ty sustains itself with farming and forestry activities, fish-
ing, and tourism. Twelve percent of HCPS cases originate
in this area.
Sample Design
The sample included 847 persons, enough to give us
accurate point seroprevalence given estimated seropreva-
lence rates of 7%, 3%, and 5% for the Andean, Central,
and Coastal areas, respectively, as derived from previous
studies in the country (27) and the distribution of HCPS
cases in the region. The sample was designed to be repre-
sentative of each community according to information
available from the National Statistics Agency about popu-
lation sex, ethnicity, age distribution, and rural-urban pro-
portion. Persons were contacted in their homes with a pre-
determined plan that included the selection of the housing
blocks to be studied, the home at which to start enrollment,
and how to proceed thereafter. Only one person was
828 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure. Geographic IX Region, Chile, and hantavirus seropreva-
lence in the tested communities of the IX Region. HCPS,
Hantavirus cardiopulmonary syndrome.
HCPS

enrolled in each household contacted. That person was
informed of the study, signed an informed consent, and
agreed to answer a questionnaire and donate a sample of
blood. Only persons >
15 years of age who were free of any
current febrile respiratory illness were asked to enroll. The
epidemiologic questionnaire, previously validated in the
community, requested information about sex, age, race,
residency, labor activity, rodent exposure at home or work,
cardiorespiratory disease history, hospital admissions, and
direct contact with HCPS patients.
Nine communities were included, three per geographic
area. Curacautín, Melipeuco, and Pucón (Figure) were
chosen as representative of the Andean area (n=277);
Victoria, Temuco, and Loncoche from the Central area
(n=279); and Purén, Carahue, and Toltén from the Coastal
area (n=291). The Andean and Coastal areas were consid-
ered rural; the Central area was considered to be a compos-
ite of rural and urban. Persons were classified as Mapuches
when they had at least one surname of Mapuche deriva-
tion. The study was approved by the Catholic University
Ethics Committee.
Collection, Processing, and Analysis of Samples
Blood samples were collected, transported, and cen-
trifuged the same day. Serum samples were kept frozen at
–20°C and sent for analysis to the Laboratory of Virology
at the Pontificia Universidad Católica de Chile, Santiago,
Chile. An enzyme-linked immunosorbent assay (ELISA)
was performed to detect hantavirus-specific immunoglob-
ulin (Ig) G antibodies in all samples.
ELISA was performed as described (33,34) by using
recombinant antigen for the Sin Nombre strain (produced
in Escherichia coli and provided by Centers for Disease
Control and Prevention [CDC]), which crossreacts with all
American hantaviruses. Briefly, serial dilutions (1:100,
1:400, 1:1,600, and 1:6,400) of patient serum samples
were incubated for 1 h at 37°C in antigen-coated, 96-well
plates. Peroxidase-coupled anti-human IgG was used as
secondary antibody and incubated 1 h at 37°C. After sub-
strate reaction, plates were read at 414 nm.
The net absorbance values are results of the hemiplates
absorbance subtraction with and without antigen. To be
considered positive, the net absorbance values for a sam-
ple had to be >0.2 in the 1:100 and 1:400 dilutions, and the
sum of all net absorbance values had to be >0.95. Positive
serum samples were tested twice and verified to be posi-
tive at CDC.
A confirmatory strip immunoblot assay (SIA) was per-
formed an all positive samples (B.H.). Four recombinant N
antigens from Sin Nombre, Puumala, Rio Mamoré, and
Seoul hantaviruses were fixed onto a nitrocellulose mem-
brane by vacuum. Serum samples (1:200) were incubated
with the strips and alkaline phosphatase-coupled anti-
human IgG added. Reactivity was estimated visually, and
each band given an intensity value on a four-point scale, as
previously described (35,36). We regarded a sample as
positive for antibodies when reactive to both Sin Nombre
and Rio Mamoré on the basis of the presumption that anti-
bodies to Andes virus will always react to closely related
Rio Mamoré and generally will crossreact with the other
sigmodontine rodent-borne virus, Sin Nombre. A sample
was regarded as confirmed seropositive when the serum
was reactive in the ELISA and confirmed by the SIA.
Fisher exact test was used for the analysis of independent
variables. A p value <0.05 was considered significant.
Results
Sample Population Characteristics
A total of 829 persons were included in the study, with
271, 272, and 286 from the Andean, Central, and Coastal,
respectively. The age range was 15–88 years of age (mean
39.4 years of age); 47.6% were men, and 18.3% were of
Mapuche origin. In all, 73.3% of the sampled population
was considered rural. Men worked in farming, forestry, or
both in 61.8% of the cases; 62.2% of the women were
housewives. A history of exposure to rodents, their excre-
ta, or both at home was reported in 88.0% of enrolled per-
sons; 54.6% reported exposure at work.
Serology
Six (0.72%) samples were hantavirus antibody-positive
by both ELISA and SIA. The samples came from persons
who lived in the Andean area, giving a seroprevalence of
2.15% (6/271) for this area, significantly higher than the
other two regions studied (p=0.0001). The seropositive
cases belonged to each of the three counties studied; how-
ever, relative frequencies varied: Curacautín, 2.6%
(2/132); Melipeuco, 6.1% (2/33); and Pucón, 2.0%
(2/100). All three counties have previously had HCPS
cases reported (Figure). As shown in Table 1, four case-
patients were male, and two were Mapuche. The case-
patients ranged from 28 to 76 years of age, with a mean of
52.8 years of age. Seropositivity could not be associated
with sex (p=0.43), race (p=0.30), or age (p=0.18). Five of
the case-patients worked in farming or forestry, although
only forestry had a significant association with seroposi-
tivity (p=0.018), meaning the risk for infection was 10
times higher (relative risk 9.72; confidence interval 1.15 to
82.44). Five of the case-patients said they had been
exposed to rodents or their excreta either at home or work.
This exposure, however, did not reach statistical signifi-
cance (p=0.1 and p=1.0, respectively). The sixth case-
patient was not exposed to rodents; however, he had been
working in a large shed, the woods, and a sawmill; he also
had been weeding. Finally, none of the six antibody-posi-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 829
RESEARCH

tive persons had previous contact with HCPS patients, his-
tory of pneumonia, or hospital admission.
Discussion
This study shows serologic evidence of past infection
by hantavirus in the Chilean population. The hantavirus
associated with symptomatic infections in Chile is Andes
virus, identified in patient blood samples by reverse tran-
scription-polymerase chain reaction (RT-PCR) and recent-
ly isolated from human blood (12,37).
Our results show the global seroprevalence to han-
tavirus in the adult general population in the IX Region to
be 0.72% (ranging from 0 to 6%, depending on the com-
munity). Seroprevalence is significantly higher in the
Andes rural area, consistent with the observed elevated
incidence of HCPS disease in this area of Region IX
between 1997 and 2000 (Table 2). This geographic distri-
bution is probably related to the great tracts of native for-
est, where Chusquea quila, a bamboo-like shrub that pro-
tects and feeds the carrier rodent, is abundant. Moreover,
the increasing development of the forestry industry plus an
increase in the rodent population caused by the favorable
climatic condition because of the El Niño effect have
caused humans and mice to interact closely.
We did not find an association between seroprevalence
and reported exposure to rodents, probably because this
type of exposure is frequent in all groups in the region,
occurring both at work and at home. The exposure to
rodents or their excreta is necessary but apparently not suf-
ficient to acquire the infection. Presumably an exposure to
a specific reservoir mouse or virus carrier, human behav-
ior, and biologic factors are important.
Forestry is associated with a higher risk for infection.
This labor, whether in the woods or at sawmills, is per-
formed almost exclusively by men; both men and women
share farm work in most instances. This finding may
explain why a higher proportion of HCPS develops in men
(75% of reported cases). However, any apparent associa-
tion between hantavirus exposure and a particular occupa-
tion does not necessarily implicate these occupations as a
risk factor. In fact, occupation may be a marker for living,
sleeping, or housing conditions that constitute the proxi-
mate risk factor for exposure. Indoor exposure to rodents
is common in patients with HCPS (38).
On the basis of these results, we argue that infections by
hantavirus follow a gradient of exposure to the virus in the
830 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 1. Relationship between independent variables and
seropositivity to hantaviruses, IX Region, Chile
Variables
Seropositive
(total)
p value
RR (95% CI)
a
Sex
Male
4 (395)
0.43
Female
2 (435)
Age
15–44 y of age
2 (550)
0.18
>44 y of age
4 (280)
Race
Mapuche
2 (151)
0.30
Other
4 (679)
Area
Andean
6 (271)
0.0001
a
NA
b
Central
0 (273)
Coastal
0 (286)
Risk activity or labor
Agrarian
c
No activity
1 (271)
Two or more
4 (341)
0.38
Foresting
d
No activity
1 (279)
With activity
5 (135)
0.018
9.72
(1.15 to 82.44)
Exposure to rodents
Peridomestic
5 (731)
0.10
Laboral
5 (453)
1.00
a
p <0.05; p values were determined by using 2-tailed Fisher exact test; RR,
relative risk; CI, confidence interval.
b
Not applicable.
c
Clearing, shrubbery cutting, working in pastures or cellars.
d
Foresting, working at a sawmill.
Table 2. Incidence of hantavirus cardiopulmonary syndrome (HCPS) and seroprevalence in Chile’s IX Region according to geographic area
Region
Global
Andean (A)
Central (B)
b
Coastal
p value
a
(A vs. B)
RR (95% CI)
a
Disease
Cases
34
29
1
4
Population
781,242
127,974
424,278
229,190
0.0001
29.6 (11.46 to 76.47)
Cumulative Incidence
(1997-2001) (x10
6
)
c
4.35
22.66
0.23
1.74
Infection
Seropositive
6
6
0
0
Sample
829
271
272
286
Prevalence (%)
0.72
2.15
0
0
0.00013
NA
d
a
p values were determined using 2-tailed Fisher exact test; RR, relative risk; CI, confidence interval.
b
B is Central and Coastal regions combined.
c
Chile’s Department of Health, March 2001.
d
Not applicable.

IX Region. Our population has a different epidemiologic
profile to those of the aborigines of Paraguay (40%) and
North Argentina (17%) in South America, which have low
seroprevalence, similar to that described for North
American populations, as reported by Vincent (13) and
Ferrer (26). This lower seroprevalence could be because of
a greater pathogenicity and clinical severity in infections
by the prototypical (southern) form of Andes virus, similar
to Sin Nombre virus.
Less pathogenic hantaviruses may cause a greater
amount of asymptomatic infections, as seen for HFRS in
Europe and Asia (seroprevalence 7.9% to 10%, death rate,
0.1% to 15%) (39) and some American hantaviruses
(Laguna Negra in Bolivia and Paraguay, Choclo in
Panama, Oran and Lechiguanas in Argentina) for which
seroprevalence is high and case-fatality ratios are <30%
(9,13,15). This finding is in contrast to the findings of han-
taviruses with severe clinical syndromes and high death
rate (i.e., Sin Nombre and Andes viruses, both of which
have been associated with few subclinical infections). The
lack of subclinical infections can be caused by a variation
in virulence or by different genotypes of the hosts, which
give them greater resistance to infection and disease.
A slightly disproportionate fraction of seropositive
samples (33%) were from Mapuche (18.3% of those sam-
pled were Mapuche). Moreover, when both hantavirus
seroprevalence and the HCPS incidence rates in this
region are considered, more infection but less disease (not
significant) is found in the Mapuche population (Table 3).
Future epidemiologic studies should address this finding
and use a larger sample to evaluate possible associations
between racial origin and the incidence rates of infection
and disease.
Acknowledgments
We thank Thomas G. Ksiazek for thoughtful technical assis-
tance in hantavirus serologic analysis, Jovita Mardones and
Ligia Sanhueza for providing assistance in collection and pro-
cessing of samples, Gustavo Jimenez for helping to critically
revise the manuscript, and Julio Valdivia for providing statistical
analysis.
Funding was provided by Child Health Foundation,
Alabama, Birmingham and a grant from Universidad de la
Frontera, Temuco, Chile and was supported in part by U.S. Public
Health Service grants U19 AI45452 and D43 TW01133.
Dr. Täger Frey is an assistant professor in the Pediatrics
Department at Universidad Austral de Chile. Her interests
include infectious diseases.
References
1. Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem.
Emerg Infect Dis 1997;3:95–104.
2. Wells R, Young J, Williams R, Armstrong L, Busico K, Khan A, et al.
Hantavirus transmission in the United States. Emerg Infect Dis
1997;3:361–5.
3. Smadel JE. Epidemic hemorrhagic fever. Am J Public Health
1953;43:1327–30.
4. McCaughey C, Hart C. Hantaviruses. J Med Microbiol
2000;49:587–99.
5. Duchin J, Koster F, Peters C, Simpson G, Tempest B, Zaki S, et al.
Hantavirus pulmonary syndrome: a clinical description of 17 patients
with a newly recognized disease. N Engl J Med 1994;330:949–55.
6. Khan A, Khabbaz R, Armstrong L, Holman R, Bauer S, Graber J, et
al. Hantavirus pulmonary syndrome: the first 100 U.S. cases. J Infect
Dis 1996;173:1297–303.
7. Doyle T, Bryan R, Peters C. Viral hemorrhagic fevers and hantavirus
infections in the Americas. Infect Dis Clin North Am
1998;12:95–110.
8. Iversson LB, Travassos da Rosa APA, Rosa MBD, Lomar AV, Saski
M, LeDuc JW. Infeccao humana por Hantavirus no sul e sudeste do
Brasil. Rev Assoc Med Bras 1994;40:85–92.
9. Williams R, Bryan R, Mills J, Palma R, Vera I, de Velásquez F, et al.
An outbreak of hantavirus pulmonary syndrome in western Paraguay.
Am J Trop Med Hyg 1997;57:274–82.
10. Johnson A, Bowen M, Ksiasek T, Williams R, Bryan R, Mills J, et al.
Laguna Negra virus associated with HPS in western Paraguay and
Bolivia. Virology 1997;238:115–27.
11. López N, Padula P, Rossi C, Miguel S, Edelstein A, Ramírez E, et al.
Genetic characterization and phylogeny of Andes virus and variants
from Argentina and Chile. Virus Res 1997;50:77–84.
12. Toro J, Vega J, Khan A, Mills J, Padula P, Terry W, et al. An outbreak
of hantavirus pulmonary syndrome, Chile, 1997. Emerg Infect Dis
1998;4:667–702.
13. Vincent M, Quiroz E, Gracia F, Sánchez A, Ksiazek T, Kitsutani P, et
al. Hantavirus pulmonary syndrome in Panama: identification of
novel hantaviruses and their likely reservoirs. Virology
2000;277:14–9.
14. López N, Padula P, Rossi C, Lazaro ME, France-Fernández MT.
Genetic identification of a new hantavirus causing severe pulmonary
syndrome in Argentina. Virology 1996;220:223–6.
15. Lewis S, Rowe J, Morzunov S, Henria D, St. Jeor S. New hantavirus-
es causing pulmonary syndrome in central Argentina. Lancet
1997;349:998–9.
16. López N, Padula P, Rossi C, Miguel S, Edelstein A, Ramirez E, et al.
Genetic characterization and phylogeny of Andes virus and variants
from Argentina and Chile. Virus Res 1997;50:77–84.
17. Murua R. Ecología de los reservorios silvestres de hantavirus en
Chile. Rev Chil Infect 1998;15:79–83.
18. Levis S, Morzunov S, Rowe J, Enria D, Pini N, Calderon G, et al.
Genetic diversity and epidemiology of hantaviruses in Argentina. J
Infect Dis 1998;177:529–38.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 831
RESEARCH
Table 3. Incidence of hantavirus cardiopulmonary syndrome (HCPS) and seroprevalence in Chile’s IX Region according to race
Seroprevalence
Incidence
HCPS cases/population
Race
Positive/total (%)
RR (95% CI)
a
(rate x 100.000)
RR (95% CI)
a
Mapuche
2/151 (1.3)
2.23 (0.41 to 12.06)
4/205,466 (1.9)
0.37 (0.13 to 1.06)
Other
4/678 (0.6)
–
30/575,776 (5.2)
a
Relative risk (95% confidence interval). p values were determined by using two-tailed Fisher exact test; RR, relative risk; CI, confidence interval.

19. Kitsutani P, Denton R, Fritz C, Murray R, Todd R, Pape J, et al. Acute
Sin Nombre hantavirus infection without pulmonary syndrome,
United States. Emerg Infect Dis 1999;5:701–5.
20. Zavasky D, Hjelle B, Peterson M, Denton R, Reimer R. Acute infec-
tion with Sin Nombre hantavirus without pulmonary edema. Clin
Infect Dis 1999;29:664–6.
21. Weissenbacher M, Cura E, Segura E, Hortal M, Baek L, Chu Y, et al.
Serological evidence of human hantavirus infection in Argentina,
Bolivia and Uruguay. Medicina 1996;56:17–22.
22. Baró M, Vergara J, Navarrete M. Hantavirus in Chile. A review and
case analysis since 1975. Rev Méd Chile 1999;127:1513–23.
23. Parisi M, Enria D, Pini N, Sabattini M. Detección retrospectiva de
infecciones clínicas por hantavirus en la Argentina [in Spanish].
Medicina 1996;56:1–13.
24. Vitek CH, Breiman RF, Ksiazek T, Rollin P, McLaughlin J, Umland
E, et al. Evidence against person-to-person transmission of hantavirus
to health care workers. Clin Infect Dis 1996;22:824–6.
25. Zeitz PS, Butler JC, Cheek JE, Samuel MC, Childs JE, Shands LA, et
al. A case-control study of hantavirus pulmonary syndrome during an
outbreak in the southwestern United States. J Infect Dis
1995;171:864–70.
26. Ferrer J, Jonsson C, Esteban E, Galligan D, Basombrio M, Peralta-
Ramos M, et al. High prevalence of hantavirus infection in Indian
communities of the Paraguayan and Argentinean Gran Chaco. Am J
Trop Med Hyg 1998;59:438–44.
27. Valderrama R, Vega J, Terry W, Aguilar G, Gallegos N, Escobar P, et
al. Community serological survey of infection by hantavirus in the XI
Region, Aysen, Chile. In: The Fourth International Conference on
HFRS and Hantaviruses. 5–7 Mar 1998. Atlanta, GA, USA.
28. Holmes R, Boccanera R, Figueiredo L, Mancano S, Pane C.
Seroprevalence of human hantavirus infection in the ribeirao preto
region of São Paulo State, Brazil. Emerg Infect Dis 2000;6:560–1.
29. Mascarenhas-Batista AV, da Rosa ES, Ksiazek TG, da Rosa AP, Leduc
JW, Pinheiro F, et al. Anti-hantavirus antibodies in school children in
Salvador, Bahía. Rev Soc Bras Med Trop 1998;31:433–40.
30. Childs JE, Korch GW, Glass GE, LeDuc JW, Shah KV. Epizootiology
of hantavirus infections in Baltimore: isolation of a virus from
Norway rats, and characteristics of infected rat populations. Am J
Epidemiol 1987;126:55–68.
31. Korch G, Glass J, Rossi C, Le Duc J. Serologic evidence of hantavirus
infections within small mammal communities of Baltimore,
Maryland: spatial and temporal patterns an host range. Am J Trop
Med Hyg 1989;41:230–40.
32. Diglisic G, Rossi C, Doti A, Walshe D. Seroprevalence study of han-
tavirus infection in the community based population. Md Med J
1999;48:303–6.
33. Glass GE, Watson AJ, LeDuc JW, Kelen GD, Quinn TC, Childs JE.
Infection with a ratborne hantavirus in U.S. residents is consistently
associated with hypertensive renal disease. J Infect Dis
1993;167:614–20.
34. Rossi C, Ksiazek T. Enzyme-linked immunosorbent assay (ELISA).
In: Lee HW, Calisher C, Schmaljohn C, editors. Manual of hemor-
rhagic fever with renal syndrome and hantavirus pulmonary syn-
drome. Seoul: WHO Collaborating Center for Virus Reference and
Research (Hantaviruses) Asian Institute for Life Sciences; 1999. p.
87–91.
35. Hjelle B, Jenison S, Torrez-Martínez N, Herring B, Quan S, Polito A,
et al. Rapid and specific detection of Sin Nombre virus antibodies in
patients with hantavirus pulmonary syndrome by a strip inmunoblot
assay suitable for field diagnosis. J Clin Microbiol 1997;35:600–8.
36. Hjelle B, Torrez-Martínez N, Bharadwaj M. Recombinant western
blot and strip immunoblot assays for hantaviruses. In: Lee HW,
Calisher C, Schmaljohn C, editors. Manual of hemorrhagic fever with
renal syndrome and hantavirus pulmonary syndrome. Seoul: WHO
Collaborating Center for Virus Reference and Research
(Hantaviruses) Asian Institute for Life Sciences; 1999. p. 122–30.
37. Galeno H, Villagra E, Fernández J, Ramírez E, Mora J. Infección por
hantavirus en humanos: experiencia del laboratorio de referencia para
enfermedades infecciosas emergentes [in Spanish]. Rev Chil Infect
2000;17:216–9.
38. Hjelle B, Glass GE. Outbreak of hantavirus infection in the Four
Corners region of the United States in the wake of the 1997–1998 El
Nino-southern oscillation. J Infect Dis 2000;181:1569–73.
39. Clas A, Juto P, Stegmayr B, Settergren B, Wadell G, Tärnvik A, et al.
Prevalence of serum antibodies to hantaviruses in Northern Sweden
as measured by recombinant nucleocapsid proteins. Scand J Infect
Dis 1997;29:349–54.
Address for correspondence: Marlis Täger Frey, Instituto de Pediatría,
Facultad de Medicina Universidad Austral de Chile, Valdivia, Chile; fax:
56-63-229570; email: [email protected]
832 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Use of trade names is for identification only and does not imply
endorsement by the Public Health Service or by the U.S.
Department of Health and Human Services.

Clinical antimicrobial susceptibility breakpoints are
used to predict the clinical outcome of antimicrobial treat-
ment. In contrast, microbiologic breakpoints are used to
identify isolates that may be categorized as susceptible
when applying clinical breakpoints but harbor resistance
mechanisms that result in their reduced susceptibility to the
agent being tested. Currently, the National Committee for
Clinical Laboratory Standards (NCCLS) guidelines utilize
clinical breakpoints to characterize the activity of the fluoro-
quinolones against Streptococcus pneumoniae. To deter-
mine whether levofloxacin breakpoints can identify isolates
that harbor recognized resistance mechanisms, we exam-
ined 115 S. pneumoniae isolates with a levofloxacin MIC of
>2 µg/mL for first-step parC mutations. A total of 48 (59%)
of 82 isolates with a levofloxacin MIC of 2 µg/mL, a level
considered susceptible by NCCLS criteria, had a first-step
mutation in parC. Whether surveillance programs that use
levofloxacin data can effectively detect emerging resist-
ance and whether fluoroquinolones can effectively treat
infections caused by such isolates should be evaluated.
T
he emergence of Streptococcus pneumoniae resistance
to β-lactam and macrolide antimicrobial agents has led
to recommendations that fluoroquinolones with increased
activity against S. pneumoniae, such as levofloxacin, mox-
ifloxacin, and gatifloxacin, be used to treat patients at risk
for infection caused by such multidrug-resistant strains
(1–6). Fluoroquinolone resistance in S. pneumoniae is pri-
marily due to mutations in the genes encoding the target
topoisomerase enzymes, namely parC, which encodes the
A subunit of DNA topoisomerase IV, and gyrA, which
encodes the A subunit of DNA gyrase (7). Mutations in
parE and gyrB have been reported, but to a lesser extent
(8–10). Most pneumococcal isolates with reduced suscep-
tibilities to fluoroquinolones have amino acid substitutions
in either ParC alone or ParC and GyrA (11–14). Resistance
can also be mediated by active efflux (15), although the
role of efflux in contributing to resistance by the newer
fluoroquinolones is unclear (16).
The MIC of an antimicrobial agent is a value that has
been used to determine breakpoints that predict the proba-
bility of clinical success, detect resistant populations, or
both (17). Clinical breakpoints are dependent on the
antimicrobial activity and pharmacology of the drug; such
breakpoints are ascertained with the goals of eradicating
the infection and ultimately achieving clinical success with
the antimicrobial agent. In contrast, microbiologic break-
points are established to identify isolates that may be cate-
gorized as susceptible when applying clinical breakpoints
but harbor resistance mechanisms that result in their
reduced susceptibility to the agent being tested. These
microbiologic breakpoints are therefore useful in monitor-
ing the emergence of resistance. The current National
Committee for Clinical Laboratory Standards (NCCLS)
guidelines make no distinction between these two interpre-
tations of MIC, with clinical breakpoints used to character-
ize most antimicrobial agents, including the fluoro-
quinolones.
Levofloxacin has been used as a surrogate marker to
predict fluoroquinolone susceptibility in clinical laborato-
ries and surveillance studies (18). To establish whether
current levofloxacin breakpoints are also able to function
as microbiologic breakpoints, we determined the percent-
age of S. pneumoniae isolates with first-step parC muta-
tions that would go undetected by using the current
NCCLS breakpoints for levofloxacin (19).
Materials and Methods
A total of 6,076 clinical isolates of S. pneumoniae were
collected as part of a 1993–1998 surveillance program
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 833
RESEARCH
Antimicrobial Susceptibility
Breakpoints and First-Step
parC Mutations in Streptococcus
pneumoniae: Redefining
Fluoroquinolone Resistance
Sue Lim,*† Darrin Bast,*† Allison McGeer,*† Joyce de Azavedo,*† and Donald E. Low*†
*Toronto Medical Laboratories/Mount Sinai Hospital, Toronto,
Ontario, Canada; and †University of Toronto, Toronto, Ontario,
Canada

throughout Canada. All isolates were identified as S. pneu-
moniae by standard methods. The isolates were frozen at
–70°C, thawed, subcultured onto blood agar, and incubated
at 37°C in 5% CO
2
for 24 h twice before testing. In vitro
susceptibility testing was performed by broth microdilu-
tion, according to NCCLS guidelines (20,21).
Susceptibility interpretive criteria used were those pub-
lished in the NCCLS M100-S12 document (19). The non-
susceptible category was defined as those isolates with
MICs of fluoroquinolines in the intermediate and resistant
category. The parC gene of 115 isolates with a levofloxacin
MIC >2 µg/mL (82 = MIC 2 µg/mL; 8 = MIC 4 µg/mL; 10
= MIC 8 µg/mL; and 15 = MIC ≥16 µg/mL) was amplified
by polymerase chain reaction (PCR), and the nucleotide
sequence determined as previously described (9). All iso-
lates (n=33) with a levofloxacin MIC of ≥4 µg/mL, and a
random sample of 29 isolates with a levofloxacin MIC of 2
µg/mL were examined for gyrA mutations. For compara-
tive purposes, the parC gene of 14 isolates with a
ciprofloxacin MIC of 2 µg/mL, regardless of their lev-
ofloxacin MIC, was amplified and sequenced. Although
numerous single mutational events occur in parC, the focus
of this investigation was on amino acid substitutions for
Ser-79 or Asp-83, because previous studies have consis-
tently demonstrated that mutations at either of these posi-
tions are associated with decreased susceptibility (9,14).
Crude cell lysates were used as DNA templates for
PCR. After overnight growth on Columbia nutrient agar
and supplemented with 5% sheep blood, a loopful of
growth was suspended in 100 µL of lysis buffer (100 mM
NaCl, 10 mM Tris-HCl [pH 8.3], 1 mM EDTA, 1% Triton
X-100) and boiled for 10 min. Ten microliters of the super-
natant was used as the template in a 50-µL reaction vol-
ume. The quinolone-resistance–determining regions of
parC and gyrA were amplified by PCR. Primers used were
based on published sequences (7,8), and amplification
products were purified with either the QIAquick PCR
purification kit (Qiagen Inc., Mississauga, Ontario,
Canada) or the Concert Rapid PCR purification kit (Life
Technologies, Burlington, Ontario, Canada).
DNA sequencing was performed by ABI prism Big Dye
terminator cycle sequencing with the ABI 377 automated
sequencer (PE Applied Biosystems, Mississauga, Ontario,
Canada). Nucleotide and amino acid sequence compar-
isons were performed by the multiple-alignment sequence
function of Vector NTI Suite software (InforMax Inc.,
Bethesda, MD). The GenBank accession numbers for the
wild-type sequences used for comparison purposes were
Z67739 for parC and parE (22), AB010387 for gyrA, and
Z67740 for gyrB (23).
Isolates were examined for active efflux by agar dilu-
tion on Mueller-Hinton agar containing 5% sheep blood in
the presence of ciprofloxacin with or without 10 mg/mL of
reserpine (Sigma Chemical Co., St. Louis, MO) (24).
Strains for which a fourfold or greater decrease in the MIC
of ciprofloxacin existed in the presence of reserpine were
considered in this study to be positive for reserpine-inhib-
ited efflux. S. pneumoniae strain P121/1N27 and clinical
isolate BSP 823 were used as quality control strains, the
latter of which demonstrated a 16-fold decrease in the
ciprofloxacin MIC in the presence of reserpine (9).
Results
Of the 115 S. pneumoniae isolates with a levofloxacin
MIC >2 µg/mL, 78 (69%) had an amino acid substitution
in ParC (Ser-79 or Asp-83) (Table 1). Mutations in gyrA
were not found in any of the randomly selected isolates
with a levofloxacin MIC of 2 µg/mL, but were present in
three (38%) of eight isolates with a levofloxacin MIC of 4
µg/mL and in all isolates with a levofloxacin MIC ≥ 8
µg/mL (Table 2). The specific ParC amino acid substitu-
tions of the isolates and their corresponding levofloxacin
MICs are shown in Table 1. The most common substitution
was Ser-79 to Phe, accounting for 60% of all observed
amino acid substitutions. The prevalence of first-step ParC
amino acid substitutions among all strains according to
their levofloxacin and ciprofloxacin MICs is shown in
Table 3. Using the current MIC interpretive standards for
levofloxacin, 48 (59%) of 82 of isolates with a first-step
mutation fall in the susceptibility category of levofloxacin
834 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 1. ParC amino acid substitutions found in 115 Streptococcus pneumoniae isolates with levofloxacin MICs >2 µg/mL and
corresponding levofloxacin MICs
No. isolates inhibited by levofloxacin MIC (µg/mL) of
ParC amino acid
substitution
2
4
8
16
≥32
Total no. of strains
Ser79→Phe
28
4
3
9
3
47
Ser79→Tyr
7
1
3
2
1
14
Ser79→Ala
1
0
0
0
0
1
Asp83→Asn
7
0
3
0
0
10
Asp83→Gly
1
0
0
0
0
1
Asp83→Tyr
3
0
0a
0
0
3
Asp83→Val
1
0
0
0
0
1
Asp83→Ala
0
0
1
0
0
1
No. isolates/total with
amino acid substitutions
48/82 (59%)
5/8
a
(63%)
10/10
11/11
4/4
78/115 (69%)
a
One isolate with no ParC amino acid substitution found to have active efflux; two isolates had ParC amino acid substitutions at sites other than Ser79 or Asp83.

(MIC<4 µg/mL). In comparison, 4 (29%) of 14 randomly
chosen isolates with a ciprofloxacin MIC of 2 µg/mL har-
bored a first-step mutation.
Thirty-three isolates were nonsusceptible to lev-
ofloxacin (MIC>4 µg/mL); for 25, which harbored both
ParC and GyrA amino acid substitutions, the levofloxacin
MIC was ≥8 µg/mL (Table 3). For eight isolates, the lev-
ofloxacin MIC was 4 µg/mL; three (38%) of those isolates
had a substitution in GyrA (Ser-81-Phe) as well as a sub-
stitution in ParC (Ser-79-Phe, Asp-78-Asn and Ala-115-
Pro) (Table 4). In addition, two of the eight (25%) isolates
had no substitution in GyrA, but were considered positive
for reserpine-inhibited efflux, while three isolates had a
Ser-79-Phe amino acid substitution in ParC. No mutations
were found in either parE or gyrB in the isolates with a
levofloxacin MIC of 4 µg/mL.
Discussion
Before the development of fluoroquinolones such as
levofloxacin, ofloxacin was used to determine trends of
pneumococcal fluoroquinolone resistance in the United
States (25). By using this system, an increase of ofloxacin-
nonsusceptible isolates from 2.6% in 1995 to 3.8% in 1997
was reported. However, the significance of such an
increase was questioned, since ofloxacin-resistant strains
could be observed with only a single topoisomerase muta-
tion, whereas for fluoroquinolones such as levofloxacin,
multiple mutations are required for a strain to be classified
as resistant according to NCCLS breakpoints (25–27). As
a consequence, ofloxacin was replaced by levofloxacin in
1998 as a marker for fluoroquinolone nonsusceptibility,
and not surprisingly, given levofloxacin’s increased activi-
ty against S. pneumoniae, fluoroquinolone resistance rates
were only 0.2% in 1998 and 1999 (25).
Since effective surveillance depends upon the ability to
detect the emergence of resistance, the prevalence of pneu-
mococci that harbor resistance mechanisms to the fluoro-
quinolones may not be accurately represented if surveil-
lance systems that rely on levofloxacin MIC data are used
(25,28–34). We found that 59% of isolates with a lev-
ofloxacin MIC of 2 µg/mL, a level considered susceptible
according to NCCLS criteria, had a first-step mutation in
parC. Similarly, Davies et al. (12) found that of 14 strains
for which levofloxacin MICs were 2 µg/mL, 10 (71%) had
a parC mutation. Therefore, if the goal of surveillance is to
detect emerging problems, then by extension, the detection
of first-step mutations is also important and the use of cur-
rent NCCLS breakpoints to estimate fluoroquinolone
resistance is clearly inadequate. Apart from DNA sequenc-
ing, currently no accurate test can reliably identify isolates
with first-step mutations (35). Although decreasing lev-
ofloxacin breakpoints has been proposed as a solution to
this problem, we found that 8 (25%) of 32 strains for which
the levofloxacin MIC was 1 µg/mL already had first-step
mutations (data not shown). Similarly, the replacement of
levofloxacin as a surveillance indicator by another fluoro-
quinolone has also been suggested. However, use of
ciprofloxacin does not fare significantly better, with 4
(29%) of 14 isolates in the susceptible category (MIC of 4
µg/mL to define nonsusceptible isolates) harboring first-
step mutations.
In addition to causing an underestimation of the emer-
gence of fluoroquinolone resistance, the use of clinical
breakpoints has therapeutic implications, as supported by
recent reports of treatment failure when a fluoroquinolone
was used to treat an infection caused by a strain of pneu-
mococci with a first-step mutation (36,37). Clearly, a first-
step mutation is necessary for the development of subse-
quent mutations, the latter of which result in MICs that fall
within the nonsusceptible category. However, studies have
shown that upon acquisition of a first-step mutation, the
likelihood of developing a subsequent mutation is
enhanced in comparison to the development of the first-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 835
RESEARCH
Table 2. Number of isolates with ParC and GyrA amino acid
substitutions and their corresponding levofloxacin MICs
No. strains with amino acid substitutions in
MIC (µg/mL)
ParC (%)
ParC and GyrA (%)
2
48/82 (59)
0/29
a
(0)
4
5/8 (63)
3/8 (38)
8
0/10 (0)
10/10 (100)
≥16
0/15 (0)
15/15 (100)
a
29/82 isolates were randomly examined for GyrA mutations.
Table 3. The prevalence of ParC amino acid substitutions
among all strains according to their levofloxacin and
ciprofloxacin MICs
No. strains with ParC amino acid
substitution at 79 or 83
MIC (µg/mL)
Ciprofloxacin (%)
Levofloxacin (%)
2
4/14 (29)
48/82 (59)
4
24/37 (65)
5/8 (63)
8
11/12 (92)
10/10 (100)
>8
22/22 (100)
15/15 (100)
Total
62/87 (71)
78/115 (69)
Table 4. Characterization of Streptococcus pneumoniae isolates
with levofloxacin MIC 4 µg/mL
a
Amino acid substitution
Isolate
no.
In ParC
In GyrA
Change in MIC
with inhibition of
efflux
1
Ser79→Phe
None
8-fold
2
Ser79→Phe
None
No effect
3
Asp78→Asn
Ser81→Phe
No effect
4
Ala115→Pro
Ser81→Phe
No effect
5
Ser79→Phe
None
No effect
6
Ser79→Phe
None
No effect
7
None
None
4-fold
8
Ser79→Phe
Ser81→Phe
No effect
a
parE and gyrB sequencing was performed on all isolates, but no mutations were
found in the quinolone-resistance–determining region.

step mutation itself (38–40). Studies are required to deter-
mine whether isolates with one or more mutations in genes
encoding ParC, GyrA, or both, can still be effectively treat-
ed with a fluoroquinolone when that fluoroquinolone is
found to be susceptible by using current clinical break-
points. Recognizing the presence of underlying mutations
may be especially important when using these agents to
treat patients with large biomass infections such as pneu-
mococcal pneumonia.
Lastly, the acquisition of a second-step mutation
appears more likely than not to raise the MIC to >8 µg/mL
and not to 4 µg/mL as would be expected. Isolates with a
levofloxacin MIC of 4 µg/mL represented 0.1% of the total
number of isolates in our study, which is notable, consid-
ering that a levofloxacin MIC of 4 µg/mL is currently used
to define nonsusceptibility. Furthermore, these isolates
were for the most part either genotypically or phenotypi-
cally distinct from other isolates characterized (Table 4);
two had efflux mechanisms, one singly and the other con-
current with a ParC amino acid substitution, and two had
unusual substitutions in ParC (Asp78→Asn and
Ala115→Pro). The importance of this latter finding
remains to be determined.
In summary, levofloxacin susceptibility testing that
uses current MIC clinical breakpoints does not identify
most S. pneumoniae isolates with only first-step parC
mutations. This finding may not only have implications for
the ability of surveillance programs to detect emerging
resistance, but therapeutic implications as well.
Dr. Lim is a physician trained in internal medicine and cur-
rently a fellow in infectious diseases and medical microbiology
at the University of Toronto. Her research interests include
infections in the immunocrompromised host and transplant
recipients.
This work was supported by grants from Hoffman-La Roche
and the Canadian Bacterial Diseases Network.
References
1. Bartlett JG, Dowell SF, Mandell LA, File TM Jr, Musher DM, Fine A.
Practice guidelines for the management of community-acquired
pneumonia in adults. Clin Infect Dis 2000;31:347–82.
2. Niederman MS, Mandell LA, Anzueto A, Bass JB, Broughton WA,
Campbell GD, et al. Guidelines for the management of adults with
community-acquired pneumonia. Diagnosis, assessment of severity,
antimicrobial therapy, and prevention. Am J Respir Crit Care Med
2001;163:1730–54.
3. Mandell LA, Marrie TJ, Grossman RF, Chow AW, Hyland RH.
Canadian guidelines for the initial management of community-
acquired pneumonia: an evidence-based update by the Canadian
Infectious Diseases Society and the Canadian Thoracic Society. Clin
Infect Dis 2000;31:383–421.
4. Heffelfinger JD, Dowell SF, Jorgensen JH, Klugman KP, Mabry LR,
Musher DM, et al. Management of community-acquired pneumonia
in the era of pneumococcal resistance: a report from the Drug-
Resistant Streptococcus pneumoniae Therapeutic Working Group.
Arch Intern Med 2000;160:1399–408.
5. File TM Jr, Segreti J, Dunbar L, Player R, Kohler R, Williams RR, et
al. A multicenter, randomized study comparing the efficacy and safe-
ty of intravenous and/or oral levofloxacin versus ceftriaxone and/or
cefuroxime axetil in treatment of adults with community-acquired
pneumonia. Antimicrob Agents Chemother 1997;41:1965–72.
6. Petitpretz P, Arvis P, Marel M, Moita J, Urueta J. Oral moxifloxacin
vs high-dosage amoxicillin in the treatment of mild-to-moderate,
community-acquired, suspected pneumococcal pneumonia in adults.
Chest 2001;119:185–95.
7. Pan XS, Ambler J, Mehtar S, Fisher LM. Involvement of topoiso-
merase IV and DNA gyrase as ciprofloxacin targets in Streptococcus
pneumoniae. Antimicrob Agents Chemother 1996;40:2321–6.
8. Perichon B, Tankovic J, Courvalin P. Characterization of a mutation
in the parE gene that confers fluoroquinolone resistance in
Streptococcus pneumoniae. Antimicrob Agents Chemother
1997;41:1166–7.
9. Bast DJ, Low DE, Duncan CL, Kilburn L, Mandell LA, Davidson RJ,
et al. Fluoroquinolone resistance in clinical isolates of Streptococcus
pneumoniae: contributions of type II topoisomerase mutations and
efflux to levels of resistance. Antimicrob Agents Chemother
2000;44:3049–54.
10. Weigel LM, Anderson GJ, Facklam RR, Tenover FC. Genetic analy-
ses of mutations contributing to fluoroquinolone resistance in clinical
isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother
2001;45:3517–23.
11. Ho PL, Yam WC, Que TL, Tsang DN, Seto WH, Ng TK, et al. Target
site modifications and efflux phenotype in clinical isolates of
Streptococcus pneumoniae from Hong Kong with reduced suscepti-
bility to fluoroquinolones. J Antimicrob Chemother. 2001;47:655–8.
12. Davies TA, Evangelista A, Pfleger S, Bush K, Sahm DF, Goldschmidt
R. Prevalence of single mutations in topoisomerase type II genes
among levofloxacin-susceptible clinical strains of Streptococcus
pneumoniae isolated in the United States in 1992 to 1996 and 1999 to
2000. Antimicrob Agents Chemother 2002;46:119–24.
13. Jones ME, Sahm DF, Martin N, Scheuring S, Heisig P, Thornsberry
C, et al. Prevalence of gyrA, gyrB, parC, and parE mutations in clin-
ical isolates of Streptococcus pneumoniae with decreased susceptibil-
ities to different fluoroquinolones and originating from worldwide
surveillance studies during the 1997–1998 respiratory season.
Antimicrob Agents Chemother 2000;44:462–6.
14. Brueggemann AB, Coffman SL, Rhomberg P, Huynh H, Almer L,
Nilius A, et al. Fluoroquinolone resistance in Streptococcus pneumo-
niae in United States since 1994–1995. Antimicrob Agents
Chemother 2002;46:680–8.
15. Gill MJ, Brenwald NP, Wise R. Identification of an efflux pump gene,
pmrA, associated with fluoroquinolone resistance in Streptococcus
pneumoniae. Antimicrob Agents Chemother 1999;43:187–9.
16. Piddock LJ, Johnson MM, Simjee S, Pumbwe L. Expression of efflux
pump gene pmrA in fluoroquinolone-resistant and -susceptible clini-
cal isolates of Streptococcus pneumoniae. Antimicrob Agents
Chemother 2002;46:808–12.
17. Mouton JW. Breakpoints: current practice and future perspectives. Int
J Antimicrob Agents 2002;19:323–31.
18. Jones RN, Pfaller MA. Can antimicrobial susceptibility testing results
for ciprofloxacin or levofloxacin predict susceptibility to a newer flu-
oroquinolone, gatifloxacin? Report from the SENTRY Antimicrobial
Surveillance Program (1997–99). Diagn Microbiol Infect Dis
2001;39:237–43.
19. National Committee for Clinical Laboratory Standards. Performance
standards for antimicrobial susceptibility testing. Twelfth informa-
tional supplement, M100-S12. Wayne (PA): The Committee; 2002.
836 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH

20. National Committee for Clinical Laboratory Standards. Methods for
dilution antimicrobial susceptibility tests for bacteria that grow aero-
bically, 5th ed. Approved standard M7-A5. Wayne (PA): The
Committee; 2000.
21. National Committee for Clinical Laboratory Standards. Performance
standards for antimicrobial disk susceptibility tests, 7th ed. Approved
standard M2-A7. 7th ed. Wayne (PA): The Committee; 2000.
22. Pan XS, Fisher LM. Cloning and characterization of the parC and
parE genes of Streptococcus pneumoniae encoding DNA topoiso-
merase IV: role in fluoroquinolone resistance. J Bacteriol
1996:178;4060–9.
23. Munoz R, Bustamante M, De La Campa AG. Ser-127-to-Leu substi-
tution in the DNA gyrase B subunit of Streptococcus pneumoniae is
implicated in novobiocin resistance. J Bacteriol 1995:177;4166–70.
24. Brenwald NP, Gill MJ, Wise R. Prevalence of a putative efflux mech-
anism among fluoroquinolone-resistant clinical isolates of
Streptococcus pneumoniae. Antimicrob Agents Chemother
1998;42:2032–5.
25. Centers for Disease Control. Resistance of Streptococcus pneumoni-
ae to fluoroquinolones—United States, 1995–1999. MMWR Morb
Mortal Wkly Rep 2001;50:800–4.
26. Jorgensen JH, Weigel LM, Ferraro MJ, Swenson JM, Tenover FC.
Activities of newer fluoroquinolones against Streptococcus pneumo-
niae clinical isolates including those with mutations in the gyrA,
parC, and parE loci. Antimicrob Agents Chemother 1999;43:329–34.
27. Jorgensen JH, Weigel LM, Swenson JM, Whitney CG, Ferraro MJ,
Tenover FC. Activities of clinafloxacin, gatifloxacin, gemifloxacin,
and trovafloxacin against recent clinical isolates of levofloxacin-
resistant Streptococcus pneumoniae. Antimicrob Agents Chemother
2000;44:2962–8.
28. Ho PL, Yung RW, Tsang DN, Que TL, Ho M, Seto WH, et al.
Increasing resistance of Streptococcus pneumoniae to fluoro-
quinolones: results of a Hong Kong multicentre study in 2000. J
Antimicrob Chemother 2001;48:659–65.
29. Jones ME, Karlowsky JA, Blosser-Middleton R, Critchley IA,
Karginova E, Thornsberry C, et al. Longitudinal assessment of antip-
neumococcal susceptibility in the United States. Antimicrob Agents
Chemother 2002;46:2651–5.
30. McGee L, Klugman KP, Wasas A, Capper T, Brink A. Serotype 19f
multiresistant pneumococcal clone harboring two erythromycin
resistance determinants (erm(B) and mef(A)) in South Africa.
Antimicrob Agents Chemother 2001;45:1595–8.
31. Critchley IA, Blosser RS, Karlowsky JA, Yamakita J, Barth A, Sader
HS, et al. Antimicrobial resistance in respiratory pathogens isolated
in Brazil during 1999–2000. Braz J Infect Dis 2001;5:294–304.
32. Thornsberry C, Jones ME, Hickey ML, Mauriz Y, Kahn J, Sahm DF.
Resistance surveillance of Streptococcus pneumoniae, Haemophilus
influenzae and Moraxella catarrhalis isolated in the United States,
1997–1998. J Antimicrob Chemother 1999;44:749–59.
33. Schmitz FJ, Perdikouli M, Beeck A, Verhoef J, Fluit AC. Molecular
surveillance of macrolide, tetracycline and quinolone resistance
mechanisms in 1191 clinical European Streptococcus pneumoniae
isolates. Int J Antimicrob Agents 2001;18:433–6.
34. Reinert RR, Simic S, Al Lahham A, Reinert S, Lemperle M,
Lutticken R. Antimicrobial resistance of Streptococcus pneumoniae
recovered from outpatients with respiratory tract infections in
Germany from 1998 to 1999: results of a national surveillance study.
J Clin Microbiol 2001;39:1187–9.
35. Richardson DC, Bast D, McGeer A, Low DE. Evaluation of suscep-
tibility testing to detect fluoroquinolone resistance mechanisms in
Streptococcus pneumoniae. Antimicrob Agents Chemother
2001;45:1911–4.
36. Davidson R, Cavalcanti R, Brunton JL, Bast DJ, de Azavedo JC,
Kibsey P, et al. Resistance to levofloxacin and failure of treatment of
pneumococcal pneumonia. N Engl J Med 2002;346:747–50.
37. Weiss K, Restieri C, Gauthier R, Laverdiere M, McGeer A, Davidson
RJ, et al. A nosocomial outbreak of fluoroquinolone-resistant
Streptococcus pneumoniae. Clin Infect Dis 2001;33:517–22.
38. Pestova E, Millichap JJ, Noskin GA, Peterson LR. Intracellular tar-
gets of moxifloxacin: a comparison with other fluoroquinolones. J
Antimicrob Chemother 2000;45:583–90.
39. Li X, Zhao X, Drlica K. Selection of Streptococcus pneumoniae
mutants having reduced susceptibility to moxifloxacin and lev-
ofloxacin. Antimicrob Agents Chemother 2002;46:522–4.
40. Gillespie SH, Dickens A. Fluoroquinolone resistance in
Streptococcus pneumoniae: evidence that gyrA mutations arise at a
lower rate and that mutation in gyrA or parC predisposes to further
mutation. Microb Drug Resist 2003;9:17–24.
Address for correspondence: Donald E. Low, Department of
Microbiology, Rm. 1487, Mount Sinai Hospital, 600 University Ave.,
Toronto, Ontario, Canada, M5G 1X5; fax: 416-586-8746; email:
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 837
RESEARCH

Alterations in genes involved in the repair of DNA
mutations (mut genes) result in an increased mutation fre-
quency and better adaptability of the bacterium to stressful
conditions. W-Beijing genotype strains displayed unique
missense alterations in three putative mut genes, including
two of the mutT type (Rv3908 and mutT2) and ogt. These
polymorphisms were found to be characteristic and unique
to W-Beijing phylogenetic lineage. Analysis of the mut
genes in 55 representative W-Beijing isolates suggests a
sequential acquisition of the mutations, elucidating a plau-
sible pathway of the molecular evolution of this clonal fam-
ily. The acquisition of mut genes may explain in part the
ability of the isolates of W-Beijing type to rapidly adapt to
their environment.
T
uberculosis (TB) and AIDS cause more deaths in
adults worldwide than any other infectious disease.
Globally, the number of TB cases is growing at a rate of
2% per year. Resistance, especially multidrug-resistance
(MDR), is an increasing problem (1) and a growing hazard
to human health. Many outbreaks of MDR-TB, defined as
resistance to at least rifampicin and isoniazid, have been
reported, with poor response to therapy and very high dis-
ease and death rates. Some TB outbreaks have involved
patients with HIV co-infection (2,3). Although in several
instances, MDR outbreaks associated with a particular
genotype, such as the W strain, have been identified (4,5),
drug-susceptible variants of the W strain account for most
of this group of isolates characterized to date.
In 1995, the largest proportion of the Mycobacterium
tuberculosis strains from Beijing, China, shared a high
degree of similarity in IS6110 restriction fragment length
polymorphism (RFLP) patterns and identical spoligo pat-
terns (6). Subsequent molecular analyses have indicated
that the W and Beijing isolates constitute a single group of
strains designated as the W-Beijing genotype (Figure 1).
The global distribution and success of M. tuberculosis iso-
lates of the W-Beijing genotype have led to the hypothesis
that these strains may have selective advantages over other
M. tuberculosis strains. In addition to the W-MDR strain
identified in New York City, and areas in Cuba, Estonia,
Vietnam, and Russia, the W-Beijing genotype has been
significantly associated with drug resistance (7 and unpub.
data). Several studies have suggested that the W-Beijing
genotype strains are disseminating throughout the world
(7). In Vietnam, the proportion of W-Beijing strains was
71% in patients <25 years of age and 41% for those >25
years of age (8). Furthermore, W-Beijing strains have been
implicated in several TB epidemics globally, including
ones in New York, Texas, California, South Carolina, and
New Jersey in the United States (9) and South Africa,
Russia, and Spain (10). A recent study showed that 82% of
MDR strains isolated in a prison in Azerbaijan, Eastern
Europe, are of the W-Beijing genotype (11).
Ongoing research is focused on identifying the factors
responsible for the worldwide spread of the W-Beijing
strains and their ability to adapt and enhance their patho-
genicity or virulence. Identifying a possible mechanism for
increased adaptation of these bacteria to the human
immunologic host defense system or human interventions
such as anti-TB treatment is of the utmost importance.
Such mechanisms may indicate how the bacterium adapts
to the host, a prerequisite for an enhanced accumulation of
genomic mutations associated with resistance. In M. tuber-
culosis, resistance to antibiotics occurs because of genom-
ic mutations in certain genes, such as the katG gene for iso-
niazid (INH) resistance and the rpoB gene for rifampicin
resistance (12). In contrast to several other pathogens with
MDR phenotypes, plasmid or transposon-mediated mech-
anisms of resistance have not been reported in M. tubercu-
losis (13–15). Since resistance to bacteriostatic in M.
838 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Mutations in Putative Mutator
Genes of Mycobacterium
tuberculosis Strains of the
W-Beijing Family
Mina Ebrahimi Rad,* Pablo Bifani,† Carlos Martin,‡ Kristin Kremer,§ Sofia Samper,‡ Jean Rauzier,*
Barry Kreiswirth,¶ Jesus Blazquez,# Marc Jouan,* Dick van Soolingen,§ and Brigitte Gicquel*
*Institut Pasteur, Paris, France; †Institut Pasteur, Lille, France;
‡Universidad de Zaragoza, Zaragoza, Spain; §National Institute of
Public Health and the Environment, Bilthoven, the Netherlands;
¶Public Health Research Institute, Newark, New Jersey, USA; and
#National Institute of Health, Madrid, Spain

tuberculosis is exclusively due to genomic mutations, the
bacterium would benefit from an increased mutation rate.
Recent studies provided evidence for a role of mutator
phenotypes in the emergence of MDR clinical
Pseudomonas isolates (16). Such phenotypes not only
enable the bacteria to acquire resistance to antibiotics more
easily but also facilitate their adaptation to a new niche.
Bacteria can escape immune surveillance by modulating
bacterial resistance to host defense mechanisms (16–18).
This finding prompted us to investigate whether a similar
situation exists in M. tuberculosis. We have undertaken a
comprehensive comparative sequence analysis of selected
target genes to evaluate and study the presence of muta-
tions in putative genes expected to play a role in the muta-
tion frequency in such strains.
Mutated phenotypes commonly result from defects in
DNA repair (19). An in silico analysis suggested that most
mismatch repair systems (e.g., mutS, mutL, or mutH) were
missing in the M. tuberculosis genome (20). However, the
frequency of spontaneous mutations in M. tuberculosis (in
vitro cultures) is similar to that found in other bacteria-car-
rying mismatch repair systems (21), which suggests that
other DNA repair mechanisms must be present.
Hypothetical open reading frames (ORF), similar to genes
known to be responsible for the avoidance or repair of
DNA lesions resulting from the alkylation or oxidation of
nucleotides, are present in the genome of M. tuberculosis.
We searched for variations in these genes in 139 clinical
isolates to detect possible mutations that could allow an
enhanced adaptability to the host and increased resistance
to anti-TB drugs.
Methods
We searched for mut genes variation in 139 M. tubercu-
losis complex strains originating from 35 different coun-
tries. Ninety-four of these strains were selected because
they were representative strains characterized with 13 dif-
ferent genetic markers in previous studies (6,22).
This set comprised 125 M. tuberculosis strains, 1 M.
africanum, 8 M. bovis, 3 M. bovis BCG, and 2 M. microti.
Fifty-five strains had a W-Beijing genotype; 12 had an
MDR phenotype. Strains representing different branches
of the W-Beijing genotype were studied. Eight MDR M.
tuberculosis strains with a genotype other than Beijing
were included. Five M. tuberculosis strains of the W-
Beijing genotype and three strains of unrelated genotype
were obtained from the national program for surveillance
of MDR tuberculosis in Spain. Four M. tuberculosis W-
Beijing genotype strains isolated in the Netherlands and
one from Vietnam were included because they showed
spoligo patterns with fewer than nine spacers. Five other
W-Beijing genotype strains showed hybridization to an
additional spacer, as demonstrated by using the extended
set of spacers, two of which lacked hybridization to spac-
er 37. Strain W4 is part of a drug-susceptible outbreak in
New Jersey (4); W147 is a drug-resistant isolate widely
spread in Russia (23). Eleven strains were representative
of ancestral W-Beijing strains, which diverged early in the
evolution of the W-Beijing phylogenetic lineage. Finally,
29 strains of another frequently observed genotype, the
Haarlem genotype (6), were investigated.
The collection consisted of 55 W-Beijing genotype iso-
lates, 29 Haarlem genotype isolates, 8 strains of the
African genotype, 1 M. bovis strain, and 46 representatives
of other genotypes. Principal genetic grouping (PGG),
according to the polymorphism in katG and gyrA, was
known (24) for most of the isolates in this study. Seventy-
four strains belong to PGG 1, 54 to PGG 2, and 3 to PGG
3. All isolates were subjected to at least IS6110 RFLP typ-
ing and spoligotyping (6). Drug susceptibility was deter-
mined for 41 of 139 strains (Table 1 and 2). Several puta-
tive mut genes were annotated as such in the released
genome sequence of M. tuberculosis (25). In addition,
using the BLAST program (26), we identified Rv3908 as
an ORF carrying a mutT domain (27) and have since
named it mutT4.
Primers were designed to amplify putative mut genes:
mutY (5'-CCGGCGACGAATCGCTCGTT-3', 5'-
AGCTGGGACAGTCGTCGCGG-3'), mutM (5'-CTG-
GTTCGATGGTGATGACC-5', 5'-GTGCGCTCGACC-
CACAG-3'), mutT2 (5'-TCCGGATGATGATTTACCTCC-
3', 5'-TCCGCCGGGTCGGGGAC-3'), mutT1 (5'-ATCG-
TCGGCGTGCCGTG-3', 5'-GTCAGCGTCCTGCCCGG-
3'), mutT4 (5'-TCGAAGGTGGGCAA- ATCGTG-3' 5'-
TGGGGTTCGCTGGAAGTGG–3'), ogt (5'-CAGCGC-
TCGCTGGCGCC-3', 5'-GACTCAG CCGCTCGCGA-3'),
and mut T3 (5'-GTCACGTCTGTTAGGACCTC-3', 5'-
CGCGCAACGGCTGCCGG-3'). Similar primers were
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 839
RESEARCH
Figure 1. Characteristic patterns
of Mycobacterium tuberculosis
Beijing genotype strains. RFLP,
restriction fragment length poly-
morphism.

designed to amplify the rpoB gene (5'-TACGGTCGGC-
GAGCTGATCC-3', 5'- TACGG- CGTTTCGATGAACC-
3').
DNA sequencing was performed directly on the ampli-
fied fragments by using the dideoxy chain-termination
method with the Big Dye Terminator Cycle sequencing Kit
(Perkin Elmer Applied Biosystems, Courtaboeuf, France)
on a GeneAmp polymerase chain reaction (PCR) system
9600 (Perkin Elmer) and run on a DNA analysis system
model 373 or 3100 (Applied Biosystems). Sequences of
mutY, mutT2, mutT4, rpoB, mutT1, mutT3, and ogt of the
M. tuberculosis strains H37Rv, CDC1551, and MT210
were obtained from published sequences or at the TIGR
Web site (available from: URL: http://www.tigr.org/).
Results
We searched for allele variation in putative genes cod-
ing for DNA repair enzymes: mutT (which hydrolyzes 8-
oxo-deoxyguanosine triphosphate) (28), ogt (which
removes methyl groups from O6-methylguanine in DNA)
(29), mutM (formamidopyrimidine-DNA glycosylate)
(30), and mutY (specific adenine glycosylate) (31) in 12
MDR M. tuberculosis strains. Subsequently, we genotyped
for the observed single nucleotide polymorphism (SNP)
variation in 124 strains, members of the M. tuberculosis
complex, and in the three published sequences of M. tuber-
culosis strains. In total, the sampling comprises 55 W-
Beijing genotype M. tuberculosis strains, including 11
ancestral W-Beijing isolates (unpub. data ), 84 M. tubercu-
losis strains of other genotypes, and 1 M. bovis strain.
Several putative mut genes were annotated as such in
the released genome sequence of M. tuberculosis. A
BLAST search using the E. coli mutT sequences as tem-
plate identified, in addition to mutT1, mutT2, mutT3, the
hypothetical ORF Rv3908, which we have designated as
mutT4. The best matches with E. coli mutT gene were
observed for mutT2 and mutT4. Figure 2 depicts sequence
alignment of the conserved region of the different genes of
the M. tuberculosis genome showing similarity with mutT
of E. coli. The search for sequences similar to ogt, mutM,
and mutY identified a single ORF in each case. Primers
were designed for PCR amplification of all the genes men-
tioned above.
We first determined the sequences of the different
genes mentioned above in 12 MDR M. tuberculosis strains
(ZA20, ZA65, ZA67, ZA68, ZA69, ZA11, ZA16, ZA12,
ZA13, ZAA14, ZA17, and ZA19), including 5 W-Beijing
strains (ZA20, ZA65, ZA67, ZA78, and ZA69). For the
mutY, mutM, mutT1, and mutT3 putative genes, PCR
amplification was obtained in all strains tested, but
sequence analysis did not indicate any nucleotide changes
at these loci except for the same silent SNP in mutT3 in
strains with a Haarlem genotype. We confirmed these find-
ings by sequencing mutT1, mutT3, mutM, and mutY in a
collection of 26 MDR strains from North Africa. No vari-
ation was observed in mutT1 or mutM. Only one strain had
a major variation in mutY. All Haarlem strains carried one
characteristic silent mutation in mutT3 and one character-
istic mutation in ogt (Ser 15 replaced by Thr). These defin-
ing SNPs were also observed for all Haarlem strains of this
study. No other variations were observed in mutT1, mutT3,
mutM, or mutY. However, comparative sequence analysis
of H37Rv, CDC1551, and the five MDR–W-Beijing iso-
lates indicated polymorphisms in mutT2, mutT4, and ogt.
These mutations in mutT4, mutT2, and ogt were also found
in the W-Beijing strain 210 (TIGR) but not in MDR strains
other than those belonging to the W-Beijing genotype. We
therefore decided to extend this investigation and look for
mutations in these three genes in a collection of M. tuber-
culosis complex isolates, including well-defined branches
of the W-Beijing phylogenetic lineage (Table 1 and 2).
In 43 of 55 strains with a W-Beijing genotype, either
susceptible to bacteriostat or MDR, we found a mutation in
mutT4. Codon 48 (CGG) of the annotated ORF had been
changed to GGG, resulting in the amino acid substitution
of Arg by Gly (Table 1 and 2). All 11 W-Beijing isolates
known to be closely related to the ancestral W-Beijing
strain (AM, HI, N16, DU2, DV, LB2, KY, IK, 122(C11),
113, and 107(LB)) were found to have the wild-type geno-
type as all other 84 isolates with a genotype other than W-
Beijing.
Thirty-nine of 43 W-Beijing strains with the mutation
in mutT4 carried an additional mutation in mutT2 and in
ogt. The mutT2 mutation constitutes a change in codon 58
(GGA to CGA), resulting in an amino acid substitution of
Gly by Arg. The active site of the E. coli MutT enzyme
comprises amino acids 53, 56, 57, and 98. Therefore, a
mutation Gly to Arg at position 58 may have a important
effect on enzyme activity and lead to a mutator phenotype.
All 39 W-Beijing isolates carrying the mutT2 polymor-
phism at codon 58 also displayed a concurrent silent muta-
840 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure 2. MutT proteins’ sequences alignment. Mycobacterium
tuberculosis Rv2985(MutT1), Rv1160(MutT2), Rv0413(MutT3),
and Rv3908(MutT4) were selected from the M. tuberculosis
genome because of their annotation or after a BLAST analysis.
These sequences were compared to Escherichia coli mutT by
using alignments available from: URL: http://www.biochem.uthsc-
sa.edu/~barnes/mutt.html. The detected region of similarity is
shown here. # absolutely conserved residues; * residues that are
strongly conserved and that define the mutT or nudix motif.

tion in codon 12 (Gly GGG to GGA Gly) of the ogt gene.
Of four possible codons encoding for glycine, GGG and
GGA had the lowest relative synonymous codon usage
(RSCU) in genes with high expression levels (0.20 and
0.17, respectively, compared to 1.32 and 2.31 for GGU and
GGC). For genes with low expression levels, the RSCU
values are 0.92, 0.37, 0.65, and 2.06 for GGG, GGA, GGU,
and GGC, respectively (32).
The five W-Beijing isolates with a mutation in mutT4
and a wild-type mutT2 gene did not contain the ogt silent
mutation on codon 12 either. Instead, they all shared a din-
ucleotide substitution in codon 37 (ACC to CTC) of ogt,
resulting in amino acid substitution of Arg to Leu. These
five W-Beijing isolates of 43 with the mutT4 mutations,
without the mutT2 (codon 58) or the ogt (codon 12) muta-
tions, differed molecularly from all other W-Beijing iso-
lates in their spoligotype pattern and accompanying dele-
tion flanking the DR locus. Four of five were isolated from
Dutch patients in the Netherlands; the fifth originated from
a patient in Vietnam. The Vietnamese isolate (no. 94)
shared >95% IS6110 pattern similarity with the Dutch iso-
late 115 when standard RFLP analysis was used. Overall,
the five isolates were closely related to each other accord-
ing to IS6110 profiling (>90% similarity). Spacer 37 in the
DR locus of Dutch isolates 114 and 139 was absent, while
sample 115 was missing spacers 37 and 38, and 111 had a
deletion of spacers 38 and 39 but not spacer 37, suggesting
that these isolates may belong to a different sublineage. A
tentative phylogeny of the W-Beijing strains analyzed in
this study is proposed in Figure 3. Seven of nine MDR W-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 841
RESEARCH
Table 1. Characteristics of Mycobacterium tuberculosis complex strains originating from 35 different countries
Strains
Genotype
No.
isolates
Country of
isolation
Group
mutT2
mutT4
ogt
ZA20/65
W-Beijing
2
Spain
1
wt
Arg CGG 48 GGG Gly
Arg CGC 37 Leu CTC
ZA67-69
W-Beijing
3
Spain
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
ZA11/16
Haarlem
2
Spain
2
wt
wt
Thr ACC 15 Ser AGC
ZA12-14/17
other
4
Spain
nd
wt
wt
wt
ZA19
M. bovis
1
Spain
wt
wt
wt
ZA15
other
1
Spain
nd
wt
wt
wt
ZA60-62
W-Beijing
3
Spain
1
wt
Arg CGG 48 GGG Gly
Arg CGC 37 Leu CTC
CDC1551
1
USA
2
wt
wt
wt
H37Rv
1
USA
3
wt
wt
wt
MT210
W-Beijing
1
USA
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
20
W-Beijing
1
Mongolia
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
30
W-Beijing
1
South Africa
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
34
W-Beijing
1
Malaysia
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
43
W-Beijing
1
China
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
44
W-Beijing
1
Thailand
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
45
W-Beijing
1
Malaysia
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
91/102-6
W-Beijing
6
Vietnam
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
110/116/119/12
4-5/140-2
W-Beijing
8
the Netherlands
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
133
W-Beijing
1
South Africa
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
W4/10/126/129
W-Beijing
4
USA
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
W99
W-Beijing
1
Singapore
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
W147
W-Beijing
1
Russia
1
Gly GGA 58 CGA Arg
Arg CGG 48 GGG Gly
Gly GGG 12 Gly GGA
94
W-Beijing
1
Vietnam
1
wt
Arg CGG 48 GGG Gly
Arg CGC 37 Leu CTC
111
W-Beijing
1
South Korea
1
wt
Arg CGG 48 GGG Gly
Arg CGC 37 Leu CTC
115
W-Beijing
1
the Netherlands
1
wt
Arg CGG 48 GGG Gly
Arg CGC 37 Leu CTC
5107(HG1)
W-Beijing
1
USA
1
wt
Arg CGG 48 GGG Gly
Arg CGC 37 Leu CTC
114, 139
W-Beijing
1
the Netherlands
1
wt
Arg CGG 48 GGG Gly
wt
166(HD6)
W-Beijing
1
USA
1
wt
Arg CGG 48 GGG Gly
wt
165(001)
W-Beijing
1
USA
1
wt
wt
Arg CGC 37 Leu CTC
107(LB)
W-Beijing
1
USA
1
wt
wt
wt
113
W-Beijing
1
the Netherlands
1
wt
wt
wt
122(CI1)
W-Beijing
1
USA
1
wt
wt
wt
IK/KY/LB2/DV
/DU2/HI
W-Beijing
6
Russia
1
wt
wt
wt
N16
W-Beijing
1
USA
1
wt
wt
wt
AM
W-Beijing
1
USA
1
wt
wt
wt
a
wt, wild-type alleles (identical to H37Rv strain); nd, not determined

Beijing strains carried missense mutations in two muT
genes (mutT2 and mutT4), and two had a missense muta-
tion in both mutT4 and ogt (Table 1 and 2).
No mutations in mutT4 or in mutT2 were observed in
any of the 84 M. tuberculosis complex strains, including
19 strains of PGG1, 54 strains of PGG2, and 2 strains of
PGG3; the strains originated from 29 countries and were a
genotype other than W-Beijing. A Thr15Ser mutation was
observed in 24 of 29 strains of the Harlem family. No other
change was observed in ogt.
Resistance to rifampicin in MDR strains was correlated
with mutations in the rpoB gene. The three tested MDR W-
Beijing strains isolated in Spain, with the mutations at the
mutT2 and mutT4 loci, harbored a different mutation in the
rpoB gene. These strains were isolated from patients who
had emigrated from Eastern Europe to Spain (ZA67,
842 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 2. Characteristics of Mycobacterium tuberculosis complex strains originating from 35 different countries
Strains
Genotype
No. isolates
Country of isolation
Group
mutT2
mut T4
ogt
AU
Haarlem
1
USA
2
wt
wt
Thr ACC 15 Ser AGC
3,5,22,32,39,48,50
,52-3,55
Haarlem
10
Argentina
2
wt
wt
Thr ACC 15 Ser AGC
8
Haarlem
1
Vietnam
2
wt
wt
Thr ACC 15 Ser AGC
13/28
Haarlem
2
Sri Lanka
2
wt
wt
Thr ACC 15 Ser AGC
51
Haarlem
1
the Netherlands
2
wt
wt
Thr ACC 15 Ser AGC
57/59
Haarlem
2
Czech republic
2
wt
wt
wt
84
Haarlem
1
Czech Republic
2
wt
wt
Thr ACC 15 Ser AGC
86/143/145
Haarlem
3
Bolivia
2
wt
wt
Thr ACC 15 Ser AGC
87
Haarlem
1
USA
2
wt
wt
Thr ACC 15 Ser AGC
99
Haarlem
1
Italy
2
wt
wt
Thr ACC 15 Ser AGC
123
Haarlem
1
Czech Republic
2
wt
wt
Thr ACC 15 Ser AGC
144/146-7
Haarlem
3
Bolivia
2
wt
wt
wt
Apr-35
Africa
2
Rwanda
2
wt
wt
wt
37
Africa
1
Uganda
2
wt
wt
wt
40/120
Africa
2
Burundi
2
wt
wt
wt
72
Africa
1
Central African Republic
2
wt
wt
wt
97
Africa
1
Uganda
2
wt
wt
wt
121
Africa
1
Central African Republic
2
wt
wt
wt
2
BCG
1
the Netherlands
1
wt
wt
wt
6/47/73/130
M. bovis
4
the Netherlands
1
wt
wt
wt
12
Other
1
Tunisia
3
wt
wt
wt
15/31
Other
2
Iran
2
wt
wt
wt
16
Other
1
Canada
2
wt
wt
wt
17
Other
1
Greenland
2
wt
wt
wt
18
Other
1
USA
2
wt
wt
wt
19/36/74
Other
2
India
1
wt
wt
wt
25/62
M. microti
2
UK
1
wt
wt
wt
26
Other
1
Zimbabwe
2
wt
wt
wt
27
Other
1
Ethiopia
2
wt
wt
wt
38/42
Other
2
Tahiti
2
wt
wt
wt
41/46
Other
2
Chile
2
wt
wt
wt
49
Other
1
Tanzania
1
wt
wt
wt
56
Other
1
Curacao
2
wt
wt
wt
64
Other
1
Honduras
2
wt
wt
wt
65/112
Other
2
the Netherlands
1
wt
wt
wt
71
BCG
1
Japan
1
wt
wt
wt
76/101/126
M. bovis
3
Argentina
1
wt
wt
wt
83
BCG
1
Russia
1
wt
wt
wt
89/95
Other
2
Spain
2
wt
wt
wt
96
Other
1
the Netherlands
3
wt
wt
wt
98
Other
1
Ecuador
2
wt
wt
wt
100
M. africanum
1
the Netherlands
1
wt
wt
wt
108
Other
1
China
2
wt
wt
wt
118
Other
1
Honduras
2
wt
wt
wt
a
nd, not determined; wt, wild-type alleles (identical to H37Rv strain).
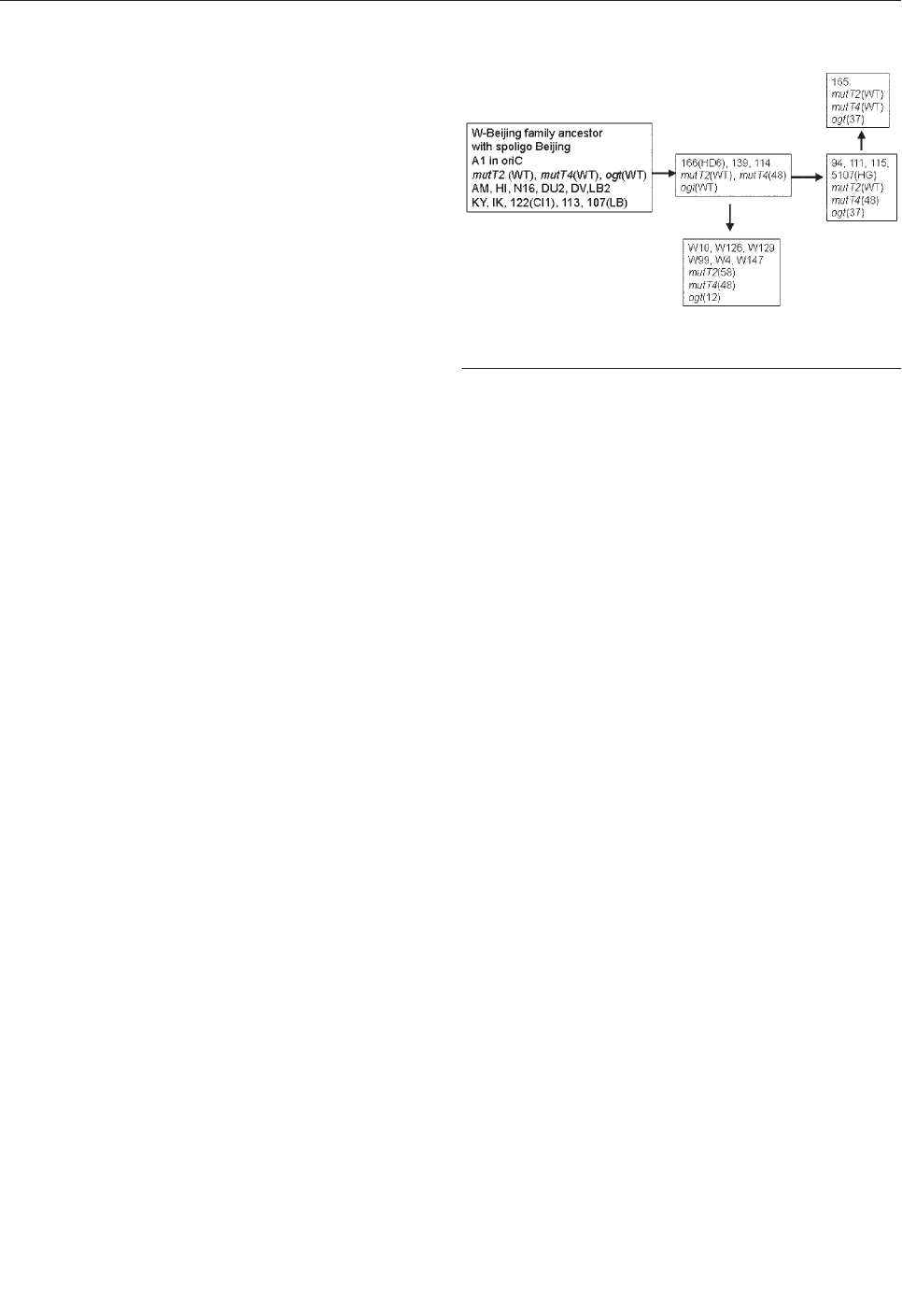
ZA68, and ZA69). Analysis of the IS6110 RFLP of the
respective isolates showed a difference of a single band.
These findings suggest that the three strains may be relat-
ed. The acquisition of the three different mutations in the
rpoB gene leading to rifampicin resistance must have
occurred after the acquisition of mutations in the putative
nucleotide repair enzyme genes mutT4 and mutT2.
Discussion
Our results show that M. tuberculosis strains of the W-
Beijing genotype acquired missense mutations in DNA
repair genes. These M. tuberculosis W-Beijing genotype
strains are genetically highly conserved and widespread.
DNA repair genes have been previously shown to be asso-
ciated with mutator phenotypes in other microorganisms.
The success of this group of strains may result in part from
mutations in DNA repair enzymes, which might provided
a true selective advantage for these bacteria to adapt and
persist, including through the acquisition of resistance to
anti-TB drugs. Mutations in the DNA repair genes might
be the evolutionary answer of the TB bacillus to increase
adaptation to hosts. This adaptation will lead to increasing
trends in the TB epidemic in the coming decades. The
World Health Organization considers MDR and resistance
as a problem of local rather than of global importance (1).
If the relative contribution of W-Beijing genotype strains
to the current worldwide TB epidemic is increasing as sug-
gested (7), this approach should be revised. In areas with
an increasing problem with MDR-TB, such as Estonia and
Russia, W-Beijing genotype strains are predominantly
associated with MDR cases (33). In Germany, the relative
proportion of W-Beijing strains among isolates from resist-
ant cases has increased from 12% in 1995 to 35% in 2000
(unpub. data). The latter observation may indicate an
increasing influence of W-Beijing strains on the worldwide
TB epidemic.
We identify polymorphisms in M. tuberculosis in genes
that might result in a mutator phenotype and therefore a
plausibly better adaptation of the bacilli to a hostile envi-
ronment (34). Forty-three of 55 W-Beijing isolates ana-
lyzed were found to have a unique mutation on the ORF
Rv3908. This ORF contains a MutT domain and is denot-
ed here as mutT4. Thirty-nine of 43 W-Beijing strains car-
ried an additional and identical mutation in a second puta-
tive gene of the mutT family, mutT2, and an identical silent
mutation in ogt.
The W-Beijing phylogenetic lineage probably acquired
the mutation on codon 48 of the mutT4 only once and
before other mutations associated with the mutator genes
we describe. This mutation clearly distinguishes ancestral
W-Beijing isolates from contemporary W-Beijing strains.
The 11 W-Beijing isolates that did not have the character-
istic mutT4 mutation on codon 48, consist of a collection
of isolates known to be ancestral within this phylogenetic
lineage, as determined by various other molecular tech-
niques (unpub. data).
Nine of W-Beijing strains with the wild-type mutT2
gene had a characteristic mutation on codon 37 of the ogt
gene, which suggests that these isolates constitute a branch
of the W-Beijing family that diverged after the acquisition
of the mutT4 mutation but before the development of the
nucleotide substitution on mutT2. One strain carries the
mutation 37 in ogt but no mutation in mutT4, a reversion
that might have occurred after a transient mutator pheno-
type.
A mutation in mutT2 was always associated with a
mutation in muT4. A first mutation may have occurred in
mutT4 and thereafter a second mutation either in mutT2 or
ogt was acquired. As observed for other bacterial popula-
tions, mutator phenotypes may be transient in many cases
to limit deleterious effects (35). Identifying these muta-
tions may aid in the identification of mut genes in M.
tuberculosis. These mutations associated with mutator
genes provide a reliable tool for the identification of W-
Beijing isolates and thus a useful marker for strains
endowed with capacity to yield epidemics. The biologic
consequences of these mutations and function of these
DNA repair genes are currently been investigated in the
laboratory.
Nine MDR strains with a W-Beijing genotype were
among strains carrying two missense mutations in putative
mutator genes. Phylogenetically unrelated M. tuberculosis
MDR isolates had no mutations within the DNA repair
genes investigated in this study. Our data support the idea
that M. tuberculosis strains of the W-Beijing genotype may
have adapted to hostile environments, including exposure
to anti-TB drugs, because of a succession of alterations of
DNA repair enzymes. Other genes involved in other DNA
repair mechanisms or in the fidelity of DNA replication
may also be involved and remain to be investigated.
The acquisition of mutator alleles was described as an
adaptive response of bacteria to a succession of different
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 843
RESEARCH
Figure 3. Schematic representation of a plausible pathway to
explain the accumulation of mutations in mut genes.

environments (18,35,36). After infecting a host, M. tuber-
culosis has to adapt to different environments such as alve-
olar macrophages and dendritic cells and subsequently to
granuloma containing inactivated macrophages or to acti-
vated macrophages after induction of the acquired immune
responses. In addition, the bacilli have to adapt to the
caseous media with low oxygen concentration in the cen-
ter of tubercles and to different types of tissues during dis-
semination of the disease. Such variable growth conditions
might select for mutations in M. tuberculosis strains, as
described in other bacterial populations exposed to differ-
ent environmental challenges. Mutations and selection
might occur with an increased frequency caused by the
toxic radicals produced in phagocytic cells.
However, a mutator phenotype is often transient.
Otherwise a continual accumulation of mutations would
lead to deleterious effects and loss of fitness. No difference
in the frequency of spontaneous mutations, resulting in a
rifampin resistance phenotype, was observed for W-
Beijing strains (37). We suggest that a transient mutator
phenotype allowed a better adaptation of W-Beijing
strains. Subsequent compensatory mutations occurred to
reverse the mutator phenotype. An alternative hypothesis
would be the existence of a higher mutation rate in specif-
ic conditions (i.e., in mutagenic radicals inside phago-
cytes). The accumulation of mutations leading to antibiot-
ic resistance in W-Beijing strains may be a consequence of
the appearance of strains with a better adaptation to the
hosts. MDR strains would be easily selected when patients
with strains that have adapted better received inadequate
anti-TB regimens.
Acknowledgments
We thank Maxime Schwartz and Lluis Quintana-Murci for
many helpful discussions and Xavier Nouvel for help with the
manuscript.
This work received support from the European Commission
(grant QLK2-CT-2000-00630), and from Louis D. French
Academy of Sciences award.
Dr. Rad is a molecular biologist. Her expertise includes
DNA amplification, sequencing, and cloning.
References
1. Raviglione MC, Gupta R, Dye CM, Espinal MA. The burden of drug-
resistant tuberculosis and mechanisms for its control. Ann N Y Acad
Sci 2001;953:88–97.
2. Farmer P, Kim JY. Community based approaches to the control of
multidrug resistant tuberculosis: introducing “DOTS-plus.” BMJ
1998;317:671–4.
3. Telzak EE, Sepkowitz K, Alpert P, Mannheimer S, Medard F, el-Sadr
W, et al. Multidrug-resistant tuberculosis in patients without HIV
infection. N Engl J Med 1995;333:907–11.
4. Bifani PJ, Mathema B, Liu Z, Moghazeh SL, Shopsin B, Tempalski
B, et al. Identification of a W variant outbreak of Mycobacterium
tuberculosis via population-based molecular epidemiology. JAMA
1999;282:2321–7.
5. Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissem-
ination of the Mycobacterium tuberculosis W-Beijing family strains.
Trends Microbiol 2002;10:45–52.
6. Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans
PW, Martin C, et al. Comparison of methods based on different
molecular epidemiological markers for typing of Mycobacterium
tuberculosis complex strains: interlaboratory study of discriminatory
power and reproducibility. J Clin Microbiol 1999;37:2607–18.
7. Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D.
Worldwide occurrence of Beijing/W strains of Mycobacterium tuber-
culosis: a systematic review. Emerg Infect Dis 2002;8:843–9.
8. Anh DD, Borgdorff MW, Van LN, Lan NT, van Gorkom T, Kremer
K, et al. Mycobacterium tuberculosis Beijing genotype emerging in
Vietnam. Emerg Infect Dis 2000;6:302–5.
9. Bifani PJ, Plikaytis BB, Kapur V, Stockbauer K, Pan X, Lutfey ML,
et al. Origin and interstate spread of a New York City multidrug-
resistant Mycobacterium tuberculosis clone family. JAMA
1996;275:452–7.
10. Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Garcia
I, Cabrera P, et al. Epidemiological evidence of the spread of a
Mycobacterium tuberculosis strain of the Beijing genotype on Gran
Canaria Island. Am J Respir Crit Care Med 2001;164:1165–70.
11. Pfyffer GE, Strassle A, van Gorkum T, Portaels F, Rigouts L, Mathieu
C, et al. Multidrug-resistant tuberculosis in prison inmates,
Azerbaijan. Emerg Infect Dis 2001;7:855–61.
12. Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial
agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber
Lung Dis 1998;79:3–29.
13. Van Rie A. Analysis for a limited number of gene codons can predict
drug resistance of Mycobacterium tuberculosis in a high incidence
community. J Clin Microbiol 2001;39:636–41.
14. Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, et
al. Detection of rifampicin-resistance mutations in Mycobacterium
tuberculosis. Lancet 1993;341:647–50.
15. Finken M, Kirschner P, Meier A, Wrede A, Bottger EC. Molecular
basis of streptomycin resistance in Mycobacterium tuberculosis:
alterations of the ribosomal protein S12 gene and point mutations
within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol
1993;9:1239–46.
16. Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequen-
cy of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung
infection. Science 2000;288:1251–4.
17. Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, et al.
Costs and benefits of high mutation rates: adaptive evolution of bac-
teria in the mouse gut. Science 2001;291:2606–8.
18. Richardson AR, Stojiljkovic I. Mismatch repair and the regulation of
phase variation in Neisseria meningitidis. Mol Microbiol
2001;40:645–55.
19. Horst JP, Wu TH, Marinus MG. Escherichia coli mutator genes.
Trends Microbiol 1999;7:29–36.
20. Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis.
What have we learnt from the genome sequence? Mol Microbiol
1998;29:1331–9.
21. David HL, Newman CM. Some observations on the genetics of iso-
niazid resistance in the tubercle bacilli. Am Rev Respir Dis
1971;104:508–15.
22. Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C.
Automated high-throughput genotyping for study of global epidemi-
ology of Mycobacterium tuberculosis based on mycobacterial inter-
spersed repetitive units. J Clin Microbiol 2001;39:3563–71.
844 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH

23. Kurepina N, Ramaswamy S, Shashkina EF, Sloutsky AM, Blinova
LN, Mishustin SP, et al. The sequence analysis of the pncA gene
determining the PZA-resistance in the predominant M. tuberculosis
strains isolated in the Tomsk penitentiary system, Western Siberia,
Russia. In: 32nd World Conference on Lung Health of the
International Union against Tuberculosis and Lung Disease. Paris,
France; 2001.
24. Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN,
Whittam TS, et al. Restricted structural gene polymorphism in the
Mycobacterium tuberculosis complex indicates evolutionarily recent
global dissemination. Proc Natl Acad Sci U S A 1997;94:9869–74.
25. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al.
Deciphering the biology of Mycobacterium tuberculosis from the
complete genome sequence. Nature 1998;393:537–44.
26. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W,
et al. Gapped BLAST and PSI-BLAST: a new generation of protein
database search programs. Nucleic Acids Res 1997;25:3389–402.
27. Fujii Y, Shimokawa H, Sekiguchi M, Nakabeppu Y. Functional sig-
nificance of the conserved residues for the 23-residue module among
MTH1 and MutT family proteins. J Biol Chem 1999;274:38251–9.
28. Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent
mutagenic substrate for DNA synthesis. Nature 1992;355:273–5.
29. Margison GP, Cooper DP, Potter PM. The E. coli ogt gene. Mutat Res
1990;233:15–21.
30. Boiteux S, O’Connor TR, Laval J. Formamidopyrimidine-DNA gly-
cosylase of Escherichia coli: cloning and sequencing of the fpg struc-
tural gene and overproduction of the protein. EMBO J
1987;6:3177–83.
31. Au KG, Clark S, Miller JH, Modrich P. Escherichia coli mutY gene
encodes an adenine glycosylase active on G-A mispairs. Proc Natl
Acad Sci U S A 1989;86:8877–81.
32. Andersson GE, Sharp PM. Codon usage in the Mycobacterium tuber-
culosis complex. Microbiology 1996;142:915–25.
33. Kruuner A, Hoffner SE, Sillastu H, Danilovits M, Levina K, Svenson
SB, et al. Spread of drug-resistant pulmonary tuberculosis in Estonia.
J Clin Microbiol 2001;39:3339–45.
34. Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B.
Mutators, population size, adaptive landscape and the adaptation of
asexual populations of bacteria. Genetics 1999;152:485–93.
35. Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH,
Godelle B. Role of mutator alleles in adaptive evolution. Nature
1997;387:700–2.
36. Rainey BP, Moxon ER. Microbiology. When being hyper keeps you
fit. Science 2000;288:1186–7.
37. Werngren J, Hoffner SE. Drug-susceptible Mycobacterium tuberculo-
sis Beijing genotypes does not develop mutation-conferred resistance
to rifampin at an elevated rate. J Clin Microbiol. In press 2003.
Address for correspondence: Brigitte Gicquel, Unité de Génétique
Mycobactérienne, Institut Pasteur, 28 rue du Dr. Roux, 75015 Paris,
France; fax: 33145688843; email: bgicquel@pasteur.fr
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 845
RESEARCH
Search ppast iissues oof EEID aat wwww.cdc.gov/eid

During 5,230 trapping nights, 672 small mammals
were trapped in the areas where most hantavirus pul-
monary syndrome (HPS) cases occur in Uruguay. Yellow
pygmy rice rats (Oligoryzomys flavescens) were the only
rodents that showed evidence of antibodies to hantavirus,
with a seroprevalence of 2.6%. The rodents were trapped
in all the explored environments, and most of the seropos-
itive rodents were found in habitats frequented by humans.
Nucleotide sequences were obtained from four HPS case-
patients and four yellow pygmy rice rats of the M genome
segment. Sequence comparison and phylogenetic analysis
showed that rodent-borne viruses and viruses from three
HPS case-patients form a well-supported clade and share
a 96.4% identity with the previously characterized Central
Plata hantavirus. These results suggest that yellow pygmy
rice rat (O. flavescens) may be the host for Central Plata, a
hantavirus associated with HPS in the southern area of
Uruguay.
T
he family Bunyaviridae consists of five genera.
Viruses in the Hantavirus genus are unique among
them because all members (except Thottapalayam virus)
are rodent-borne. Viruses in the other four genera are
arthropod-borne. Hantavirus pulmonary syndrome (HPS)
was first identified in the United States in 1993. The dis-
covery of the outbreak was followed by the identification
of Sin Nombre virus (SNV) as the primary etiologic agent
of HPS (1). Since these findings, many countries in the
Americas have identified cases and outbreaks of this syn-
drome, and several other related viruses (New World han-
taviruses) have been recognized (2–9).
New World hantaviruses are carried by different species
of sigmodontine and arvicoline rodents (Muridae). Indeed,
genetic diversity and geographic distribution of these
viruses are related to the genetic diversity, geographic dis-
tribution, and phylogenetic history of their rodent hosts. In
South America, studies of the correlation between rodent
hosts and indigenous hantaviruses are complicated by the
great diversity of sigmodontine rodents in this area. Also,
the sympatric distributions between the different species of
sigmodontine rodents in South America provide opportu-
nities for spillover infections and host-switching events
(10).
In Uruguay, the first evidence of the circulation of these
viruses came from a study of serum specimens collected
from blood donors between 1985 and 1987 that showed a
seroprevalence of 1%, as measured by indirect fluorescent
antibody (IFA) test using Hantaan and Seoul antigens (11).
Since 1997, the Ministerio de Salud Pública, through the
Departamento de Laboratorios,
began the surveillance and
diagnosis of HPS. In 2000 Padula et al. (12) reported par-
tial sequences (G1 and G2 glycoprotein) derived from two
HPS cases that occurred in Uruguay in 1997 and 1999.
These viruses clustered within a previously reported line-
age named Central Plata.
Knowledge about small mammal communities and
habitat preferences is limited in Uruguay. However, some
studies about systematic distribution, reproduction, and
cytogenetic aspects have been published (13–17).
Research regarding the distribution and habitat preferences
of the Muridae family in Uruguay is currently being con-
ducted (18–20).
The purpose of this study was to identify the carrier
rodents of hantavirus in Uruguay and their potential associ-
ation with HPS cases, to determine the prevalence of infec-
tion in different habitats, and to begin to genetically charac-
terize the hantaviruses recovered from these rodents.
846 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Yellow Pygmy Rice Rat
(Oligoryzomys flavescens) and
Hantavirus Pulmonary Syndrome
in Uruguay
Adriana Delfraro,* Mario Clara,* Lorena Tomé,* Federico Achaval,* Silvana Levis,†
Gladys Calderón,† Delia Enria,† Mario Lozano,‡ José Russi,§ and Juan Arbiza*
*Facultad de Ciencias de la Universidad de la República,
Montevideo, Uruguay; †Instituto Nacional de Enfermedades
Virales Humanas “Dr. Julio I. Maiztegui,” Pergamino, Buenos
Aires, Argentina; ‡Universidad Nacional de Quilmes, Bernal,
Argentina; and §Ministerio de Salud Pública, Montevideo,
Uruguay

Material and Methods
HPS Case Identification
National surveillance for HPS was reinforced when a
case definition was established by the Ministerio de Salud
Pública in 1997. An HPS case was suspected in a previous
healthy person with an acute febrile illness (temperature
>38°C), associated with dyspnea, acute respiratory distress
syndrome with pulmonary noncardiogenic edema, or inter-
stitial bilateral infiltrates, hypotension or shock, elevated
leukocyte count, and thrombocytopenia (21). A case of
HPS was confirmed when, in addition to clinical illness,
circulating specific hantavirus immunoglobulin (Ig) M
was detected.
Human serum samples were tested for the presence of
IgM and IgG antibodies with an enzyme-linked
immunosorbent assay (ELISA) developed by MRL
(Hantavirus ELISA IgM and Hantavirus ELISA IgG, MRL
Diagnostics, Cypress, CA). The test was used to screen
patients, and in every case, the results were confirmed by
retesting the specimens by an in-house enzyme immunoas-
say with a recombinant nucleocapsid antigen specific to
Andes virus, according to the procedure developed by
Padula et al. (12).
Site Selection
Rodent sampling was conducted at the most likely sites
of infection for known HPS case-patients and included the
places where the person had lived or worked in the 6
weeks before onset of symptoms and nearby natural habi-
tats. The trapping sites were classified as 1) domestic and
peridomestic, including all sites in the immediate vicinity
of houses, sheds, gardens, road borders, and fence lines,
and 2) rural natural ecosystems and agro-ecosystems,
including representative habitats of each area such as open
fields, cultivated areas, wetlands, shrublands, brook bor-
ders, natural forests, and artificial woods (planted by
humans) (Table 1).
The trapping expeditions were performed in the follow-
ing areas: Puntas de Valdéz (34°32′S/56°36′W) and
Piedritas (34º20′S/55°39′W) (one expedition each);
Cerrillos (34°38′S/56°19′W), Melilla (34º44′S/56°16′W),
and Sauce (34°35′S/56°08′W) (two expeditions each)
(Figure 1). The geographic area covered by the trapping
expeditions corresponded to areas where 16 HPS cases
occurred in Montevideo and Canelones, two cases
occurred in San José, and one case occurred in Florida.
The other 19 cases were dispersed in the southern half of
Uruguay, and for some of them, the probable site of infec-
tion was not clearly identified.
The landscape of Canelones, rural Montevideo, Florida,
and San José shows cultivated areas, stubble areas, shrub-
lands in the territories abandoned by rural people, range
lands, natural and artificial woodlands, small wetlands,
and small brooks. In recent years, many rural inhabitants
have migrated to the cities; abandoned farmlands have thus
been transformed into shrublands.
Small-Mammal Trapping and Processing
Small mammals were trapped in six expeditions in the
above-mentioned areas from May 1997 until September
2001. Each trapping site was sampled with Sherman live-
capture traps (model LFATDG 23 cm x 8 cm x 8.5 cm)
(Sherman Traps Inc., Tallahassee, FL). The number of
traps depended on the available area for trap placement at
each trapping site. The traps were placed at 5-m intervals
in line transects, along the different environments at the
trapping site. The traps were set out in the late afternoon
and checked in the early morning for the next two
mornings. The animals were trapped and sampled
according to established biosafety guidelines (22). Each
animal was anesthetized, and blood was collected from the
retroorbital sinus. The animals were humanely killed, and
their size, mass, sex, and reproductive status were
recorded. Samples of liver, kidney, lung, and brain were
extracted and stored in liquid nitrogen for further
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 847
RESEARCH
Table 1. Trapping efficiency/environment/species in the total of captures
a
Env
TN
C
E%
Md
Of
Od
St
Aa
No
Hb
Cl
Mm
Rr
Ca
NW
288
27
9.4
-
1
7
10
6
2
-
1
-
-
-
RB
1,010
172
17.0
-
55
4
24
11
14
-
1
63
-
-
PD
1,420
44
3.1
1
19
1
7
-
2
-
-
11
-
3
WE
265
14
5.2
-
2
-
8
-
2
2
-
-
-
-
BB
680
57
8.4
1
11
1
23
4
10
-
-
7
-
-
AG
190
50
26.3
-
9
6
9
2
6
-
1
17
-
-
SH
1,198
268
22.4
1
93
2
117
11
13
-
-
30
-
1
AW
179
40
22.3
1
4
1
-
-
3
-
-
29
2
-
T
5,230
672
12.8
4
194
22
198
34
52
2
3
157
2
4
Sp%
0.6
28.9
3.3
29.5
5.1
7.7
0.3
0.4
23.4
0.3
0.6
a
Env, environments; TN, trapping nights; C, captures; E, efficiency; NW, natural woods; RB, road borders; PD, peridomestic; WE, wetlands; BB, brook borders; AG,
agroecosystems; SH, shrublands; AW, artificial woods; T, totals; Sp%, species %; Md, Monodelphis dimidiate; Of, Oligoryzomys flavescens; Od, O. delticola; St,
Scapteromys tumidus; Aa, Akodon azarae; No, Necromys obscurus; Hb, Holochilus brasiliensis; Cl, Calomys laucha; Mm, Mus musculus; Rr, Rattus rattus; Ca, Cavia
aperea.
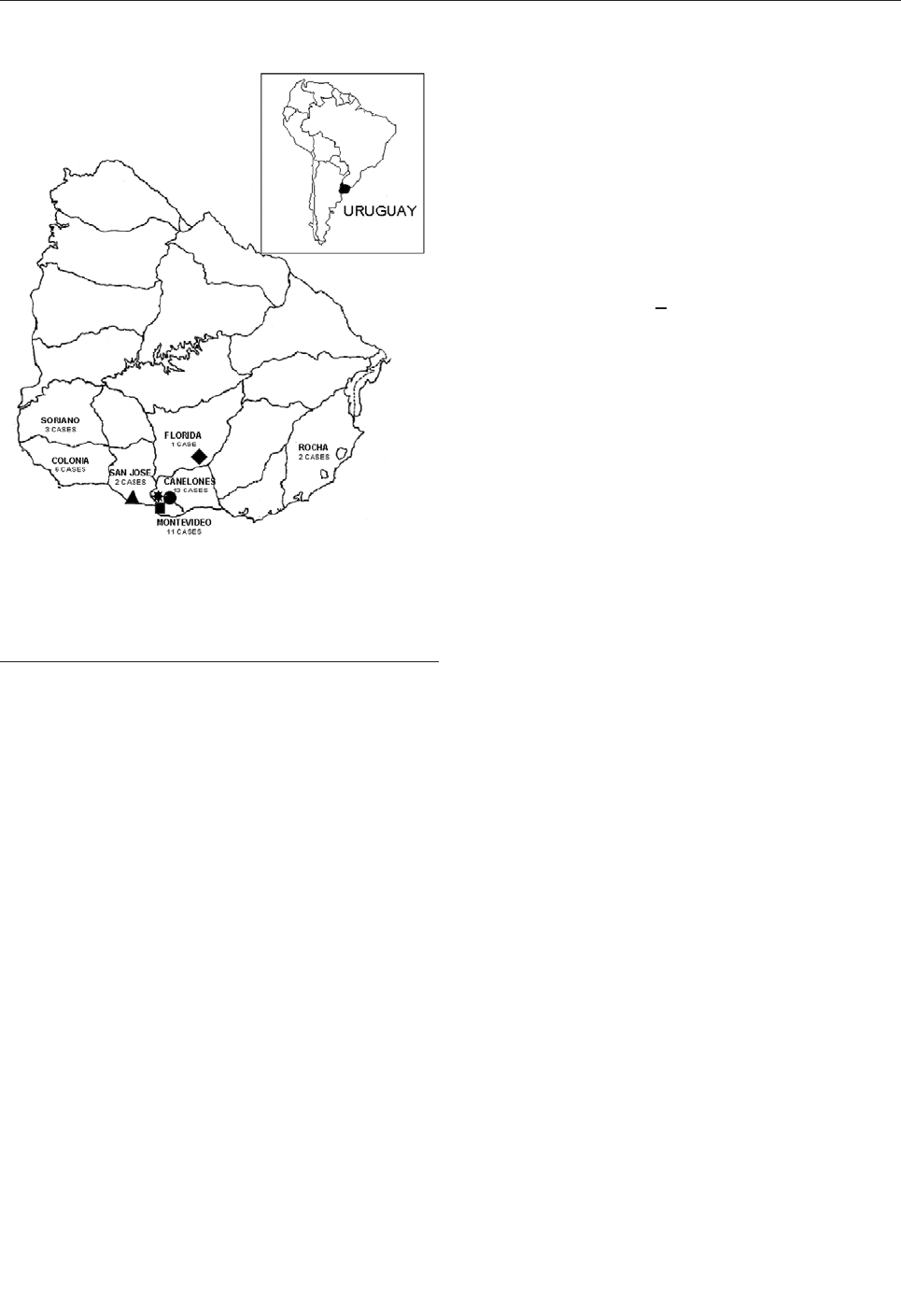
processing. The individual animals were tentatively
identified in the field by external characteristics, and the
carcasses were kept in 10% formalin. Identification was
confirmed by cranial measurements and dental
examination at the laboratory. All specimens were
deposited at the collection of the Sección Zoología
Vertebrados, Facultad de Ciencias, Universidad de la
República, Montevideo, Uruguay.
ELISA
Serologic testing of rodents was performed by IgG
ELISA. Briefly, the IgG ELISAs were performed by coat-
ing polyvinyl chloride microtiter plates (Dynex
Technologies, Chantilly, VA) overnight at 4°C with a
Lechiguanas virus (LECV) antigen (inactivated, 3 M rad
gamma-ray irradiation detergent-extracted lysate of Vero-
E6 infected cells, with a 100% infection index controlled
by indirect immunofluorescence). An uninfected Vero E6
cell culture antigen was used to determine the specificity
of mouse antibodies. Unbound antigen was removed by
washing three times with phosphate-buffered saline (PBS)-
Tween 20, 0.1% (Sigma-Aldrich, St. Louis, MO). After
blocking with PBS-Tween 20, 0.1%-dry milk 5% (37°C, 1
h), sera diluted fourfold, beginning with 1:100, were added
to react with the antigen-coated wells. Bound antigen was
measured by the use of a hyperimmune mouse ascitic fluid
and by using goat anti–Peromyscus leucopus IgG (H+L)
and goat-anti rat IgG (heavy- and light-chain–specific;
Kirkegaard & Perry Laboratories, Gaithersburg, MD) con-
jugated to horseradish peroxidase. Optical densities (ODs)
at 405 nm were recorded on a microplate spectrophotome-
ter (Labsystems Multiskan EX; Thermo Labsystems,
Finland, Vartaa, Finland), and the ODs of the uninfected
antigen-coated well were subtracted from that of its corre-
sponding viral antigen to yield the adjusted OD. A serum
dilution was considered positive if OD was >0.2 U after
adjustment. A serum titer >
400 was considered positive.
Total RNA Extraction and RT-PCR
Total RNA was extracted from lung tissue of seroposi-
tive rodents and from blood clots from HPS case-patients.
Approximately 100 mg of tissue was treated with 1 mL of
TRIzol reagent (GIBCO BRL, Life Technologies,
Rockville, MD), according to manufacturer’s instructions.
An M genome segment of the G2 glycoprotein encoding
region was amplified by using reverse transcription-poly-
merase chain reaction (RT-PCR) and specific oligonu-
cleotides as previously described by Levis et al. (4).
RT was carried out using MMLV reverse transcriptase
(GIBCO BRL) and the oligonucleotide 3348(-)
(5'CTGTCCAGATTTAGTGTTCCA 3'). cDNA was then
precipitated with NaAc (ICN Biomedicals, Costa Mesa,
CA) 3 M pH 5.6, ethanol (Merck Química Argentina,
Buenos Aires, Argentina), and lineal polyacrylamide 2.5
µg/µL (ICN Biomedicals), and resuspended in 20 µLof
double distilled water. Two microliters of first-strand
cDNA was used in the PCR reaction. Two rounds of PCR
were performed by using Taq DNA polymerase (GIBCO
BRL). The first round was performed with oligonu-
cleotides 3348 (-) and 2765 (+) (5'CTGTATGTGAGTAC-
CAAG 3'), and the second round (heminested) was per-
formed with 1 µL of first-round reaction and oligonu-
cleotides 3221 (-) (5'TCAGAAGAGCAGTCAGTGT-
CATG 3') and 2765 (+), giving a 456-nucleotide (nt) frag-
ment. PCR fragments were visualized on ethidium bro-
mide 1.5% agarose gels.
Sequencing and Phylogenetic Analysis
The PCR fragments obtained from rodent and HPS
case-patient samples were purified for further sequencing
by using the Concert rapid gel extraction system (GIBCO
BRL) or QIAquick gel extraction kit (QIAGEN Inc.,
Valencia, CA). Nucleotide sequencing was conducted by
using the oligonucleotide 2765(+) and an ABI 377 Genetic
Analyser (PE Applied Biosystems, Inc., Foster City, CA).
Alignment of sequences was done by using
CLUSTALX (1.5) (23). Phylogenetic analyses and
sequence comparison were carried out with PAUP* 4.0b10
(24) and MEGA version 2.1 (25). Maximum parsimony
848 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Figure 1. Locations of capture sites and enzyme-linked
immunosorbent assay–confirmed hantavirus pulmonary syndrome
case-patients in Uruguay. Capture sites: triangle = Puntas de
Valdéz, diamond = Piedritas, circle = Sauce, star = Cerrillos,
square = Melilla.

analysis was carried out by using the heuristic search
option. Maximum parsimony trees were searched by apply-
ing the tree bisection reconnection branch-swapping algo-
rithm. A consensus tree was obtained through 50% majori-
ty rule consensus. For the distance-based approach, MOD-
ELTEST 3.06 (26) was used to establish the most suitable
model of DNA substitution that best fitted our dataset, and
a phylogenetic tree was obtained by using the neighbor-
joining algorithm. Bootstrap analysis (27) was performed
to estimate topologic accuracy of the trees (500 replicates),
and only values >70% were considered significant.
For comparison, existing sequence data from GenBank
were used: hantavirus sequences from Argentina
(GenBank accession nos. AF028023 to AF028027,
AF028029 to AF028063), Central Plata genotype from
Uruguay (GenBank accession nos. AY101184 and
AY101185), and Sin Nombre virus (L37903, isolate
NMR11); the last one was used as outgroup.
Results
HPS Cases
From April 1997 to August 2002, 38 cases of HPS were
confirmed by ELISA, with a fatality rate of 21.0%.
Twenty-four (63.2%) of these cases occurred in rural or
suburban areas of Montevideo and Canelones, 6 (15.7%)
in Colonia, 3 in Soriano (7.9%), 2 in San José (5.3%), 2 in
Rocha (5.35%), and 1 in Florida (2.6%) (Figure 1). As of
August 2002, HPS cases in Uruguay had occurred in the
southern half of the country.
Distribution of Rodents by Species and Capture Site
During 5,230 trap-nights, 672 small mammals were
collected (trap success = 12.8%). The trapped small mam-
mals belonged to two families (Muridae and Caviidae)
within the order Rodentia and one family (Didelphidae) in
the order Didelphimorphia. The mammals belonged to 11
species, 75.1% of the captured animals corresponded to the
Sigmodontinae subfamily, 23.7 % to the Murinae subfam-
ily, 0.6% to the Caviidae family, and 0.6% to the
Didelphidae family (Table 1).
Captures and percentage of trap success by habitats
were as follows: natural woodlands, 27 (9.4%) of 288;
road borders, 172 (17.0%) of 1,010, peridomestic areas, 44
(3.1%) of 1,420; wetlands,14 (5.2%) of 65; brook borders,
57 (8.4%) of 680; agroecosystems, 50 (26.3% of 190;
shrublands, 268 (22.4%) of 1,198; and in artificial wood-
lands, 40 (22.3%) of 179 (Table 1). The most common cap-
tured small mammals were the following: swamp rats
(Scapteromys tumidus), 198 (29.5%); yellow pygmy rice
rats (Oligoryzomys flavescens), 194 (28.9%); and house
mice (Mus musculus), 157 (23.4%) (Table 1). No sigmod-
ontine rodents were found inside the houses, where only
house mice and black rats (Rattus rattus) were found.
Yellow pygmy rice rats were found in areas of human dis-
turbance such as peridomestic areas, agroecosystems, road
borders, and shrublands. We found that the trapping suc-
cess in these sites was higher than in natural areas. As
shown in Table 1, yellow pygmy rice rats were found in all
of the habitats where traps were set.
Screening for Hantavirus Infection of Rodents
Serum specimens collected from rodents were screened
for IgG antibodies to LECV by ELISA. As mentioned
above, 672 small mammals were trapped in six areas
where HPS cases occurred between 1997 and 2001. Anti-
LECV antibodies were detected in five rodents (O.
flavescens) from four different locations. Absorbances
with LECV antigen of positive samples screened at 1:400
dilution were at least fourfold the absorbance of the nega-
tive control. Further titration showed that three samples
had titers >1:1,600, and one had a titer >1:6,400 (Table 2).
The proportion of positive rodents in the different locali-
ties ranged from 2.1% to 2.9%. Piedritas was the only
locality where no antibody-positive rodents were recorded
(Table 3).
Total RNA Isolation, RT-PCR, and Sequence Analysis
Total viral RNA was extracted from the lungs of the
five seropositive yellow pygmy rice rats and blood clots
from four case-patients. RT of viral RNA and PCR ampli-
fication of a 456-nt fragment of the G2
glycoprotein–encoding region of the virus M genome seg-
ment (bases corresponding to LECV 2,805–3,215) and
nucleotide sequences were obtained from four rodent sam-
ples and four human blood clots. Amplified DNA was not
recovered from the rodent sample CE155, which had the
lower antibody titer (Table 3). A 292-nt segment (LECV
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 849
RESEARCH
Table 2. Hantavirus-seropositive rodents found in the different geographic areas where captures were performed
Sample
Rodent species
Geographic area
Habitat
Antibody titer (arbitrary units)
U89
a
Oligoryzomys flavescens
Puntas de Valdéz (San José)
Road border
1,600
SA63
a
O. flavescens
Sauce (Canelones)
Peridomestic
1,600
Ce20
a
O. flavescens
Melilla (Montevideo)
Peridomestic
>6,400
Ce22
a
O. flavescens
Melilla (Montevideo)
Shrublands
1,600
Ce155
a
O. flavescens
Cerrillos (Canelones)
Shrublands
400
a
Specimens deposited at the Specimen Collection of the Sección Zoología Vertebrados, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay with
the following numbers: U89=ZVC-M2154, SA63=ZVC-M2155, Ce20=ZVC-M2156, Ce22=ZVC-M2157, and Ce155=ZVC-M2158.

2,815–3,106 G2 glycoprotein–encoding region) was used
for further comparison and phylogenetic analysis.
Sequence Comparison and Phylogenetic Analysis
For phylogenetic comparisons, a 292-nt fragment of the
M gene from lung RNA of four yellow pygmy rice rats
(GenBank accession nos. AY204677 to AY204680), as
well as clot RNA from four HPS case-patients (GenBank
accession nos. AF283896 to AF283899) was used. These
nucleotide sequences were compared with the equivalent
region of published hantavirus sequences. Phylogenetic
analysis indicated that two previously known hantavirus
genotypes are circulating in Uruguay: Central Plata and
LEC (Figure 2).
By both maximum parsimony and distance-based
analysis, the four sequences recovered from Uruguayan
yellow pygmy rice rats were closely related to each other
and formed a monophyletic group with the hantavirus
sequences derived from three HPS case-patients from
Canelones and Montevideo and two HPS case-patients
from the same geographic area, previously characterized as
Central Plata. This clade was supported by high bootstrap
values (Figure 2A,B). Comparison of these sequences at
the nucleotide level showed 96.4% identity. The most
closely related genotype was LEC, with 87.9% identity,
followed by Bermejo (85.0%), Orán (83.1%), Andes
(82.5%), and Hu39694 (82.1%). The less-related geno-
types of the Argentinean hantaviruses were Maciel (79.3%)
and Pergamino (78.6%). One viral sequence from an HPS
case-patient in Soriano clustered into LEC genotype.
Conclusion
Most HPS cases were in rural and suburban areas of
Montevideo and Canelones (24 of 38 cases) (Figure 1) in
southern Uruguay. Rodent sampling was conducted at the
most likely sites of infection for known HPS case-patients.
The trapping success rate was higher in the environ-
ments influenced by humans (agroecosystems, road bor-
ders, shrublands, artificial woods, peridomestic areas) than
in the natural areas (natural woods, wetlands, brook bor-
ders): 574 (85.4%) of trapped individual animals were cap-
tured in environments influenced by humans, and 98
(14.6%) were captured in natural environments. Swamp
rats, yellow pygmy rice rats, and house mice were the most
frequently trapped species. Of 44 (3.1%) rodents trapped
in peridomestic environments, 19 (43.2%) were yellow
pygmy rice rats (Table 1). Five seropositive yellow pygmy
rice rats were captured in modified environments: one was
captured along a road border, two were captured in perido-
mestic environments, and two were captured in shrub-
lands, at <
150 m from homes. Three of five seropositive
rodents were therefore trapped in environments frequented
by humans (road borders and peridomestic environments).
These findings could indicate an increased risk for infec-
tion for human inhabitants.
Yellow pygmy rice rats were the only rodents that
showed evidence of antibodies to hantavirus, with a preva-
lence of 2.6%. Researchers have found that hantavirus
seroprevalence in rodents may vary widely, according to
the season, geographic area, altitude, and rodent species
analyzed (29–33). We found that the percentage of
seropositive rodents (2.6%) is the same as encountered in
the central zone of Argentina (2.6%) (32), although the
habitats are not similar to the southern area of Uruguay. In
Uruguay, we found that only yellow pygmy rice rats were
antibody positive, while in central Argentina seropositive
yellow pygmy rice rats, Azara’s field mice (Akodon
azarae), dark mice (Necromys benefactus), and small
water rats (Holochilus brasiliensis) were found (2–4). In
the different locations in Uruguay, seroprevalence was
similar, ranging from 2.1% to 2.9%. In Piedritas, where no
positive rodents were found, only four yellow pygmy rice
rats were trapped. All seropositive rodents in Uruguay
were adult males, which is consistent with horizontal
transmission and in accordance with the findings of sever-
al authors (30–32).
The phylogenetic analysis on a 292-nt region of the M
segment showed that these rodent sequences clustered
together with those from five Uruguayan HPS case-
patients from the same geographic area (Canelones and
Montevideo); these data suggest that the yellow pygmy
rice rat can be considered as the putative reservoir host for
Central Plata hantavirus in this region of Uruguay. This
study also showed the circulation of LEC genotype in the
western location of Soriano, 250 km from Montevideo,
separated from the Argentinean central HPS-endemic area
by the Uruguay River. This virus shared a 99% identity at
the nucleotide level with LEC genotype.
Phylogenetic analysis shows that the genotype Central
Plata recovered from rodents and HPS case-patient from
850 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
Table 3. Small mammals trapped in the different sites, number and % of positives Monodelphis dimidiate
Area
Md
+/%
Of
+/%
Od
+/%
St
+/%
Aa
+/%
No
+/%
Hb
+/%
Cl
+/%
Mm
+/%
Rr
+/%
Ca
+/%
Total
PV
-
-/-
38
1/2.6
-
-/-
-
-/-
10
0/0
-
-/-
-
-/-
2
0/0
81
0/0
-
-/-
-
-/-
131
Ce
-
-/-
47
1/2.1
9
0/0
52
0/0
4
0/0
17
0/0
2
0/0
-
-/-
23
0/0
-
-/-
-
-/-
153
Me
-
-/-
67
2/2.9
8
0/0
37
0/0
-
-/-
17
0/0
-
-/-
1
0/0
10
0/0
-
-/-
3
0/0
297
Pi
-
-/-
4
0/0
-
-/-
8
0/0
14
0/0
-
-/-
-
-/-
-
-/-
2
0/0
-
-/-
-
-/-
28
Sa
3
0/0
42
1/2.4
1
0/0
92
0/0
5
0/0
18
0/0
-
-/-
-
-/-
12
0/0
-
-/-
-
-/-
173
Ca
1
0/0
-
-/-
-
-/-
9
0/0
1
0/0
-
-/-
-
-/-
-
-/-
29
0/0
2
0/0
1
0/0
43
Total
4
0/0
194
5/2.6
22
0/0
198
0/0
34
0/0
52
0/0
2
0/0
3
0/0
157
0/0
2
0/0
4
0/0
672
a
Md, Monodelphis dimidiate; Of, Oligoryzomys flavescens; Od, O. delticola; St, Scapteromys tumidus, Aa, Akodon azarae, No, Necromys obscurus; Hb, Holochilus brasiliensis; Cl, Calomys laucha; Mm, Mus musculus; Rr
,Rattus rattus; Ca, Cavia aperea; PV, Puntas de Valdéz; Ce, Cerrillos; Me, Melilla; Pi, Piedritas, Sa, Sauce; Ca, Canelones. O. flavescens, S. tumidus, and M. musculus were the most frequently captured rodents.
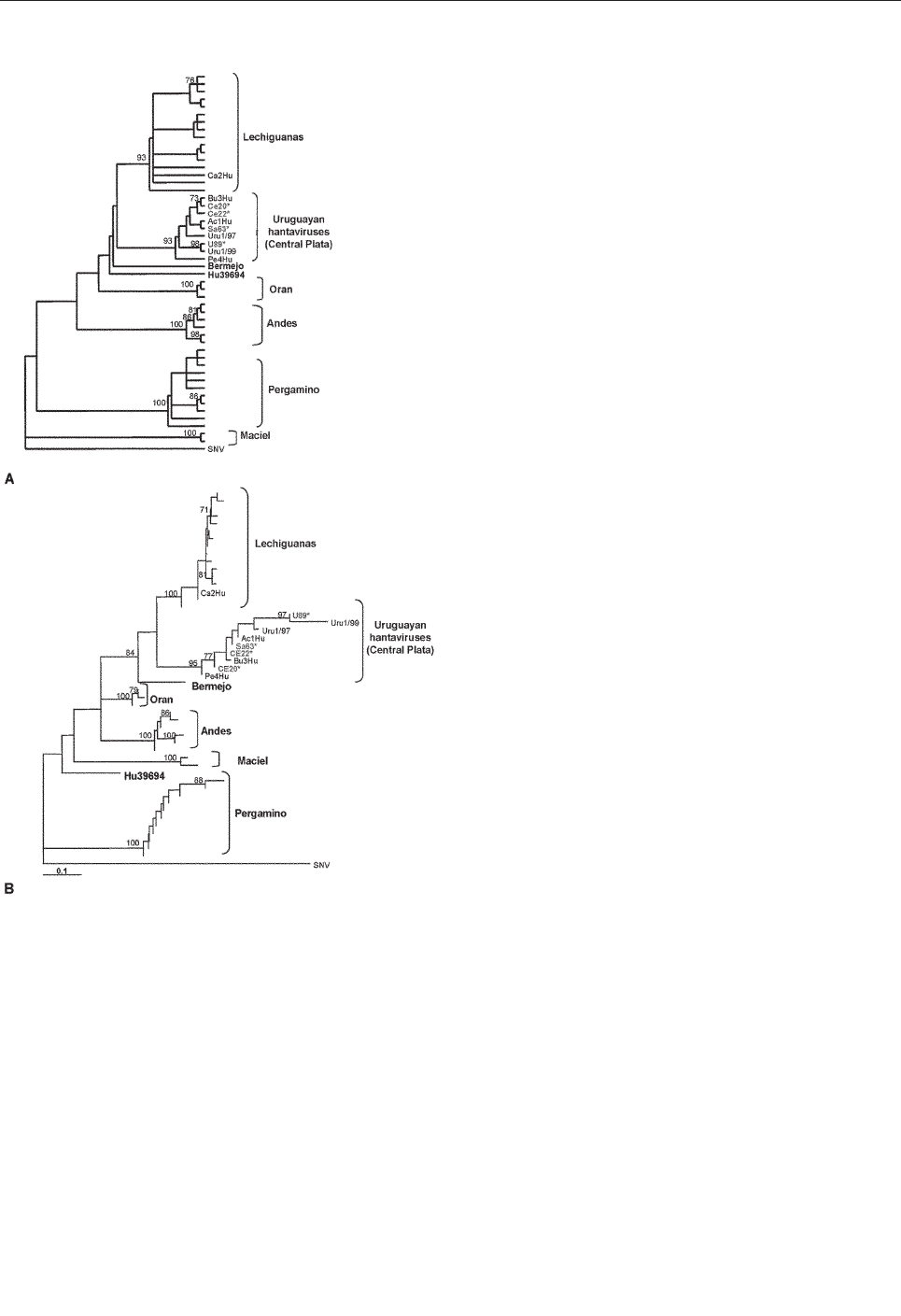
Canelones, San José, and Melilla is phylogenetically dis-
tinct from (although related to) the previously described
LEC genotype, whose reservoir host in Argentina is also
the yellow pygmy rice rat. Hantaviruses have been associ-
ated with subspecies of closely related rodents: Sin
Nombre–like hantaviruses with mice from the genus
Peromyscus (34) and Andes virus recovered in southwest-
ern Argentina and Orán virus in northwestern Argentina,
both recovered from long tailed pygmy rice rats (O. longi-
caudatus) (4). Recent studies have shown that these two
rodent populations differ with respect to their mitochondr-
ial DNA (10). This fact raises the question of whether
rodents morphologically identified as O. flavescens in
Uruguay are indeed a different subspecies of O. flavescens
in Argentina. Further experiments will be needed to iden-
tify both the interspecific and intraspecific phylogenetic
relationships of O. flavescens in these regions.
Acknowledgments
We thank Noemí Pini and Juan Cristina for critical reading
and useful suggestions; Raúl Maneyro, Juan José Porta, and Ana
Liñares for rodent trapping; W. Slenczka for contributing biosafe-
ty equipment; Leandro Jones, Mónica Galiano, and Guillermo
D´Elía for help in Modeltest implementation.
This research was supported in part by grant no. 37/2000
from the Pan American Health Organization/Red
Latinoamericana de Ciencias Biológicas (Silvana Levis, Mario
Lozano, Juan Arbiza, and Mario Clara). Mario E. Lozano is a
research career member of Consejo Nacional de Investigaciones
Cientificas y Técnicas, Argentina. A. Delfraro received a predoc-
toral training fellowship from Programa de desarrollo de las
Ciencias Básicas. L. Tome was supported by a grant from
Laboratorio Santa Elena.
Adriana Delfraro is a doctoral student in microbiology and
assistant professor in the Sección Virología of the Facultad de
Ciencias, Universidad de la República. She also worked in the
Hantavirus Program of Uruguay in the Departamento de
Laboratorios of the Ministerio de Salud Pública, Montevideo,
Uruguay, from its beginning in 1997 until 2002.
References
1. Nichol ST, Spiropoulou C, Morzunov S, Rollin P, Ksiazek G,
Feldman H, et al. Genetic identification of a Hantavirus associated
with an outbreak of acute respiratory illness. Science
1993;262:914–7.
2. López N, Padula P, Rossi C, Lazaro ME, Franze-Fernandez MT.
Genetic identification of a new hantavirus causing severe pulmonary
syndrome in Argentina. Virology 1996;220:223–6.
3. López N, Padula P, Rossi C, Miguel S, Edelstein A, Ramirez E, et al.
Genetic characterization and phylogeny of Andes virus and variants
from Argentina and Chile. Virus Res 1997;50:77–84.
4. Levis S, Morzunov S, Rowe J, Enria D, Pini N, Calderon G, et al.
Genetic diversity and epidemiology of hantaviruses in Argentina. J
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 851
RESEARCH
Figure 2. A: Maximum parsimony phylogenetic tree B: Distance-
based phylogenetic tree. The tree was built under the Tamura-Nei
model of DNA substitution with estimation of the shape parameter
of the gamma distribution (28). This model and the associated
parameters resulted from testing our dataset with the program
MODELTEST 3.06 (26). Both trees include Argentinean and
Uruguayan hantaviruses from hantavirus pulmonary syndrome
(HPS) case-patients and rodents. Hantavirus sequences from
HPS case-patients in Uruguay: Ac1Hu, Ca2Hu, Bu3Hu, Pe4Hu,
Uru1/97, Uru1/99. Hantavirus sequences from yellow pygmy rice
rats: U89, Sa63, Ce20, Ce22. Sin Nombre virus was used as out-
group.*Specimens deposited at the Collection of the Sección
Zoología Vertebrados, Facultad de Ciencias, Universidad de la
República, Montevideo, Uruguay, with the following numbers:
U89=ZVC-M2154, SA63=ZVC-M2155, Ce20=ZVC-M2156,
Ce22=ZVC-M2157, and Ce155=ZVC-M2158.

Infect Dis 1998;177:529–38.
5. Vasconcelos M, Lima V, Iversson L, Rosa M, Travassos da Rosa E,
Travassos da Rosa A, et al. Hantavirus pulmonary syndrome in the
rural area of Juquitiba, Sao Paulo, Metropolitan Area, Brasil. Rev Inst
Med Trop (S. Paulo) 1997;39:237–8.
6. Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills
J, et al. Laguna Negra virus associated with HPS in western Paraguay
and Bolivia. Virology 1997;238:115–27.
7. Johnson AM, de Souza LTM, Ferreira IB, Pereira LE, Ksiazek G,
Rollin P, et al. Genetic investigation of novel hantaviruses causing
fatal HPS in Brazil. J Med Virol 1999;59:527–35.
8. Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, Ksiazek TG, Kitsutani
PT, et al. Hantavirus pulmonary syndrome in Panama: identification
of novel hantaviruses and their likely reservoirs. Virology
2000;277:14–9.
9. Padula P, Gonzalez Della Valle M, Garcia Alai M, Cortada P, Villagra
M, Gianella A. Andes virus and first case report of Bermejo virus
causing fatal pulmonary syndrome. Emerg Infect Dis 2002 8:437–9.
10. Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of
hantaviruses and their rodent hosts. In: Schmaljohn CS, Nichol ST,
editors. Hantaviruses. Berlin: Springer-Verlag; 2001. p. 47–75.
11. Weissenbacher M, Cura E, Segura E, Hortal M, Baek L, Kyu Chu Y,
et al. Serological evidence of human hantavirus infection in
Argentina, Bolivia and Uruguay. Medicina (Buenos Aires)
1996;56:17–22.
12. Padula P, Colavecchia S, Martínez P, González Della Valle M,
Edelstein A, Miguel S, et al. Genetic diversity, distribution and
serological features of hantavirus infection in five countries in South
America. J Clin Microbiol 2000;38:3029–35.
13. Devicenzi GJ. Mamíferos del Uruguay. Anales del Museo de Historia
Natural de Montevideo 1935;4:1–96.
14. Langguth A. Contribución al conocimiento de los Cricetinae del
Uruguay (Especies halladas en los regurgitos de búho). Anais do
Segundo Congresso Latino-Americano de Zoología (Sao Paulo,
16–21/7/1962). 1965;2:327–35. San Pablo, Brasil: Sociedad
Latinoamericana de Zoología. Secretaria de Agricultura de San
Pablo.
15. Barlow JC. Observations on the biology of rodents in Uruguay. Life
Sciences Contributions of Royal Ontario Museum, 1969. Toronto:
The Museum, 1969. p. 75.
16. Brum-Zorrilla N. Investigaciones citogenéticas sobre algunas
especies de Cricetinae (Rodentia) del Uruguay. Anais do Segundo
Congresso Latino-Americano de Zoología (Sao Paulo, 16-
21/7/1962), 1965, 2:315–20. San Pablo, Brasil: Sociedad
Latinoamericana de Zoología. Secretaria de Agricultura de San
Pablo.
17. Brum-Zorrilla N, Degiovanangelo C, Di Tomaso MV. Estado actual
de los estudios cromosómicos en mamíferos del Uruguay. Actas de
las Jornadas de Zoología del Uruguay (Montevideo, 23-28/9/1985);
1985:77–8. Montevideo, Uruguay: Sociedad Zoologica del Uruguay;
1985.
18. Clara M, Achaval F. Datos preliminares sobre reservorios de
Hantavirus en Uruguay. Bol. Soc. Zool. Uruguay (2ª época). Act. V.
Jorn Zool Uruguay 1999;11:12.
19. Clara M, Achaval F. Preferencia de hábitat de algunas especies de
Muridae (Rodentia) del Uruguay. Act. VI . Jorn Zool Uruguay 2001;
33.
20. Clara M, Achaval F, Arbiza J, Delfraro A, Tomé L, Lozano M, Enría
D. Caracterización de Hantavirus y Arenavirus en sus reservorios
naturales en Uruguay. Act. VI . Jorn Zool Uruguay 2001;34.
21. Aguila J, Roucco G, Curto S, Willat G, Russi J, Gaye G, Salvatella R.
Guía para la vigilancia epidemiológica de los casos de Síndrome
Pulmonar por Hantavirus. Montevideo, Uruguay: Ministerio de Salud
Pública; 1998.
22. Mills J, Childs J, Thomas G, Ksiazek G, Peters CJ. Methods for trap-
ping and sampling small mammals for virologic testing. editors.
Atlanta: Centers for Disease Control and Prevention; 1995.
23. Thompson JD, GibsonTJ, Plewniak F, Jeanmougin F, Higgins DG.
The ClustalX windows interface: flexible strategies for multiple
sequence alignment aided by quality analysis tools. Nucleic Acids
Res 1997;24:4876–82.
24. Swofford, DL. PAUP*: Phylogenetic analysis using parsimony (and
other methods), v. 4.0b4a. 2000. Sunderland (MA): Sinauer
Associates, Inc. Publishers; 2000.
25. Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evo-
lutionary genetics analysis software. Tempe (AZ): Arizona State
University; 2001.
26. Posada D, Crandall KA. MODELTEST: testing the model of DNA
substitution. Bioinformatics Application Note 1998;14:817–8.
27. Felsenstein J. Confidence limits on phylogenies: an approach using
the bootstrap. 1985. Evolution 1985;39:783–91.
28. Tamura K, Nei M. Estimation of the number of nucleotide substitu-
tions in the control region of mitochondrial DNA in humans and
chimpanzees. Mol Biol Evol 1993;10:512–26.
29. Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S,
Maupin GO, et al. Serologic and genetic identification of Peromyscus
maniculatus as the primary rodent reservoir for a new hantavirus in
the southwestern United States. J Infect Dis 1994;169:1271–80.
30. Kuenzi AJ, Morrison ML, Swann DE, Hardy PC, Downard GT. A
longitudinal study of Sin Nombre virus prevalence in rodents, south-
eastern Arizona. Emerg Infect Dis 1999;5:113–7.
31. Mills JN, Ksiazek TG, Peters CJ, Childs JE. Long-term studies of
Hantavirus reservoir populations in the southwestern United States: a
synthesis. Emerg Infect Dis 1999;5:135–41.
32. Calderón G, Pini N, Bolpe J, Levis S, Mills, J, Segura E, et al.
Hantavirus reservoir hosts associated with peridomestic habitats in
Argentina. Emerg Infect Dis 1999;5:792–7.
33. Kuenzi AJ, Douglass RJ, White Jr D, Bond CW, Mills JN. Antibody
to Sin Nombre virus in rodents associated with peridomestic habitats
in west central Montana. Am J Trop Med Hyg 2001;64:137–46.
34. Morzunov SP, Rowe JE, Ksiazek TG, Peters CJ, St Jeor SC, Nichol,
ST. Genetic analysis of the diversity and origin of hantaviruses in
Peromyscus leucopus mice in North America. J Virol 1998;72:57–64.
Address for correspondence: Juan Arbiza, Sección Virología, Facultad de
Ciencias. Iguá 4225. CP 11400; fax: 5982-5258617; email:jarbiza@
fcien.edu.uy
852 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
RESEARCH
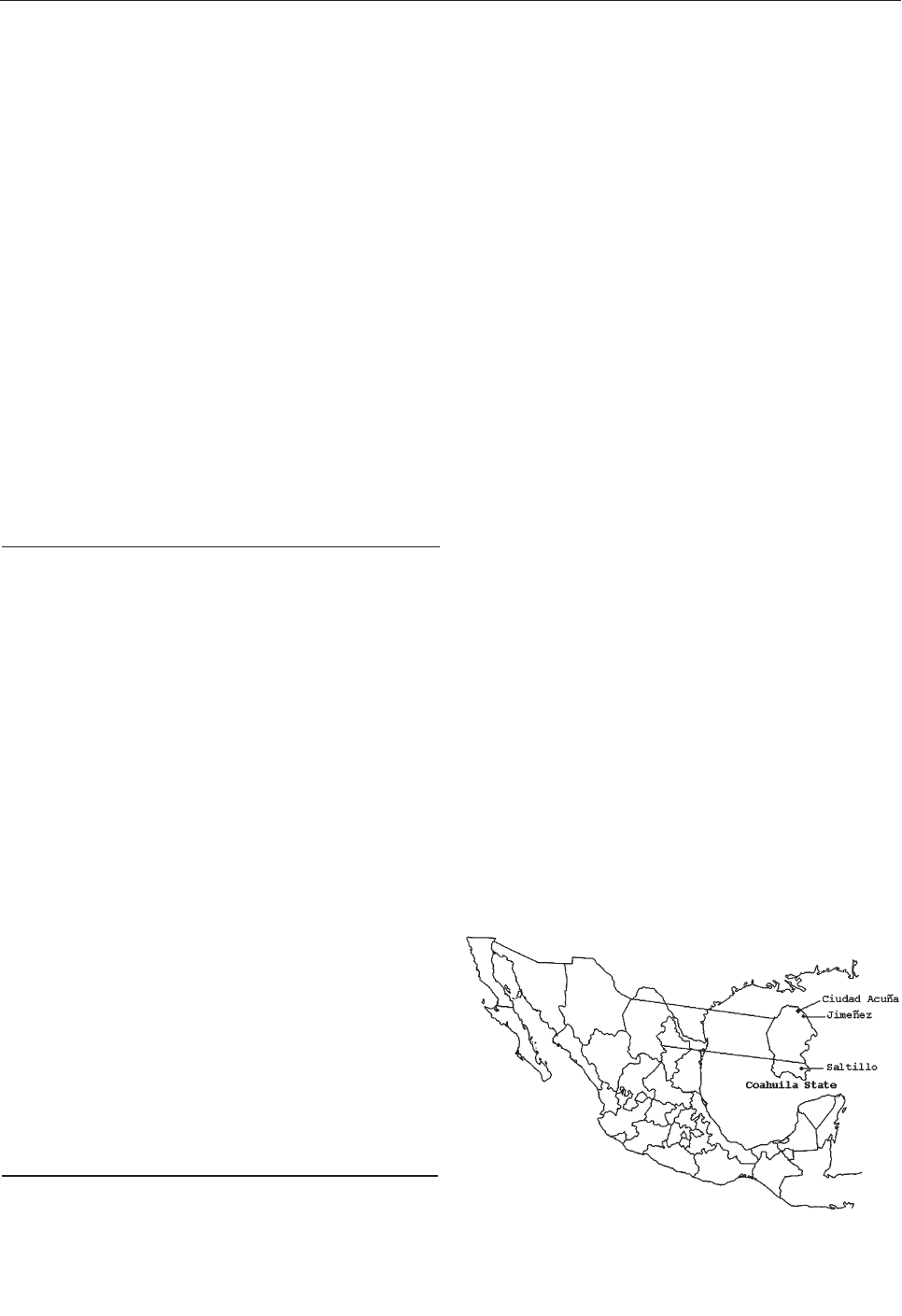
Serologic Evidence
of West Nile Virus
Infection in
Horses, Coahuila
State, Mexico
Bradley J. Blitvich,* Ildefonso Fernandez-Salas,†
Juan F. Contreras-Cordero,† Nicole L. Marlenee,*
Jose I. Gonzalez-Rojas,† Nicholas Komar,‡
Duane J. Gubler,‡ Charles H. Calisher,*
and Barry J. Beaty*
Serum samples were obtained from 24 horses in the
State of Coahuila, Mexico, in December 2002. Antibodies to
West Nile virus were detected by epitope-blocking enzyme-
linked immunosorbent assay and confirmed by plaque
reduction neutralization test in 15 (62.5%) horses. We
report the first West Nile virus activity in northern Mexico.
W
est Nile virus (WNV; family Flaviviridae, genus
Flavivirus) is a member of the Japanese encephalitis
virus complex, which also includes Japanese encephalitis
virus, Saint Louis encephalitis virus (SLEV), and Murray
Valley encephalitis virus (1). These viruses are maintained
in cycles between mosquitoes and birds (2). The principal
vectors for WNV are Culex species mosquitoes, and many
species of wild birds act as vertebrate hosts (3). Humans,
horses, and other mammals usually serve as dead-end
hosts. In humans and equines, WNV infection is usually
asymptomatic or characterized by a mild febrile illness,
although fatal meningoencephalitis or encephalitis may
occur (3–5). WNV has a broad geographic distribution,
recently including North America (4,5). The initial out-
break of WNV in North America was recognized in New
York City in August 1999. Since then, WNV’s geographic
range has increased. WNV activity has now been reported
in 44 states in the United States, the District of Columbia,
and 5 of the 10 Canadian provinces (6,7).
In anticipation of the possible emergence of WNV into
Mexico, we conducted equine infection surveillance in the
northeastern states of Mexico. Coahuila State is bordered
on the north by Texas. WNV activity has been detected in
204 (80%) of 254 Texas counties, including most counties
that border Coahuila State (8). Therefore, Coahuila State
was considered to be a likely point of incursion of WNV
into Mexico from the United States.
Case Study
We present data from a small equine serosurvey con-
ducted in Coahuila State in December 2002. A more exten-
sive equine serosurvey is currently under way and will be
described in detail elsewhere (B.J. Beaty, unpub. data). In
the present study, blood samples were taken from 24
domestic horses at study sites located in Ciudad Acuña,
Jiménez, and Saltillo (Figure). The Ciudad Acuña and
Jiménez sites are approximately 40 km apart, and both are
<15 km from the Texas border. Saltillo is located in the
southeast region of Coahuila State and is approximately
220 km from Texas. All study sites are privately owned
ranches.
The climatic conditions of the three study sites are sim-
ilar and can be described as hot, dry, and arid. The average
annual temperature ranges from 18°C to 22°C. The aver-
age rainfall is from 100 to 300 mm per year. The sites in
Ciudad Acuña and Jiménez are approximately 300 m
above sea level. The Saltillo site is situated at an elevation
of approximately 1500 m.
A horse from the Ciudad Acuña ranch died October 17,
3 days after being observed with neurologic signs. The
case was not reported immediately, and we were unable to
obtain a tissue specimen postmortem. On December 19,
with the assistance of a local veterinary practitioner, we
sampled 14 horses at this site, 5 of which had developed
neurologic disease in mid- to late October. Clinical symp-
toms included ataxia, weakness of limbs, trembling, and
anxiety. All five horses survived. The horses with clinical
signs were from 1 to 5 years of age; three were male, and
two were female. Ages and sexes of horses without clini-
cal symptoms were not documented. The veterinarian
reported a great abundance of mosquitoes in the area.
Another six horses were sampled in Jiménez and four in
Saltillo, none of which had signs of illness. According to
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 853
DISPATCHES
*Colorado State University, Fort Collins, Colorado, USA;
†Universidad Autonoma de Nuevo Leon, Nuevo Leon, Mexico;
and ‡Centers for Disease Control and Prevention, Fort Collins,
Colorado, USA
Figure. Geographic location of West Nile virus study sites in
Coahuila State, Mexico.
the owners, none of the horses had ever been outside the
State of Coahuila, and none of the horses had been vacci-
nated against WNV.
All serum samples were tested for antibodies to WNV
by epitope-blocking enzyme-linked immunosorbent assay
(ELISA). Blocking ELISAs were performed by using the
WNV-specific monoclonal antibody (MAb) 3.1112G, as
previously described (9). The ability of the Mexican horse
serum samples to block the binding of the MAb to WNV
antigen was compared to the blocking ability of horse
serum without antibody to WNV (Vector Laboratories,
Burlingame, CA). Data were expressed as relative percent-
ages by using the formula of Hall et al., (10). Previously,
we considered an inhibition value >
30% to indicate the
presence of viral antibodies (9). Recently, we have shown
that ELISAs performed with MAb 3.1112G detect WNV
antibodies in various vertebrate species, including horses
(9,11).
Fourteen serum samples were positive in blocking
ELISA that utilized MAb 3.1112G (Table). Serum from
another horse (H-16) inhibited the binding of MAb by
25%, which is close to the diagnostic criterion. Previously,
we observed that the nonspecific inhibition values for
serum samples from noninfected control birds ranged from
0% to 24.3% (9). Therefore, if we used a less stringent
threshold value of >25%, this serum could be considered
positive for WNV antibodies.
To validate the above assays, we tested serum samples
for neutralizing antibodies to WNV and SLEV by plaque
reduction neutralization assay (PRNT). Testing for neutral-
izing antibody to SLEV was important because this virus
is enzootic in the Americas and antibodies to WNV and
SLEV often cross-react. Furthermore, horses are suscepti-
ble to SLEV infection, although clinical manifestations
have not been reported (12). Viral isolates of WNV (strain
NY99-35261-11) and SLEV (strain TBH-28) were
obtained from the World Health Organization Center for
Arbovirus Reference and Research, maintained at the
Centers for Disease Control and Prevention, Division of
Vector-Borne Infectious Diseases, Fort Collins, CO.
PRNTs were performed by using Vero cells. Serum sam-
ples were tested by using a starting dilution of 1:20. Titers
were expressed as the reciprocal of serum dilutions reduc-
ing the number of plaques that were >90% (PRNT
90
).
854 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Table. Summary of serologic data for horses in Coahuila State, Mexico
a
PRNT
90
titer
c
Horse
Study site
Clinical illness
% inhibition by
ELISA
b
WNV
SLEV
PRNT diagnosis
H-1
Ciudad Acuña
No
90
>320
—d
WNV
H-2
Ciudad Acuña
No
5
—
—
Negative
H-3
Ciudad Acuña
No
0
—
—
Negative
H-4
Ciudad Acuña
No
0
—
—
Negative
H-5
Ciudad Acuña
No
90
>320
40
WNV
H-6
Ciudad Acuña
No
93
>320
—
WNV
H-7
Ciudad Acuña
No
93
>320
—
WNV
H-8
Ciudad Acuña
Yes
93
>320
—
WNV
H-9
Ciudad Acuña
Yes
86
>320
—
WNV
H-10
Ciudad Acuña
Yes
90
>320
20
WNV
H-11
Ciudad Acuña
No
89
>320
—
WNV
H-12
Ciudad Acuña
No
92
>320
20
WNV
H-13
Ciudad Acuña
Yes
91
>320
—
WNV
H-14
Ciudad Acuña
Yes
78
>320
20
WNV
H-15
Jiménez
No
82
>320
—
WNV
H-16
Jiménez
No
25
40
—
WNV
H-17
Jiménez
No
7
—
—
Negative
H-18
Jiménez
No
93
>320
20
WNV
H-19
Jiménez
No
0
20
20
Flavivirus
H-20
Jiménez
No
47
40
—
WNV
H-21
Saltillo
No
9
—
—
Negative
H-22
Saltillo
No
15
—
—
Negative
H-23
Saltillo
No
12
—
—
Negative
H-24
Saltillo
No
11
—
—
Negative
a
ELISA, enzyme-linked immunosorbent assay; PRNT, plaque reduction neutralization test; WNV, West Nile virus; SLEV, Saint Louis encephalitis virus
.
b
Inhibition values >30% are considered significant.
c
Neutralizing antibodies to WNV in selected horse serum samples were confirmed at Centers for Disease Control and Prevention-Division of Viral and Bacterial
Infectious Diseases by PRNT.
d
—, <20.

Conclusions
Overall, PRNT and ELISA data were in concordance.
Fifteen (62.5%) horses were considered to be seropositive
for WNV by PRNT because the antibody titers for WNV
were greater than or equal to fourfold higher that the cor-
responding SLEV titer (Table). These 15 were the same
serum samples that had inhibition values of >25% by
ELISA. Of these, 11 horses were from Ciudad Acuña, and
4 were from Jiménez. Evidence for WNV infections was
detected in 5 (100%) of 5 horses with clinical symptoms,
and 10 (52.6%) of 19 horses without clinical symptoms.
Therefore, the rate of asymptomatic seropositivity was
high, with 10 (66.7%) of 15 WNV-infected horses showing
no signs of illness. Similarly, 21 (58.3%) of 36 WNV-
infected horses sampled during a serosurvey in New York
in 1999 showed no clinical signs (13). However, the sam-
ple population (n=24) in the present serosurvey was
notably small, and data from our large equine serosurvey
will provide a more reliable estimate of the asymptomatic
seropositivity rate.
We were unable to detect RNA in any horse serum by
reverse-transcription polymerase chain reaction with
WNV-specific primers (14). We plan to isolate and ampli-
fy WNV RNA sequences from tissue specimens obtained
from seropositive horses, as well as from birds, in future
studies.
We are currently conducting avian infection surveil-
lance in the State of Coahuila and the neighboring states of
Tamaulipas and Nuevo Leon. Preliminary evidence sug-
gests that several birds from a region in Nuevo Leon State
have antibodies to WNV (I. Fernandez-Salas, unpub. data).
The birds were trapped in February 2003, 2 months after
we obtained samples from the horses in Coahuila State.
However, equine cases often precede the detection of
seropositive birds. For example, an equine case was the
first indication of WNV activity in 29% (660/2,289) of the
United States counties to report virus activity in 2002 (6).
In summary, we have obtained serologic evidence for
antibodies to WNV in horses in the State of Coahuila,
Mexico. In the accompanying manuscript, we report the
detection of antibodies to WNV in horses in the State of
Yucatan (15). These two reports provide the first published
evidence of WNV activity in horses in Mexico. Antibodies
to WNV, or a closely related virus, were detected in a sin-
gle bovine during a serosurvey in Chiapas, Mexico, in
mid-2001 (16). WNV will probably become endemic in
Mexico, which is a major concern to public health author-
ities in the Americas. Our findings demonstrate the impor-
tance for continued WNV surveillance in Mexico.
Acknowledgments
We thank Nicolas Fernandez-Rangel, Oscar Lorenzana, and
Leopoldo Chavarria for collecting serum specimens.
This study was supported by grant U50 CCU820510 from
the Centers for Disease Control and Prevention and in part by
grant AI45430 from the National Institutes of Health.
Dr. Blitvich is a postdoctoral scientist in the Department of
Microbiology, Immunology and Pathology at Colorado State
University, Fort Collins, CO. His research interests include the
mechanisms of vector and host interactions in arbovirus trans-
mission cycles.
References
1. Heinz FX, Collett MS, Purcell RH, Gould EA, Howard CR,
Houghton M, et al. Family Flaviviridae. Virus taxonomy. In: Van
Regenmortel CM, Fauquet CM, Bishop DHL, Carstens E, Estes MK,
Lemon S, et al., editors. 7th Report of the International Committee for
the Taxonomy of Viruses. San Diego: Academic Press; 2000. p.
859–78.
2. Burke DS, Monath TP. Flaviviruses. In: Knipe DM, Howley PM, edi-
tors. Fields virology. 4th ed. Philadelphia (PA): Lippincott Williams
and Wilkins; 2001. p. 1043–126.
3. Hayes CG. West Nile fever. In: Monath TP, editor. Vol I. The
arboviruses: epidemiology and ecology. Boca Raton (FL): CRC Press
Inc; 1988. p. 59–88.
4. Campbell GL, Marfin AA, Lanciotti RS, Gubler DJ. West Nile virus.
Lancet Infect Dis 2002;2:519–29.
5. Roehrig JT, Layton M, Smith P, Campbell GL, Nasci R, Lanciotti RS.
The emergence of West Nile virus in North America: ecology, epi-
demiology, and surveillance. Curr Top Microbiol Immunol
2002;267:223–40.
6. Centers for Disease Control and Prevention. Provisional surveillance
summary of the West Nile Virus epidemic—United States,
January–November 2002. MMWR Morb Mortal Wkly Rep
2002;51:1129–33.
7. Health Canada. Population and Public Health Branch WNV surveil-
lance updates, December 4, 2002. Available from: URL:
http://www.hc-sc.gc.ca/pphb-dgspsp/wnv-vwn/mon_e.html#sitrep
8. Texas Health Department. West Nile virus in Texas. Available from:
URL: http://www.tdh.state.tx.us/zoonosis/diseases/Arboviral/west
Nile/default.asp
9. Blitvich BJ, Marlenee NL, Hall RA, Calisher CH, Bowen RA,
Roehrig JT, et al. Epitope-blocking enzyme-linked immunosorbent
assays for the detection of serum antibodies to West Nile virus in mul-
tiple avian species. J Clin Microbiol 2003;41:1041–7.
10. Hall RA, Broom AK, Hartnett AC, Howard MJ, Mackenzie JS.
Immunodominant epitopes on the NS1 protein of MVE and KUN
viruses serve as targets for a blocking ELISA to detect virus-specific
antibodies in sentinel animal serum. J Virol Methods
1995;51:201–10.
11. Blitvich BJ, Bowen RA, Marlenee NL, Hall RA, Bunning ML, Beaty
BJ. Epitope-Blocking enzyme-linked immunosorbent assays for the
detection of West Nile virus antibodies in domestic mammals. J Clin
Micorbiol. 2003;41:2676–9.
12. Tsai TF, Mitchell CJ. St. Louis encephalitis. In: Monath TP, editor.
Vol IV. The arboviruses: epidemiology and ecology. Boca Raton
(FL): CRC Press; 1988. p. 113–43.
13. United States Department of Agriculture Animal and Plant Health
Inspection Service (APHIS). Summary of West Nile virus in the
United States. Available from: URL: www.aphis.usda.gov/
vs/ep/WNV/summary.html. 2000.
14. Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage
HM, et al. Rapid detection of West Nile virus from human clinical
specimens, field-collected mosquitoes, and avian samples by a
TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 2000;
38:4066–71.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 855
DISPATCHES

15. Loroño-Pino MA, Blitvich BJ, Farfán-Ale JA, Puerto FI, Blanco JM,
Marlenee NL, et al. Serologic evidence of West Nile virus infection
in horses, Yucatan State, Mexico. Emerg Infect Dis 2003;9:857–9.
16. Ulloa A, Langevin SA, Mendez-Sanchez JD, Arredondo-Jimenez JI,
Raetz JL, Powers AM, et al. Serologic survey of domestic animals for
zoonotic arbovirus infections in the Lacandón Forest Region of
Chiapas, Mexico. Vector Borne Zoonotic Dis. In press 2003.
Address for correspondence: Barry J. Beaty, Arthropod-borne and
Infectious Diseases Laboratory, Department of Microbiology,
Immunology and Pathology, College of Veterinary Medicine and
Biomedical Sciences, Colorado State University, Fort Collins, Colorado,
80523, USA; fax: 970-491-8323; email: [email protected]
856 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
The opinions expressed by authors contributing to this journal do
not necessarily reflect the opinions of the Centers for Disease
Control and Prevention or the institutions with which the authors
are affiliated.
S
S
e
e
a
a
r
r
c
c
h
h
p
p
a
a
s
s
t
t
i
i
s
s
s
s
u
u
e
e
s
s
o
o
f
f
E
E
I
I
D
D
a
a
t
t
w
w
w
w
w
w
.
.
c
c
d
d
c
c
.
.
g
g
o
o
v
v
/
/
e
e
i
i
d
d

Serologic Evidence
of West Nile Virus
Infection in
Horses, Yucatan
State, Mexico
María A. Loroño-Pino,*† Bradley J. Blitvich,*
José A. Farfán-Ale,† Fernando I. Puerto,†
José M. Blanco,‡ Nicole L. Marlenee,*
Elsy P. Rosado-Paredes,†
Julian E. García-Rejón,† Duane J. Gubler,§
Charles H. Calisher,* and Barry J. Beaty*
Serum samples were obtained from 252 horses in the
State of Yucatan, Mexico, from July to October 2002.
Antibodies to West Nile virus were detected by epitope-
blocking enzyme-linked immunosorbent assays in three
(1.2%) horses and confirmed by plaque reduction neutral-
ization test. We report the first West Nile virus activity in the
State of Yucatan.
W
est Nile virus (WNV) is a member of the Japanese
encephalitis virus complex within the genus
Flavivirus, family Flaviviridae (1). The virus is transmit-
ted in natural cycles mainly between mosquitoes and birds,
with humans and horses serving as incidental hosts (2).
WNV was first isolated in 1937 from the blood of a febrile
adult human in the West Nile District of Uganda (3). This
virus has since been reported in Africa, the Middle East,
Asia, southern Europe, Australia, and, more recently,
North America (4,5). The initial outbreak of WNV in
North America was recognized in New York City in
August 1999, with deaths reported in humans, horses, and
numerous species of birds. Since then, the geographic dis-
tribution of WNV in North America has greatly increased.
WNV activity has now been reported in 44 states and the
District of Columbia in the United States and in 5 of the 10
Canadian provinces (6,7).
In response to the incursion and rapid spread of WNV
in North America, we established equine and avian infec-
tion surveillance in Yucatan State, Mexico, in March 2000.
Yucatan State is a likely point of incursion of this virus into
Latin America because this area is a principal landfall for
many species of birds that migrate from the northeastern
and midwestern United States (8).
To determine whether WNV had already reached this
part of Mexico, we obtained blood samples from 252
domestic horses in 14 study sites from July to October
2002 (Table 1). The age distribution of the horses was 3
months to 25 years, and the mean age was 8.2 years. One
hundred and fifty-one horses were male, and 101 were
female. All study sites were on privately owned ranches,
where the horses were primarily used to perform heavy
labor and herd cattle. According to the owners, none of the
horses had ever been outside the State of Yucatan.
Furthermore, none of the horses had been vaccinated
against WNV.
The climate and topography of the study sites are simi-
lar. The climate can be described as tropical. The average
annual rainfall in each study site ranges from 600 to 1,100
mm, and the average annual temperature is 26°C. The
average elevation is approximately 17 m.
All serum samples were screened for antibodies to fla-
viviruses by hemagglutination inhibition (HI) assays and
epitope-blocking enzyme-linked immunosorbent assays
(ELISAs) at the Universidad Autonoma de Yucatan in
Merida. HI assays were performed by using Saint Louis
encephalitis virus (SLEV) antigen as previously described
(9). This antigen recognizes cross-reactive HI antibodies to
WNV and to other flaviviruses. To preclude nonspecific HI
reactions, samples were treated with kaolin, then adsorbed
with goose erythrocytes, according to standard methods
(9). Epitope-blocking ELISAs were performed by using
the flavivirus group-reactive monoclonal antibody (MAb),
6B6C-1, or the WNV-specific MAb, 3.1112G as previous-
ly described (10). The ability of the Mexican horse serum
samples to block the binding of MAbs to WNV antigen
was compared to the blocking ability of horse serum with-
out antibody to WNV (Vector Laboratories, Burlingame,
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 857
DISPATCHES
*Colorado State University, Fort Collins, Colorado, USA;
†Universidad Autonoma de Yucatan, Merida, Yucatan, Mexico;
‡Universidad Autonoma de Yucatan, Xmatkuil, Yucatan, Mexico;
and §Centers for Disease Control and Prevention, Fort Collins,
Colorado, USA
Table 1. Study sites and numbers of horses sampled per site,
State of Yucatan, Mexico
Study site
Global Positioning
System location
No. (%) of
horses bled
Acanceh
20° 48' 46" N, 89° 27' 14" W
8 (3.2)
Caucel
21° 00' 53" N, 89° 42' 25" W
1 (0.4)
Hobonil
20° 00' 54" N, 89° 01' 15" W
26 (10.3)
Hunucma
21° 00' 55" N, 89° 52' 28" W
7 (2.8)
Mani
20° 23' 11" N, 89° 23' 37" W
1 (0.4)
Merida
20° 58' 04" N, 89° 37' 18" W
63 (25.0)
Molas
20° 48' 57" N, 89° 37' 55" W
5 (2.0)
Progreso
21° 17' 04" N, 89° 39' 48" W
31 (12.3)
Sierra Papacal
21° 07' 16" N, 89° 43' 41" W
14 (5.6)
Timucuy
20° 48' 34" N, 89° 30' 51" W
5 (2.0)
Tixkokob
21° 00' 08" N, 89° 23' 37" W
15 (6.0)
Tizimin
21° 08' 32" N, 88° 09' 03" W
49 (19.4)
Uman
20° 49' 38" N, 89° 41' 08" W
26 (10.3)
Xbec
21° 14' 54" N, 88° 49' 29" W
1 (0.4)
Total
252 (100)

CA). Data were expressed as relative percentages, and
inhibition values >
30% were considered to indicate viral
antibodies. Recent studies in this laboratory have shown
that epitope-blocking ELISA provides a rapid and reliable
serologic technique for the detection of WNV antibodies in
various vertebrate species, including horses (10,11).
Six horses had evidence of flavivirus infection by HI
assay or ELISA (Table 2). Serum samples from three of
these horses (H-117, H-126, and H-252) were positive in
the ELISA that used the WNV-specific MAb. H-117 (7-
year-old stallion) and H-126 (2-year-old stallion) were
both sampled at the Tizimin study site. Neither horse
showed signs of illness at the time of serum collection or
during the 7 months that followed. Furthermore, neither
horse had a history of WNV-like illness. H-252 was a 3-
year-old stallion from Caucel that exhibited neurologic and
muscular symptoms at the time of sampling; it was eutha-
nized several hours later. We were not able to obtain tissue
specimens from this horse postmortem. Of the 252 horses
sampled, the only other horse to exhibit signs of clinical
illness was H-60, which had signs consistent with gastroin-
testinal illness.
Serum samples positive for flavivirus antibodies by HI
assay or ELISA were tested by plaque reduction neutral-
ization assay (PRNT) to identify the infecting virus.
PRNTs were conducted in the BSL-3 facilities at Colorado
State University. Serum sample results shown to be nega-
tive by HI assay and ELISA were not tested. PRNTs were
done by using WNV (strain NY99-35261-11), SLEV
(strain TBH-28), Ilhéus virus (ILHV, original strain), and
Bussuquara virus (BSQV, strain BeAn-4073). Virus stocks
were obtained from the World Health Organization Center
for Arbovirus Reference and Research, maintained at the
Centers for Disease Control and Prevention, Division of
Vector-Borne Infectious Diseases, Fort Collins, CO. We
tested serum samples for neutralizing antibodies to SLEV
because the virus is enzootic in the Americas and because
antibodies to WNV and SLEV often cross-react.
Furthermore, horses are susceptible to natural SLEV infec-
tions, although clinical manifestations have not been
reported (12). ILHV and BSQV are also present in the
Americas, although neither virus is known to naturally
infect horses (2). PRNTs were performed by using Vero
cells. Serum samples were tested by using a starting dilu-
tion of 1:20. Titers were expressed as the reciprocal of
serum dilutions yielding >90% reduction in the number of
plaques (PRNT
90
).
Neutralizing antibodies to WNV were detected in three
horses (Table 3). The PRNT-positive horses were H-117,
H-126, and H-252, which exhibited PRNT
90
antibody titers
of 320, >2,560, and 160, respectively. The SLEV, ILHV,
and BSQV antibody titers of the three horses were all <20.
Therefore, we considered H-117, H-126, and H-252 to be
seropositive for WNV because the PRNT
90
antibody titers
for WNV were more than fourfold higher than the other
flaviviruses tested. Overall, the PRNT and ELISA data
were in concordance; all serum samples that contained
neutralizing antibodies to WNV were positive in the assay
that used MAb 3.1112G (Tables 2 and 3). However, H-252
was negative in the assay that used MAb 6B6C-1, although
the percent inhibition value was close to the diagnostic cri-
terion. The three other horses (H-60, H-134, and H-141)
that were positive for flavivirus antibodies by HI assay or
ELISA did not have neutralizing antibodies to WNV. H-60
had a low SLEV PRNT
90
titer, suggesting it had been
infected with SLEV or a closely related virus. H-134
exhibited a HI titer of 10 but was negative by the other
serologic tests, suggesting that the HI antigen had reacted
nonspecifically. H-141 was positive by HI assay and
ELISA but had no neutralizing antibodies to any flavivirus
tested; thus, the identity of the infecting virus was not
determined.
We obtained serologic evidence for antibodies to WNV
in Yucatan State, Mexico. The mode of entry of this virus
into Yucatan State is not known; however, the virus may
have been brought in by birds migrating from the north.
858 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Table 2. Summary of horses with HI assay or epitope-blocking ELISA antibodies to flaviviruses
a
% inhibition by
blocking ELISA
b
Horse
Sampling date
Study site
Age (y)
Sex
Clinical symptoms
Outcome
HI assay
titer
3.1112G
c
6B6C-1
d
H-60
July 2, 2002
Merida
8
Male
Gastrointestinal
(recurrent colic)
Survived
10
0
59
H-117
July 5, 2002
Tizimin
7
Male
None
Survived
10
84
93
H-126
July 5, 2002
Tizimin
2
Male
None
Survived
40
87
93
H-134
July 5, 2002
Tizimin
3
Female
None
Survived
10
11
0
H-141
July 5, 2002
Tizimin
10
Male
None
Survived
80
5
47
H-252
Oct. 15, 2002
Caucel
3
Male
Neurologic and
muscular symptoms
Euthanized
20
64
25
a
HI, hemagglutination-inhibition; ELISA, enzyme-linked immunosorbent assay.
b
Inhibition values >30% are considered significant.
c
MAb 3.1112G is WNV-specific.
d
MAb 6B6C-1 is flavivirus group–reactive.

We have also detected antibodies to WNV in certain
species of migratory birds, which supports this hypothesis.
Data from the avian surveillance studies conducted in
Yucatan State will be described separately (J.A. Farfán-
Ale, unpub. data). We plan to isolate and amplify viral
sequences from migratory and resident birds, as well as
from specimens from other seropositive animals, to deter-
mine the origin of the WNV strain in Yucatan State. We
also provide serologic evidence for WNV infection in
horses in Coahuila State (13). These two reports provide
the first published evidence of WNV activity in horses in
Mexico. Neutralizing antibodies to WNV have also been
detected in a bovine in Chiapas, Mexico, in mid-2001,
indicating that the animal had been infected with WNV or
a closely related virus (14). WNV may become endemic in
this country, which demonstrates the importance for con-
tinued WNV surveillance in Mexico, and elsewhere in the
south.
Acknowledgments
We thank Eduardo Sierra-Lira and Edwin J. Gutierrez-Ruiz
for collecting the serum specimens and Luis F. Flores-Flores and
Richard Bowen for technical assistance.
This study was supported by grant U50 CCU820510 from
the Centers for Disease Control and Prevention and in part by
grant AI45430 from the National Institutes of Health.
Ms. Loroño-Pino is a Ph.D. student in the Department of
Microbiology, Immunology and Pathology at Colorado State
University, Fort Collins, CO. Previously, she was the chief of the
Laboratorio de Arbovirologia, Centro de Investigaciones
Regionales Dr. Hideyo Noguchi, Universidad Autonoma de
Yucatan, Mexico. Her research interests include the mechanisms
of West Nile virus and dengue virus pathogenesis.
References
1.Heinz FX, Collett MS, Purcell RH, Gould EA, Howard CR,
Houghton M, et al. Family Flaviviridae. Virus taxonomy. In: Van
Regenmortel CM, Fauquet CM, Bishop DHL, Carstens E, Estes MK,
Lemon S, et al., editors. 7th report of the International Committee for
the Taxonomy of Viruses. San Diego (CA): Academic Press; 2000. p.
859–78.
2. Burke DS, Monath TP. Flaviviruses. In: Knipe DM, Howley PM, edi-
tors. Fields virology. 4th ed. Philadelphia (PA): Lippincott Williams
and Wilkins; 2001. p. 1043–126.
3. Smithburn KC, Hughes TP, Burke AW, Paul JH. A neurotropic virus
isolated from the blood of a native of Uganda. Am J Trop Med Hyg
1940;20:471–92.
4. Campbell GL, Marfin AA, Lanciotti RS, Gubler DJ. West Nile virus.
Lancet Infect Dis 2002;2:519–29.
5. Roehrig JT, Layton M, Smith P, Campbell GL, Nasci R, Lanciotti RS.
The emergence of West Nile virus in North America: ecology, epi-
demiology, and surveillance. Curr Top Microbiol Immunol
2002;267:223–40.
6. Centers for Disease Control and Prevention. Provisional surveillance
summary of the West Nile Virus epidemic—United States,
January–November 2002. MMWR Morb Mortal Wkly Rep
2002;51:1129–33.
7. Health Canada. Population and Public Health Branch WNV surveil-
lance updates, December 4, 2002. Available from: URL:
http://www.hc-sc.gc.ca/pphb-dgspsp/wnv-vwn/mon_e.html#sitrep
8. Howell SNG, Webb S. A guide to the birds of Mexico and northern
Central America. Oxford: Oxford University Press; 1995.
9. Calisher CH, Beaty BJ, Chandler LJ. Arboviruses. In: Lennette E,
editor. Laboratory diagnosis of viral infections. 3rd ed. New York:
Marcel Dekker Inc; 1999. p. 305–32.
10. Blitvich BJ, Marlenee NL, Hall RA, Calisher CH, Bowen RA,
Roehrig JT, et al. Epitope-blocking enzyme-linked immunosorbent
assays for the detection of serum antibodies to West Nile virus in mul-
tiple avian species. J Clin Microbiol 2003;41:1041–7.
11. Blitvich BJ, Bowen RA, Marlenee NL, Hall RA, Bunning ML, Beaty
BJ. Epitope-blocking enzyme-linked immunosorbent assays for the
detection of West Nile virus antibodies in domestic mammals. J Clin
Microbiol. 2003;41:2676–9.
12. Tsai TF, Mitchell CJ. St. Louis encephalitis. In: Monath TP, editor.
Vol. IV, The arboviruses: epidemiology and ecology. Boca Raton
(FL): CRC Press; 1988. p. 113–43.
13. Blitvich BJ, Fernandez-Salas I, Contreras-Cordero JF, Marlenee NL,
Gonzalez-Rojas JI, Komar N, et al. Serologic evidence of West Nile
virus infection in horses, Coahuila State, Mexico. Emerg Infect Dis
2003;9;853–6.
14. Ulloa A, Langevin SA, Mendez-Sanchez JD, Arredondo-Jimenez JI,
Raetz JL, Powers AM, et al. Serologic survey of domestic animals for
zoonotic arbovirus infections in the Lacandón Forest region of
Chiapas, Mexico. Vector Borne Zoonotic Dis. In press 2003.
Address for correspondence: Barry J. Beaty, Arthropod-borne and
Infectious Diseases Laboratory, Department of Microbiology,
Immunology and Pathology, College of Veterinary Medicine and
Biomedical Sciences, Colorado State University, Fort Collins, CO 80523,
USA; fax: 970-491-8323; email: [email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 859
DISPATCHES
Table 3. Neutralizing antibody titers to West Nile, Saint Louis
encephalitis, Ilhéus, and Bussuquara viruses in serum samples
from six horses
a
PRNT
90
titer
Horse
WNV
SLEV
ILHV
BSQV
H-60
–
b
20
c
–
b
–
H-117
320
–
–
b
–
H-126
>2,560
–
b
–
–
H-134
–
–
–
–
H-141
–
–
–
–
H-252
160
–
–
–
a
WNV, West Nile virus; SLEV, Saint Louis encephalitis virus; ILHV,
Ilhéus virus; BSQV, Bussuquara virus; PRNT, plaque reduction
neutralization test; –, <20.
b
PRNT
80
titer: 20.
c
PRNT
80
titer: 40.
All material published in Emerging Infectious Diseases is in the
public domain and may be used and reprinted without special
permission; proper citation, however, is appreciated.

Serologic Evidence
of West Nile Virus
Transmission,
Jamaica, West
Indies
Alan P. Dupuis II,* Peter P. Marra,†
and Laura D. Kramer*
In spring 2002, an intensive avian serosurvey was ini-
tiated in Jamaica, Puerto Rico, and Mexico. We collected
>1,600 specimens from resident and nonresident neotropi-
cal migratory birds before their northerly migrations. Plaque
reduction neutralization test results indicated specific neu-
tralizing antibodies to West Nile virus in 11 resident species
from Jamaica.
W
est Nile virus (WNV) is maintained in nature
between birds and Culex species mosquitoes (1,2).
Unlike other viruses maintained in bird and mosquito
transmission cycles (for example, St. Louis encephalitis,
western equine encephalomyelitis, and eastern equine
encephalomyelitis), the WNV strain responsible for the
current epizootic in the Western Hemisphere is associated
with a high number of avian deaths (3). Although crows
and other corvids appear to be especially susceptible to
disease (4), WNV has been documented in >190 bird
species, including neotropical migratory species and exot-
ic zoo specimens (5).
Birds have been implicated in spreading WNV during
migratory events in Europe, Asia, Africa, and the Middle
East (6–9). Because of the apparent ease of infecting a
multitude of avian hosts, WNV can potentially be intro-
duced during annual migratory events in the Western
Hemisphere. Predicting the incursion of WNV into the
tropics is complicated by our incomplete knowledge
regarding geographic connectivity in populations of
migratory birds between winter and summer. We do not
know where most species of migratory birds from North
America spend their winter, where wintering birds spend
their summer, or the routes they use while in transit.
Evidence from mark-and-recapture efforts suggests that
birds from the northeastern United States tend to winter in
the southeastern United States and Greater Antilles (e.g.,
Puerto Rico, Jamaica) whereas birds from the western
United States migrate to Mexico and Central America
(10–12). In response to the potential introduction of WNV
in tropical America during the fall migrations, we estab-
lished a network of monitoring sites on the overwintering
grounds of neotropical migratory birds in Jamaica, Puerto
Rico, and Mexico.
The Study
The primary goal of the monitoring system was to
obtain a large number of blood specimens from birds
belonging to as many species as possible across the
Caribbean. At seven study sites in Jamaica, Puerto Rico,
and the Yucatan Peninsula of Mexico, we erected 15 mist
nets (12-m) daily for 3 to 4 weeks during January through
March 2002. We collected 1,619 avian blood specimens,
which represented 98 species, 25 families, and eight
orders. In Jamaica, 542 samples were collected from
Westmoreland, Manchester, and St. Catherine Parishes;
649 samples were collected at Roosevelt Roads Naval
Station in Puerto Rico; and 430 samples were collected
from the states of Yucatan and Campeche in Mexico (Table
1). At the time of capture, all migratory birds were banded
with an aluminum U.S. Fish and Wildlife Service band and
3–5 breast feathers were removed for isotope analysis.
Resident birds had outer rectrices cut. All birds were eval-
uated for age and gender if possible, bled with microcapil-
lary tubes from the brachial vein, and released. Blood was
added immediately to BA-1 medium, consisting of M199
medium with Hank’s salts, 1% bovine albumin, TRIS base
(tris [hydroxymethyl] aminomethane), sodium bicarbon-
ate, 20% fetal bovine serum (FBS), and antibiotics. The
samples were placed in a cooler on ice packs until storage
in a –20°C freezer. Samples were sent on dry ice or hand-
carried on ice packs to the Arbovirus Laboratories,
Wadsworth Center, New York State Department of Health
for serologic analysis and virus isolation attempts.
Specimens were screened at 1:100 for antibodies
against flaviviruses by using an indirect enzyme-linked
immunosorbent assay (ELISA) (13). Samples with a P/N
ratio >2.0 were tested further by a plaque reduction neu-
tralization test (PRNT) for Ilhéus virus (ILHV), St. Louis
encephalitis virus (SLEV), and WNV, as described (14).
The particular virus strains used for the PRNTs were ILHV
(original), SLEV 59268 Parton, and WNV (3100365), an
isolate from a pool of Culex sp. mosquitoes collected in
Staten Island, New York. The indirect ELISA was chosen
to screen the samples in order to take advantage of its abil-
ity to detect antibodies against a wide range of flavivirus-
es. PRNT was used as a confirmatory assay to differentiate
among recognized flaviviruses (15,16). Briefly, serial dilu-
tions of test samples were mixed with an equal amount of
virus suspension containing 200 PFU/0.1 mL and incubat-
ed at 37°C for 1 h. We then added 0.05 mL of each virus-
860 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
*New York State Department of Health, Slingerlands, New York,
USA; and †Smithsonian Environmental Research Center,
Edgewater, Maryland, USA
diluted blood sample onto 1 well of a 12-well tissue culture
plate containing confluent monolayers of African green
monkey kidney cells (Vero). The plate was incubated for 1
h at 37°C, after which an agarose overlay was added and
incubation was continued. When virus plaques became
visible, we added a second overlay containing neutral red
and counted plaques. The antibody titer reported is the
dilution of serum that inhibited 90% of the test virus inocu-
lum. For virus isolation attempts using confluent Vero cell
monolayers, 0.1 mL of each serum specimen was added
onto one well of a six-well tissue culture plate, incubated
for 1 h, rinsed with phosphate-buffered saline, and then the
cells were refed with minimum essential medium contain-
ing 2% FBS. Cells were monitored once a day for 5 days
for cytopathic effect. Cell cultures showing any abnormal
cell morphology were then blind passaged after 5 days.
ELISA results indicated 34 of 1,413 serum specimens
tested from the three study sites contained immunoglobu-
lin G antibody against a flavivirus. Of the 34 reactive sam-
ples representing 20 bird species, 27 were collected in
Jamaica (26 residents, 1 migrant), 5 in Puerto Rico (3 res-
idents, 2 migrants), and 2 in Mexico (1 resident, 1
migrant). PRNTs on the Jamaican bird samples indicated
18 WNV infections, 3 SLEV infections, 5 undetermined
flavivirus infections (positive results for two or more
viruses without a fourfold difference in antibody titer); one
additional reactive serum sample was negative for the
three viruses tested. Results on the serum samples collect-
ed in Puerto Rico indicated one WNV infection in a migra-
tory bird and one SLEV infection in a resident bird; three
additional reactive serum samples were negative for all
three viruses tested. In Mexico, we found evidence of
WNV infection in one migrant bird and SLEV infection in
one resident bird (Table 2). Virus isolation attempts were
negative for all 1,603 specimens tested (16 samples
destroyed). Negative isolation results were not entirely
unexpected, considering that birds are viremic for a short
period of time (17) and maintaining a proper cold chain
(i.e., temperatures) to preserve virus is difficult when
working in the tropics.
Conclusions
We detected neutralizing antibodies to WNV in resident
birds from two parishes in Jamaica. This detection marks
the earliest evidence of WNV introduction into the
neotropics; WNV antibodies have been demonstrated in
birds and horses in Mexico (late 2002, spring 2003)
(18,19) and detected in resident birds from the Dominican
Republic (spring 2003) (20). This evidence of WNV in the
neotropics may be an important development in the spread
of the virus. The tropics provide all the necessary compo-
nents (i.e., high temperatures, dense avian populations, and
large numbers of Culex sp. mosquitoes) to maintain an
enzootic focus. Furthermore, the climates of Mexico and
the Caribbean are suitable for year-round transmission of
the virus. No dead birds have been reported in Jamaica, but
surveillance activity there is less intensive than in the
United States; the study sites, being rural in nature, are not
conducive to observing dead birds. Another contributing
factor to the lack of reports of dead birds may be the rapid
decomposition of dead birds as a result of the heat, humid-
ity, and detritivore foragers, such as ants.
Arbovirus activity, particularly of flaviviruses, is well
documented in the Caribbean and Mexico. Dengue and
yellow fever viruses are recurring public health threats in
these areas. SLEV is endemic to Mexico and has been iso-
lated from mosquitoes and one Northern mockingbird
(Mimus polyglottos) nestling in Jamaica (21). This disease
is still active in the region, and its known range may have
expanded into Puerto Rico, considering the one seroposi-
tive Caribbean elaenia (Elaenia martinica) sampled during
this study, although the antibody may have resulted from
infection with yet another flavivirus. Neutralizing antibod-
ies to WNV in migratory birds collected in Mexico and
Puerto Rico, coupled with the apparent absence of anti-
body to WNV in the resident bird population, indicate that
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 861
DISPATCHES
Table 1. Number of birds collected at seven field sites in the Caribbean
Field site
Migratory birds
Resident birds
Total
Flavivirus positives (%)
a
Westmoreland Parish, Jamaica
156
232
388
18 (4.6)
Manchester Parish (site 1), Jamaica
2
21
23
1 (4.4)
Manchester Parish (site 2), Jamaica
24
83
107
4 (3.7)
St. Catherine Parish, Jamaica
12
12
24
4 (16.7)
Yucatan, Mexico
70
102
172
2 (1.2)
Campeche, Mexico
114
144
258
0 (0.0)
Mexico totals
184
246
430
2 (0.5)
Puerto Rico (one collection site) totals
391
256
647
5 (1.1)
b
Jamaica totals
194
348
542
27 (5.0)
Totals
769
850
1,619
34 (2.4)
c
a
Based on screening enzyme-linked immunosorbent assay (ELISA) results.
b
Results based on 441 samples suitable for ELISA (206 samples were blood clots only).
c
Results based on 1,413 samples tested by ELISA.

infection likely occurred in an enzootic area of the United
States, but this observation shows that individual birds
from at least three species of neotropical migratory birds
have survived WNV infection and may serve as hosts for
spreading the virus. The results from this study suggest
that WNV now appears to be established in Jamaica, on the
basis of the neutralizing antibodies to WNV found in the
resident bird population.
Acknowledgments
We thank Brian Gibbons, Karin Roux, Javier Salgado-Ortiz,
Joe Smith, and Jenn Barg for collecting field samples and Robert
Reitsma, Kim Kent, and Donna Young for tremendous logistical
and laboratory support.
This work was supported by Grant Agreement Number
U50/CCU320544-01 from the Centers for Disease Control and
Prevention (CDC) and the National Science Foundation (NSF),
Grant Number DEB 0089565. Its contents are solely the respon-
sibility of the authors and do not represent the official views of
CDC or NSF.
Mr. Dupuis is an assistant research scientist in the Arbovirus
Laboratories, Wadsworth Center, New York State Department of
Health. His research focuses on the role of birds in the mainte-
nance of arbovirus transmission cycles and dispersal of the virus.
References
1. Hayes CG. West Nile fever. In: Monath TP, editor. The arboviruses:
epidemiology and ecology. Vol. V. Boca Raton (FL): CRC Press, Inc.;
1989. p. 59–88.
862 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Table 2. Ninety percent plaque reduction neutralization titers to West Nile virus, St. Louis encephalitis virus, and Ilhéus virus in
enzyme-linked immunosorbent assay reactive bird specimens
a
Field site
Species
ILHV
SLEV
WNV
Interpretation
White-chinned thrush
(Turdus aurantius)
b
40
80
640
WNV
White-chinned thrush (T. aurantius)
b
<40
<40
320
WNV
Jamaican elaenia (Myiopagis cotta)
b
<40
160
640
WNV
Bananaquit (Coereba flaveola)
b
<40
80
160
Flavivirus
Loggerhead kingbird (Tyrannus caudifasciatus)
b
<40
<40
320
WNV
Bananaquit (C. flaveola)
b
<40
<40
160
WNV
White-chinned thrush (Turdus aurantius)
b
<40
40
>1,280
WNV
Bananaquit (C. flaveola)
b
<40
<40
80
WNV
Northern mockingbird (Mimus polyglottos)
b
<40
<40
320
WNV
Caribbean dove (Leptotila jamaicensis)
b
<40
>1,280
80
SLEV
Jamaican elaenia (Myiopagis cotta)
b
<40
<40
160
WNV
Bananaquit (C. flaveola)
b
<40
40
80
Flavivirus
White-chinned thrush (Turdus aurantius)
b
<40
40
640
WNV
Jamaican elaenia (Myiopagis cotta)
b
<40
80
<40
SLEV
Common ground-dove (Columbina passerina)
b
<40
<40
160
WNV
Caribbean dove (Leptotila jamaicensis)
b
<40
<40
<40
Negative
Black-faced grassquit (Tiaris bicolor)
b
<40
<40
320
WNV
Westmoreland Parish, Jamaica
Jamaican vireo (Vireo modestus)
b
<40
<40
80
WNV
Manchester Parish (site 1), Jamaica
Caribbean dove (Leptotila jamaicensis)
b
<40
<40
80
WNV
Black-faced grassquit (Tiaris bicolor)
b
<40
<40
160
WNV
Jamaican oriole (Icterus leucopteryx)
b
<40
160
>1,280
WNV
White-eyed thrush (Turdus jamaicensis)
b
<40
<40
320
WNV
Manchester Parish (site 2), Jamaica
Orangequit (Euneornis campestris)
b
<40
<40
40
Flavivirus
Greater Antillean grackle (Quiscalus niger)
b
40
160
160
Flavivirus
Loggerhead kingbird (Tyrannus caudifasciatus)
b
80
320
320
Flavivirus
Golden warbler
(Dendroica petechia sp. )
c
<40
80
<40
SLEV
St. Catherine Parish, Jamaica
Northern waterthrush (Seiurus noveboracensis)
d
<40
40
320
WNV
Caribbean dove (Leptotila jamaicensis)
b
<40
>1,280
80
SLEV
Yucatan State, Mexico
Yellow warbler (Dendroica petechia)
d
<40
40
320
WNV
Prairie warbler (Dendroica discolor)
d
<40
<40
<40
Negative
Golden warbler
(Dendroica petechia sp. )
c
<40
<40
<40
Negative
Pearly-eyed thrasher (Margarops fuscatus)
b
<40
<40
<40
Negative
Caribbean elaenia (Elaenia martinica)
b
<40
80
<40
SLEV
Roosevelt Roads Naval Station,
Puerto Rico
Black and white warbler (Mniotilta varia)
d
<40
40
>1,280
WNV
a
ILHV, Ilhéus virus; SLEV, St. Louis encephalitis virus; WNV, West Nile virus.
b
Resident bird.
c
Resident yellow warbler subspecies.
d
Migrant bird.

2. Kramer LD, Bernard KA. West Nile virus in the western hemisphere.
Curr Opin Infect Dis 2001;14:519–25.
3. Bernard KA, Maffei JG, Jones SA, Kauffman EB, Ebel GD, Dupuis
AP II, et al. West Nile virus infection in birds and mosquitoes, New
York State, 2000. Emerg Infect Dis 2001;7:679–85.
4. Bernard KA, Kramer LD. West Nile virus activity in the United
States, 2001. Viral Immunol 2001;14:319–38.
5. U.S. Department of the Interior, U.S. Geological Survey, National
Wildlife Health Center. Species found positive for WNV in surveil-
lance efforts. [Cited April 5, 2003.] Available from: URL:
http://www.nwhc.usgs.gov/research/west_nile/wnvaffected.html
6. Malkinson M, Banet C, Weisman Y, Pokamunski S, King R, Drouet
MT, et al. Introduction of West Nile virus in the Middle East by
migrating white storks. Emerg Infect Dis 2002;8:392–7.
7. Malkinson M, Banet C. The role of birds in the ecology of West Nile
virus in Europe and Africa. Curr Top Microbiol Immunol
2002;267:309–22.
8. Ernek E, Kozuch O, Nosek J, Teplan J, Folk C. Arboviruses in birds
captured in Slovakia. J Hyg Epidemiol Microbiol Immunol
1977;21:353–9.
9. Rappole JH, Derrickson SR, Hubalek Z. Migratory birds and spread
of West Nile virus in the Western Hemisphere. Emerg Infect Dis
2000;6:319–28.
10. Johnson LS, Wise J. Wintering grounds of North American house
wrens as revealed by band recoveries. J Field Ornithol
2000;71:501–5.
11. Webster MS, Marra PP, Haig SM, Bensch S, Holmes RT. Links
between worlds: unraveling migratory connectivity. Trends in
Ecology and Evolution 2002;17:76–82.
12. Elphick J. The atlas of bird migration. New York: Random House;
1995.
13. Ebel GD, Dupuis AP, Nicholas D, Young D, Maffei J, Kramer LD.
Detection by enzyme-linked immunosorbent assay of antibodies to
West Nile virus in birds. Emerg Infect Dis 2002;8:979–82.
14. Lindsey HS, Calisher CH, Matthews JH. Serum dilution neutraliza-
tion test for California group virus identification and serology. J Clin
Microbiol 1976;4:503–10.
15. DeMadrid AT, Porterfield JS. The flaviviruses (group B arboviruses):
a cross-neutralization study. J Gen Virol 1974;23:91–6.
16. Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS,
Westaway EG, et al. Antigenic relationships between flaviviruses as
determined by cross-neutralization tests with polyclonal antisera. J
Gen Virol 1989;70:37–43.
17. Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, et
al. Experimental infection of North American birds with the New
York 1999 strain of West Nile virus. Emerg Infect Dis
2003;9:311–22.
18. West Nile virus, equine, 2002—Mexico (Tamaulipas). ProMed mail
archive no. 20030325.0739. [cited 25 March 2003]. Available from:
URL: http://www.promedmail.org
19. West Nile virus, birds—Mexico (North). ProMed mail archive no.:
20030315.0640. [Cited 15 March 2003]. Available from: URL:
http://www.promedmail.org
20. West Nile virus, birds—Dominican Republic. ProMed mail archive
no.: 20030315.0645. [Cited 15 March 2003]. Available from: URL:
http://www.promedmail.org
21. Belle EA, King SD, Griffiths BB, Grant LS. Epidemiological inves-
tigation for arboviruses in Jamaica, West Indies. Am J Trop Med Hyg
1980;29:667–75.
Address for correspondence: Alan P. Dupuis II, Griffin Laboratory, 5668
State Farm Rd., Slingerlands, NY 12159, USA; fax: (518) 869-4530;
email: [email protected].us
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 863
DISPATCHES
The print journal is available at no charge to public health professionals
YES, I would like to receive Emerging Infectious Diseases.
Please print your name and business
address in the box and return by fax to
404-371-5449 or mail to
EID Editor
CDC/NCID/MS D61
1600 Clifton Road, NE
Atlanta, GA 30333
Moving? Please give us your new address (in the box) and print the number of your old
mailing label here_________________________________________
Full text free online at
www.cdc.gov/eid

Sulfa Resistance
and Dihydropteroate
Synthase Mutants
in Recurrent
Pneumocystis
carinii Pneumonia
Aimable Nahimana,*
1
Meja Rabodonirina,†
Jannik Helweg-Larsen,‡ Isabelle Meneau,*
Patrick Francioli,* Jacques Bille,*
and Philippe M. Hauser*
Failure of sulfa or sulfone prophylaxis is associated
with mutations in Pneumocystis carinii gene coding for
dihydropteroate synthase (DHPS). The DHPS genotype
was analyzed in AIDS patients who had two separate
episodes of P. carinii pneumonia. The results suggest that
DHPS mutations can be selected de novo within patients
by the pressure of a sulfa or sulfone drug.
C
o-trimoxazole, the antifolate drug combination of
trimethoprim and sulfamethoxazole, is the drug of
choice for the prophylaxis and treatment of Pneumocystis
carinii pneumonia (PCP), a life-threatening disease in
immunosuppressed patients. Trimethoprim is an inhibitor
of dihydrofolate reductase, whereas sulfamethoxazole
inhibits dihydropteroate synthase (DHPS). The antipneu-
mocystis activity is believed to be due mainly to sul-
famethoxazole (1). Dapsone is a sulfone drug, also fre-
quently used, that targets DHPS. Widespread use of sulfa
and sulfone drugs to prevent and treat PCP in recent years
has correlated with an increase of the prevalence of muta-
tions in the gene coding for DHPS (2,3). The most frequent
DHPS mutations occur at nucleotide positions 165 and
171, which lead to an amino acid change at positions 55
(Thr to Ala) and 57 (Pro to Ser). These mutations are locat-
ed in the sulfa-binding site and may appear as either a sin-
gle or double mutation in the same isolate. Similar muta-
tions in other microbial pathogens confer sulfa resistance
(4,5). In P. carinii, DHPS mutations are associated with
failure of sulfa or sulfone prophylaxis (1,6) and decreased
survival of the patient at 3 months after PCP (2). However,
patients harboring P. carinii types with DHPS mutations
are most often successfully treated with high-dose co-tri-
moxazole (6). Because a standardized culture system for P.
carinii does not exist, the level of sulfa resistance con-
ferred by these mutations cannot be determined with in
vitro susceptibility tests. A key issue is whether the recent
emergence of DHPS mutations is a result of P. carinii
transmission between patients or arises from selection
within patients by the pressure of a sulfa or sulfone drug,
two possibilities that are not mutually exclusive. To inves-
tigate the latter possibility, we analyzed patients who had
had two separate episodes of PCP.
The Study
P. carinii DNA was extracted from bronchoalveolar
lavage specimens by using the Qiamp Blood Kit (QIA-
GEN GmbH, Hilden, Germany). Bronchoalveolar lavage
specimens from 13 patients with recurrent PCP episodes
were collected from four European hospitals (Lyon,
France; Copenhagen, Denmark; Lausanne, Switzerland;
and La Chaux-de-Fonds, Switzerland). To determine the
prevalence of the different P. carinii molecular types, we
analyzed bronchoalveolar lavage specimens from 310 PCP
patients from two Swiss hospitals (Lausanne, 111 patients;
Zurich, 64 patients) and Lyon’s hospital (135 patients).
Specific information on demographic and clinical charac-
teristics, chemoprophylaxis, and treatment regimens were
obtained from the medical charts. P. carinii infecting
humans (now named P. jiroveci [7]) was typed by using the
multilocus method developed in our laboratory as previ-
ously described (8–10). In this method, four variable
regions of the P. carinii genome are amplified by poly-
merase chain reaction (PCR), followed by the detection of
polymorphisms using single-strand conformation poly-
morphism (SSCP). A P. carinii type is defined by a combi-
nation of four alleles corresponding to the four genomic
regions. If a specimen harbored two alleles of one or more
of the four genomic regions, the patient was considered to
be co-infected with two or more P. carinii types (9). This
typing system has been validated and the stability of its
markers assessed; its index of discriminatory power has
been estimated to be 0.93 (10). The full length of the
DHPS gene was amplified by PCR as described previous-
ly (1). PCR products (765 bp) were cloned, and both
strands were sequenced (5 clones per sample). The five
clones had identical sequences for all samples, except for
those from patients 3 and 4, which contained a mixture of
DHPS sequences.
864 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
*Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland;
†Université Claude-Bernard, Lyon, France; and ‡Hvidovre
Hospital, Copenhagen, Denmark
1
Aimable Nahimana contributed to the design of the study, ana-
lyzed the samples by polymerase chain reaction and DNA
sequencing, and wrote the draft of the manuscript. Meja
Rabodonirina and Jannik Helweg-Larsen reviewed medical charts
and provided bronchoalveolar lavage specimens. All authors con-
tributed to the analysis of data and writing of the paper. Philippe M.
Hauser initiated and supervised all aspects of the study.

Thirteen patients with two separate PCP episodes
were analyzed (Table). All patients had recovered
between episodes. The intervals between the episodes
ranged from 4 to 25 months. All patients had AIDS and
all, except patients 8 and 9, were men, with a median age
of 35 (range 23–51) and median CD4 cell count of 9.5
cells/µL (range 0–98). Some patients were co-infected
with two different P. carinii types, as shown by PCR-
SSCP multilocus typing method (patients 4, 5, 8, 11, and
13) or DHPS genotyping (patients 3 and 4). In seven
(54%) patients (patients 1–7), the same PCR-SSCP type
was observed in both episodes of PCP; six (46%) patients
(patients 8–13) had different types in the first and second
episodes. This rate of genotype switch is similar to that
reported in previous studies, in which such a change was
observed in approximately half of recurrent episodes
(11–14). The importance of a genotype switch remains
uncertain. Indeed, the switch might be due to a de novo
infection or to the reactivation of a genotype not detect-
ed in the first episode because of the compartmentaliza-
tion of different co-infecting P. carinii types in the lungs
(15).
A second episode of PCP could result either from reac-
tivation of organisms that caused the first episode or from
de novo infection with a new P. carinii type acquired from
an exogenous source. In seven patients (patients 1–7),
reactivation was strongly suggested by the detection of
identical SSCP types in both episodes of PCP. An alterna-
tive explanation could be de novo infection in the second
episode by the same P. carinii PCR-SSCP type as that
which caused the first episode. However, the prevalence of
the types observed in the seven “reactivation” cases was
low in Lyon and Switzerland during the study period
(types no. 2, 5, and 7 represented 7%, 6%, and 10%,
respectively, of Lyon’s isolates; type 6 represented only
3.5% of the Swiss isolates [Figure]). Thus, reinfection with
these specific types was unlikely. All Danish patients (1, 3,
and 6) were infected with type 6. Although no prevalence
data for SSCP genotypes in Denmark are available, no
indication of possible contact between these patients, over-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 865
DISPATCHES
Table. Pneumocystis carinii DHPS and PCR-SSCP genotyping in AIDS patients with recurrent pneumonia
Patient no.
City
b
Age
Date of episode 1/
date of episode 2/
interval (mo)
CD4/mm
3
Prophylaxis at
PCP episode
c
Treatment
Outcome of
treatment
P. carinii PCR-
SSCP type
DHPS genotype
d
1
Co
29
7/16/1993
9
D
CO → P
e
Success
6
WT
6/8/1994 (11)
0
P
CO
Success
6
M1
2
Ly
36
1/31/1994
58
D
A
Success
7
M2
5/18/1995 (16)
16
CO
A
Success
7
M3
3
Co
51
8/19/1994
0
No
CO → C/P
e
Success
6
WT/M1
12/23/1994 (4)
0
P
T
Success
6
M1
4
Ly
32
11/23/1994
75
No
CO
Success
2, 5
WT/M3
3/23/1995 (4)
35
No
CO
Death
f
2, 5
M3
5
Ly
28
4/19/1995
70
No
A
Success
7, 8
WT
3/1/1996 (11)
98
CO
P
Success
7
M3
6
Co
35
11/16/1995
2
D
P → CO
e
Success
6
M1
5/6/1996 (6)
1
D
CO
Success
6
M1
7
CF
41
2/3/1998
7
CO
P
Success
6
M3
7/22/1998 (5)
7
P
C/P
Success
6
M3
8
La
28
11/24/1990
53
No
T
Success
6, 10
WT
7/29/1991 (8)
18
No
CO
Success
7
WT
9
Co
25
12/8/1992
0
No
CO
Success
5
WT
11/5/1993 (11)
0
No
CO
Success
7
WT
10
Co
35
3/22/1993
10
No
CO → P
e
Success
18
WT
10/28/1994 (7)
0
P
CO → P
e
Death
f
6
WT
11
Ly
23
3/30/1994
22
No
CO → A
e
Success
4, 7
M3
3/28/1995 (12)
26
P
D+T → A
e
Success
5
M3
12
Ly
46
9/21/1994
61
No
CO
Success
15
WT
10/21/1996 (25)
16
P
P+A
Success
3
WT
13
Ly
43
10/12/1994
50
No
CO
Success
1, 2
M2
3/25/1996 (17)
5
PM/SD
P+A
Success
1, 3
M2
a
PCP, Pneumocystis carinii pneumonia; DHPS, dihydropteroate synthase; PCR, polymerase chain reaction; SSCP, single-strand conformation polymorphism.
b
Co, Copenhagen (Denmark); CF, La Chaux-de-Fonds (Switzerland); La, Lausanne (Switzerland); Ly, Lyon (France).
c
A, atovaquone; CO, cotrimoxazole; C/P, clindamycin/primaquine; D,
dapsone; D+T, dapsone and trimethoprim; P, pentamidine; P+A,
pentamidin
e and atovaquone;
PM/SD (pyrimethamine/sulfadoxine inhibitors of dihydrofolate reductase (DHFR) and DHPS, respectively); T, trimetrexate (an inhibitor of DHFR).
d
WT, wild type (Thr55 Pro57); M1, mutant 1 (Ala
55 Pro57); M2, mutant 2 (Thr55 Ser57); M3, muta
nt 3 (Ala55 Ser57 double mutant).
e
Switch of molecules because of toxicity for patients 3, 6, and 11 and because of toxicity and treatment failure for patients 1
and 10.
f
Caused by PCP.
lap in hospitalization dates, or similar zip codes for home
address suggested transmission of type 6 between these
patients.
A change of P. carinii DHPS genotype between the two
episodes was observed in three reactivation cases, either
from wild type in the first episode to DHPS mutations in
the second one (patients 1 and 5) or from DHPS with a sin-
gle mutation (at position 57) in the first episode to a dou-
ble mutation in the second one (patient 2). In two patients
(3 and 4), the DHPS mutant strain was selected out of a
mixture of wild-type and DHPS mutant strains. Because
both episodes of each patient were caused by the same P.
carinii types and because all patients received co-trimoxa-
zole or dapsone as treatment, maintenance therapy, or
both, these results strongly suggest that selection of P.
carinii DHPS mutations occurred within the patients. The
results of tests on patients 3 and 4 isolates highlight the
fact that some patients may harbor genetically different
strains of P. carinii and that the mutant strain may be read-
ily selected when drug pressure is exerted. In the two
remaining patients (6 and 7), the P. carinii DHPS mutant
found in the bronchoalveolar lavage specimen from the
second episode was already present in the first episode.
The wild-type DHPS allele was more frequently
observed in the six reinfection cases than in the reactiva-
tion cases (8 wild-type alleles among 12 genotypes versus
4 among 16, Table). This finding is probably related to the
fact that, with the exception of the second episode of
patient 13, patients who were reinfected had no prophylax-
is or did not receive sulfa drugs for prophylaxis.
In all the second episodes caused by reactivation,
mutant DHPS strains were observed (7/7), compared to
only two of six second episodes caused by reinfection
(Table). This observation suggests an association between
mutant DHPS and second episodes attributable to reactiva-
tion (p<0.02, Fisher exact test).
Conclusions
Our study suggests that P. carinii DHPS mutants may
be selected in vivo (within a given patient) under the pres-
sure of co-trimoxazole or dapsone and that DHPS muta-
tions may be associated with reactivation of P. carinii.
Whether DHPS mutations are induced by the pressure of
the drug or preexisting and selected out by the pressure of
the drug remains to be determined. Physicians should be
alert to the increased risk for drug resistance during recur-
rence of PCP infection, although the impact of DHPS
mutations on retreatment with sulfa or sulfone drugs
remains to be determined. De novo selection of P. carinii
DHPS strongly favors the hypothesis that P. carinii is
developing sulfa and sulfone resistance.
Acknowledgments
We thank R. Lienhard for the samples he kindly provided
and S. Picot for storage of the specimens. We also thank A.
Telenti for critical reading of the manuscript, as well as J.L.
Touraine, D. Peyramond, and C. Trepo for access to patients’
charts in Lyon.
Financial support for this study was provided by the Swiss
National Program on AIDS Research grant 3345- 65407, Swiss
National Fund for Scientific Research grant 32-56715.99; Centre
de Coordination de la Lutte contre les Infections Nosocomiales
Sud-Est et Hospices Civils de Lyon; the Swiss Federal Office for
Education and Science for participation in EUROCARINII proj-
ect, Framework V Program, European Commission; and a North-
South fellowship from Lausanne University (supporting A.N.).
Dr. Nahimana obtained his bachelor’s degree and master’s
degree in microbiology at the University of Lausanne. The pres-
ent work was submitted by A. Nahimana as partial fulfillment for
a PhD degree at the University of Lausanne and was performed
under the supervision of P. Hauser.
References
1. Ma L, Borio L, Masur H, Kovacs H. Pneumocystis carinii dihy-
dropteroate synthase but not dihydrofolate reductase gene mutations
correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J
Infect Dis 1999;180:1969–78.
2. Helweg-Larsen J, Benfield TL, Eugen-Olsen J, Lundgren JD,
Lundgren B. Effects of mutations in Pneumocystis carinii dihy-
dropteroate synthase gene on outcome of AIDS-associated P. carinii
pneumonia. Lancet 1999;354:1347–51.
3. Kazanjian P, Armstrong W, Hossler PA, Burman W, Richardson J,
Lee CH, et al. Pneumocystis carinii mutations are associated with
duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J
Infect Dis 2000;182:551–7.
4. Olliaro P. Mode of action and mechanisms of resistance for anti-
malarial drugs. Pharmacol Ther 2001;89:207–19.
5. Sköld O. Sulfonamide resistance: mechanisms and trends. Drug
Resist Updat 2001;32:1608–14.
6. Navin TR, Beard CB, Huang L, del Rio C, Lee S, Pieniazek NJ, et al.
Effects of mutations in Pneumocystis carinii dihydropteroate syn-
thase gene on outcome of P carinii pneumonia in patients with HIV-
1: a prospective study. Lancet 2001;358:545–9.
866 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Figure. Frequency distribution of Pneumocystis carinii types
observed in different locations. Each type, co-infecting or not, was
considered as one isolate.

7. Stringer JR, Beard CB, Miller RF, Wakefield AE. A new name
(Pneumocystis jiroveci) for Pneumocystis from humans. Emerg Infect
Dis 2002;8:891–6.
8. Hauser PM, Francioli P, Bille J, Telenti A, Blanc DS. Typing of
Pneumocystis carinii f. sp. hominis by single-strand conformation
polymorphism of four genomic regions. J Clin Microbiol
1997;35:3086–91.
9. Nahimana A, Blanc DS, Francioli P, Bille J, Hauser PM. Typing of
Pneumocystis carinii f. sp. hominis by PCR-SSCP to indicate a high
frequency of co-infection. J Med Microbiol 2000;49:753–8.
10. Hauser PM, Blanc DS, Sudre P, Senggen Manoloff E, Nahimana A,
Bille J, et al. Genetic diversity of Pneumocystis carinii in HIV-posi-
tive and negative patients as revealed by PCR-SSCP typing. AIDS
2001;15:461–6.
11. Tsolaki AG, Miller RF, Underwood AP, Banerji S, Wakefield AE.
Genetic diversity at the internal transcribed spacer regions of the
rRNA operon among isolates of Pneumocystis carinii from AIDS
patients with recurrent pneumonia. J Infect Dis 1996;174:141–56.
12. Keely SP, Baughman RP, Smulian AG, Dohn MN, Stringer JR. Source
of Pneumocystis carinii in recurrent episodes of pneumonia in AIDS
patients. AIDS 1996;10:881–8.
13. Keely SP, Stringer JR. Sequences of Pneumocystis carinii f. sp.
hominis strains asssociated with recurrent pneumonia vary at multi-
ple loci. J Clin Microbiol 1997;35:2745–7.
14. Hughes WT. Current issues in the epidemiology, transmission, and
reactivation of Pneumocystis carinii. Semin Respir Infect
1998;13:283–8.
15. Helweg-Larsen J, Lundgren B, Lundgren JD. Heterogeneity and
compartmentalization of Pneumocystis carinii f. sp. hominis geno-
types in autopsy lungs. J Clin Microbiol 2001;39:3789–92.
Address for correspondence: Philippe M. Hauser, Centre Hospitalier
Universitaire Vaudois, 1011 Lausanne, Switzerland; fax: +41 21 314 40
60; email: Philippe.Hauser@chuv.hospvd.ch
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 867
DISPATCHES
Search ppast iissues oof EEID aat wwww.cdc.gov/eid

VIM- and IMP-Type
Metallo-β-lactamase–
Producing
Pseudomonas spp.
and Acinetobacter
spp. in Korean
Hospitals
Kyungwon Lee,* Wee Gyo Lee,† Young Uh,‡
Gyoung Yim Ha,§ Jihyun Cho,¶ Yunsop Chong,*
and the Korean Nationwide Surveillance of
Antimicrobial Resistance Group
1
We determined the occurrence of acquired metallo-β-
lactamase (MBL)–producing bacteria in Korean hospitals.
Among the isolates nonsusceptible to imipenem that were
collected from 28 hospitals from 2000 to 2001, 44 (11.4%)
of 387 Pseudomonas spp. and 38 (14.2%) of 267
Acinetobacter spp. produced MBL and had alleles of
bla
VIM-2
or bla
IMP-1
. MBL-producing isolates were detected
in 60.7% of the hospitals.
C
arbapenems are often used as a last resort for treating
serious infections attributable to multidrug-resistant
gram-negative bacilli because these drugs are stable even
to extended-spectrum and AmpC β-lactamases. However,
gram-negative bacilli with acquired metallo-β-lactamase
(MBL), IMP-1, emerged and spread during the early 1990s
in Japan (1). IMP-1 and its variants were then detected in
other countries (2).
Another type of acquired MBL, VIM-1, was first
reported in Pseudomonas aeruginosa in Italy (3), followed
by reports of VIM-2 in France and Greece. VIM-2 was
detected in P. aeruginosa in a Korean hospital isolated as
early as 1995 (4). The occurrence of the VIM enzyme has
continued to evolve: VIM-3 was reported in Taiwan (5),
and VIM-4 in the United States (6).
The bla
IMP
and bla
VIM
genes are horizontally transfer-
able because they are inserted in integrons, and some of
these integrons are located on conjugative plasmids (7).
Because of its ability to spread, carbapenem resistance
related to IMP and VIM β-lactamase production has
become a serious concern (8). Laboratory personnel and
physicians must consider the therapeutic and infection-
control implications of not detecting carbapenemase-pro-
ducing bacteria (9). A large number of VIM-2–producing
Pseudomonas spp. have been detected in a Korean hospi-
tal since 1995 (4), but the occurrence of MBL-producing
isolates has not been studied at other Korean hospitals,
despite the high prevalence of carbapenem-resistant P.
aeruginosa and Acinetobacter spp. (10). The aim of our
study was to determine the occurrence of acquired MBL-
producing P. aeruginosa and Acinetobacter spp. among
isolates collected by Korean Nationwide Surveillance of
Antimicrobial Resistance Group hospitals. The MBL types
produced and the sources of the MBL-positive isolates
were also investigated. In addition, pulsed-field gel elec-
trophoresis (PFGE) patterns were compared to determine
intra- and inter-hospital spread of resistant strains.
The Study
Nonduplicate, imipenem-resistant isolates of 387
Pseudomonas spp. and 267 Acinetobacter spp. were col-
lected from 2000 to 2001 from 28 hospitals in the Korean
Nationwide Surveillance of Antimicrobial Resistance
Group hospitals located in six cities or provinces. The
identification of the species and the imipenem susceptibil-
ity were confirmed at the coordinating laboratory by using
conventional tests (11) or ATB 32 GN system (bioMerieux,
Marcy-l’Etoile, France) and by using the disk diffusion
test (12), respectively.
MBL production was screened by using the Hodge test
and the imipenem-EDTA double disk synergy test (13).
The bla
IMP-1
and bla
VIM-2
alleles were detected by poly-
merase chain reaction (PCR), and three of the positive iso-
lates were confirmed by sequencing, as described previ-
ously (4). XbaI-digested genomic DNA of P. aeruginosa
isolates was separated by PFGE using the CHEF-DR-II
system (Bio-Rad Laboratories, Hercules, CA) (4). The pat-
tern was analyzed visually and by using UVIBand and
Map software (UVItec Ltd., Cambridge, UK).
Some of the Pseudomonas and Acinetobacter isolates
collected were not fully resistant to imipenem but showed
intermediate resistance when retested. Among the isolates
not susceptible to imipenem, 44 (11.4%) of 387
Pseudomonas spp. (42 P. aeruginosa and 2 P. putida) and
38 (14.2%) of 267 Acinetobacter spp. were considered
MBL producers on the basis of positive results by the
868 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
1
In addition to the listed authors, this group includes the following:
Jung Oak Kang, Moon Yeon Kim, Nam Yong Lee, Mi-Na Kim,
Myungshin Kim, Kyung Soon Song, Ki Sook Hong, In Ki Paik, Hye
Soo Lee, Sook-Jin Jang, Ae Ja Park, Sung Ha Kang, Won Keun
Song, Insoo Rheem, Eui-Chong Kim, Yeon Joon Park, Jong Hee
Shin, Myungseo Kang, Young-Kyu Sun, Hee Joo Lee, Hwan-Sub
Lim, Jong Wook Lee, and Bo-Moon Shin.
*Yonsei University College of Medicine, Seoul, Korea; †Ajou
University School of Medicine, Suwon, Korea; ‡Yonsei University
Wonju College of Medicine, Wonju, Korea; §College of Medicine of
Dongguk University, Kyongju, Korea; and Wonkwang University
College of Medicine, Iksan, Korea

Hodge test and imipenem-EDTA double disk synergy test
(Table 1). MBL-producing Pseudomonas spp. and
Acinetobacter spp. were detected in 11 (52.4%) of 21 and
10 (41.7%) of 24 hospitals that were located in four of five
and five of six cities or provinces, respectively. We detect-
ed the bla
VIM
allele by PCR from all 42 isolates of MBL-
producing P. aeruginosa and 2 isolates of P. putida. The
bla
VIM-2
and bla
IMP-1
alleles were detected in 27 (71.1%)
and 11 (28.9%) of 38 Acinetobacter isolates, respectively
(Table 2). Nucleotide sequencing for three representative
PCR-positive isolates confirmed the presence of the
bla
VIM-2
gene in one isolate each of P. aeruginosa and
Acinetobacter spp., and the bla
IMP-1
gene in one isolate of
Acinetobacter spp.
The MBL-producing strains were isolated mainly from
intensive-care unit patients (31.7%) and other inpatients
(50.0%); five (6.1%) were from emergency service and
other outpatients (Table 3). Overall, MBL-producing iso-
lates were mainly obtained from specimens of sputum
(50.0%) and urine (29.3%). However, the proportion of
MBL-producing isolates was relatively higher among
urine isolates: 17.3% for Pseudomonas spp. and 29.2% for
Acinetobacter spp. We obtained one MBL-producing
Acinetobacter isolate from each of the following specimen
types: blood, spinal fluid, pleural fluid, and venous
catheter tip (Table 4).
The PFGE of the XbaI-digested genomic DNA of 39
isolates of P. aeruginosa showed 22 patterns (data not
shown). Six isolates from one hospital had an identical pat-
tern. Thirteen isolates (33.3%) belonged to another identi-
cal pattern—six from one hospital, two from each of two
hospitals, and one from each of three hospitals, which were
located in a city and two provinces.
Conclusions
In this study, >10% of all imipenem-nonsusceptible iso-
lates of Pseudomonas spp. and Acinetobacter spp. were
attributable to MBL production (Table 1), and these MBL-
producing isolates were detected in 62.5% of the partici-
pating hospitals. Our finding indicates that MBL-produc-
ing P. aeruginosa is more prevalent in Korea than in other
countries (2) and that MBL-producing Acinetobacter spp.
is increasing. The percentage of hospitals with MBL-pro-
ducing isolates might have been higher if a larger number
of imipenem-nonsusceptible isolates had been collected
for this study.
VIM-2 was the only type of acquired MBL identified
initially in Korea. VIM-2–producing P. aeruginosa was
isolated at almost the same time in Europe (7) and Korea
(4). However, IMP-1–producing isolates were rare until
2000 in Korea. Only one and three IMP-1-positive P.
aeruginosa and Acinetobacter spp., respectively, have
been isolated at the coordinating laboratory (4, unpub.
data). In our study, 11 (28.9%) of 38 MBL-positive isolates
of Acinetobacter spp. were IMP producers (Table 2). This
increase suggests the possible introduction of IMP-produc-
ing strains of Acinetobacter spp. from Japan, where 28 iso-
lates of bla
IMP-1
-positive Acinetobacter baumannii were
reported in a hospital as early as 1994 to 1996 (14).
Rasmussen and Bush (15) predicted that an increase of
MBL-producing organisms was inevitable, given the more
frequent use of carbapenems. Imipenem has been used for
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 869
DISPATCHES
Table 1. Detection of metallo-β-lactamase–producing isolates among imipenem-nonsusceptible isolates of Pseudomonas spp. and
Acinetobacter spp.
No. hospitals (%)
No. isolates (%)
Organism
City/province
Tested
Positive
Tested
Positive
Seoul
11
a
4 (36.4)
144
12 (8.3)
Kyungki
2
2 (100)
40
6 (15.0)
Kangwon
2
1 (50.0)
57
2 (3.5)
Chulla
4
4 (100)
108
24 (22.2)
Kyungsang
2
0 (0)
38
0 (0)
Pseudomonas spp.
Tota l
21
11 (52.4)
387
44 (11.4)
Seoul
11
a
4 (36.4)
107
12 (11.2)
Kyungki
3
0 (0)
29
0 (0)
Kangwon
3
2 (25.0)
41
8 (19.5)
Chulla
3
1 (12.5)
25
13 (52.0)
Kyungsang
2
1 (50.0)
53
1 (1.9)
Chungchung
2
2 (100)
12
4 (33.3)
Acinetobacter spp.
Tota l
24
10 (41.7)
267
38 (14.2)
a
Four were tertiary-care hospitals.
Table 2. Detection of bla
VIM-2
and bla
IMP-1
allele from metallo-β-
lactamase–producing Pseudomonas spp. and Acinetobacter
spp. by polymerase chain reaction
No. isolates (%)
Organism
Tested
bla
VIM-2
positive
bla
IMP-1
positive
Pseudomonas aeruginosa
42
42 (100)
0 (0)
P. putida
2
2 (100)
0 (0)
Acinetobacter spp.
38
27 (71.1)
11 (28.9)
Tota l
82
71 (86.6)
11 (13.4)

only 9 years in Korea, but the imipenem-resistance rate of
P. aeruginosa has rapidly risen from 6% in 1996 to 19% in
2001. A study by the Korean Nationwide Surveillance of
Antimicrobial Resistance Group showed that the mean
imipenem-resistance rates of P. aeruginosa in 1997 did not
differ substantially depending on hospital size, (i.e., 17%
in medium hospitals [<1,000 beds] and 18% in large hos-
pitals [>1,000 beds]). The mean resistance rates to imipen-
em were not lower than those to ceftazidime in 2000, i.e.,
21% versus 18% in large hospitals and 20% versus 19% in
medium hospitals (data not shown).
Acinetobacter spp. are also common nosocomial
pathogens with multidrug resistance. The imipenem resist-
ance rate of this organism isolated in Korea was found to
be much lower than that of P. aeruginosa, but its resistance
rate rose from 4% in the first quarter to 20% in the third
quarter of 2002 at the coordinating laboratory (data not
shown).
In our study, MBL-producing Pseudomonas spp. and
Acinetobacter spp. were isolated mainly from sputum and
urine specimens, and most (81.7%) isolates were from
inpatients and intensive-care unit patients. Therefore,
proper treatment of respiratory secretions and urine from
intensive-care unit patients is considered an important
aspect of preventing the spread of MBL-producing organ-
isms. The presence of P. aeruginosa isolates with identical
PFGE patterns among those collected not only from cer-
tain hospitals but also from different hospitals suggests
that clonal spread is at least a part of the cause of intra- and
inter-hospital dissemination of MBL-producing isolates.
The presence of VIM-2-producing Serratia marcescens,
Enterobacter cloacae, and Achromobacter xylosoxidans
subsp. denitrificans (unpub. data) in other hospitals also
suggests horizontal transfer of the resistance determinants.
Cornaglia et al. reported that five of seven patients
infected with MBL-producing P. aeruginosa died,
although the cause of death was difficult to establish with
certainty (16). Clinical studies on the infection are rare
because isolation of MBL-producing gram-negative bacil-
li increased only recently. We anticipate difficulties in
treating patients infected with MBL-producing gram-neg-
ative bacilli, which can hydrolyze, in vitro, all available β-
lactams, except aztreonam for which clinical efficacy is
unknown.
Our study indicates the urgent need for action to pre-
vent further spread of MBL-producing organisms.
Previous experiences with penicillin-nonsusceptible pneu-
mococci, methicillin-resistant Staphylococcus aureus, van-
comycin-resistant Enterococcus faecium, and extended-
spectrum β-lactamase–producing Klebsiella pneumoniae
indicate that once resistant bacteria can become wide-
spread they cannot be controlled (10). Our first task is to
detect MBL producers among clinical isolates (9).
Although the National Committee for Clinical Laboratory
Standards document (12) does not contain procedures for
detection, simple procedures are available (13).
The prevalence of bla
VIM-2
allele-positive P. aeruginosa
and bla
IMP-1
allele-positive Acinetobacter spp. is increasing
possibly because of clonal and horizontal spread of the
resistance determinant in Korean hospitals. Sputum and
urine from inpatients and intensive-care unit patients were
found to be the main sources of MBL-producing isolates.
Laboratories not only in Korea but also in other countries
with carbapenem-resistant organisms must be prepared to
screen MBL-producing isolates to determine the clinical
impact and prevent further spread of MBL-producing
organisms.
870 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Table 3. bla
VIM-2
and bla
IMP-1
allele-positive Pseudomonas spp. and Acinetobacter spp. isolated by service
No. isolates (%)
Organism
Outpatient
Inpatient
Intensive-care unit
Others
Tota l
Pseudomonas spp.
3 (6.8)
a
26 (59.1)
11 (25.0)
4 (9.1)
44 (100)
Acinetobacter spp.
2 (5.2)
b
15 (39.5)
15 (39.5)
6 (15.8)
38 (100)
Tota l
5 (6.1)
41 (50.0)
26 (31.7)
10 (12.2)
82 (100)
a
Two were emergency service patients, and one was a urology patient.
b
One was an emergency service patient, and one was a pediatric patient.
Table 4. bla
VIM-2
and bla
IMP-1
allele-positive Pseudomonas spp. and Acinetobacter spp. isolated by source
No. (%) of isolates with metallo-â-lactamase
Pseudomonas spp.
Acinetobacter spp.
Tota l
Source
Tested
Positive
Tested
Positive
Tested
Positive
% positive by
source
Sputum
200
22 (11.0)
143
19 (13.3)
343
41 (12.0)
50.0
Urine
98
17 (17.3)
24
7 (29.2)
122
24 (19.7)
29.3
Wound
49
2 (4.1)
71
7 (9.9)
120
9 (7.5)
10.9
Other
a
18
3 (16.7)
29
5 (17.2)
47
8 (17.0)
9.8
Tota l
387
44 (11.4)
267
38 (14.2)
654
82 (12.5)
100
a
Others included one Acinetobacter isolate of specimens from blood, spinal fluid, pleural fluid, and a venous catheter tip.

Acknowledgments
We thank Jong Hwa Yum and Dongeun Yong for detecting
the metallo-β-lactamase genes and Yonghee Suh for screening
the MBL producers.
Dr. Lee is director of the Research Institute of Bacterial
Resistance and a professor in the Department of Laboratory
Medicine, Yonsei University College of Medicine, Seoul, Korea.
He is the Korean coordinator in the World Health
Organization/Centers for Disease Control and Prevention
External Quality Assurance Scheme, and he is the organizer of
the Korean Nationwide Surveillance of Antimicrobial Resistance
Group. His research interests include antimicrobial resistance of
bacteria and its mechanisms.
References
1. Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, et
al. Molecular characterization of an enterobacterial metallo-β-lacta-
mases found in a clinical isolates of Serratia marcescens that shows
imipenem resistance. Antimicrob Agents Chemother 1994;38:71–8.
2. Gibb AP, Tribuddharat C, Moore RA, Louie TJ, Krulicki W,
Livermore DM, et al. Nosocomial outbreak of carbapenem-resistant
Pseudomonas aeruginosa with a new bla
IMP
allele, bla
IMP-7
.
Antimicrob Agents Chemother 2002;46:255–8.
3. Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G,
Fontana R, et al. Cloning and characterization of bla
VIM
, a new inte-
gron-borne metallo-β-lactamase gene from a Pseudomonas aerugi-
nosa clinical isolate. Antimicrob Agents Chemother 1999;
43:1584–90.
4. Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, et al. bla
VIM-2
cassette-containing novel integrons in metallo-β-lactamase–produc-
ing Pseudomonas aeruginosa and Pseudomonas putida isolates dis-
seminated in a Korean hospital. Antimicrob Agents Chemother
2002;46:1053–8.
5. Yan J-J, Hsueh P-R, Ko W-C, Luh E-T, Tsai S-H, Wu H-M, et al.
Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and
identification of VIM-3, a novel variant of the VIM-2 enzyme.
Antimicrob Agents Chemother 2001;45:2224–8.
6. Tolman MA, Rolston K, Jones RN, Walsh TR. Molecular characteri-
zation of VIM-4, a novel metallo-beta-lactamase isolated from Texas:
report from the cancer surveillance program (2001). Proceedings of
the 42nd Interscience Conference on Antimicrobial Agents and
Chemotherapy. Abstract C1-1851. San Diego, California; 2002.
7. Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo T-D, et al.
Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lac-
tamase and its plasmid- and integron borne gene from a Pseudomonas
aeruginosa clinical isolate in France. Antimicrob Agents Chemother
2000;44:891–7.
8. Richet HM, Mohammed J, McDonald LC, Jarvis WR, INSPEAR.
Building communication networks: International Network for the
Study and Prevention of Emerging Antimicrobial Resistance. Emerg
Infect Dis 2001;7:319–22.
9. Molland ES, Black JA, Ourada J, Reisbid MD, Hanson ND, Thomson
KS. Occurrence of newer β-lactamases in Klebsiella pneumoniae iso-
lates from 24 U.S. hospitals. Antimicrob Agents Chemother
2002;46:3837–42.
10. Lee K, Lee HS, Jang SJ, Park AJ, Lee MH, Song WK, et al.
Antimicrobial resistance surveillance of bacteria in 1999 in Korea
with a special reference to resistance of enterococci to vancomycin
and gram-negative bacilli to third generation cephalosporin, imipen-
em, and fluoroquinolone. J Korean Med Sci 2001;16:262–70.
11. Kiska DL, Gilligan PH. Pseudomonas. In: Murray PR, Baron EJ,
Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical
microbiology, 7th ed. Washington: American Society for
Microbiology; 1999. p. 517–25.
12. National Committee for Clinical Laboratory Standards. Performance
standards for antimicrobial susceptibility testing. Eleventh informa-
tional supplement. NCCLS document M100-S11. Wayne (PA): The
Committee, 2001.
13. Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified
Hodge test and EDTA-disk synergy tests to screen metallo-β-lacta-
mase-producing strains of Pseudomonas and Acinetobacter species.
Clin Microbiol Infect 2001;7:88–91.
14. Takahashi A, Yomoda S, Kobayashi I, Kubo T, Tsunoda M, Iyobe S.
Detection of carbapenemase-producing Acinetobacter baumannii in a
hospital. J Clin Microbiol 2000;38:526–9.
15. Rasmussen BA, Bush K. Carbapenem-hydrolyzing β-lactamases.
Antimicrob Agents Chemother 1997;41:223–32.
16. Cornaglia G, Mazzariol A, Lauretti L, Rossolini GM, Fontana R.
Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa
producing VIM-1, a novel transferable metallo-β-lactamase. Clin
Infect Dis 2000;31:1119–25.
Address for correspondence: Yunsop Chong, Department of Laboratory
Medicine and Research Institute of Bacterial Resistance, Yonsei
University College of Medicine, 134 Shinchon-dong Seodaemun-ku,
Seoul, Korea 120-752; fax: 82-2-313-0908; email: whonetkor@
yumc.yonsei.ac.kr
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 871
DISPATCHES

Leishmaniasis in
Germany
Gundel Harms,* Gabriele Schönian,*
and Hermann Feldmeier†
In 2000, a reference center was created to systemati-
cally record leishmaniases in Germany. We analyzed 58
cases of leishmaniases imported during a 2-year period.
These findings will serve as a baseline for the sandfly vec-
tor’s anticipated northward move because of global warm-
ing and as an advisory for immunocompromised persons
traveling to leishmaniasis-endemic areas.
L
eishmaniases compose a spectrum of protozoal dis-
eases currently endemic in 88 countries in Asia,
Africa, the Americas, and southern Europe. The geograph-
ic distribution of leishmaniases has widened, and the dis-
ease is reported in areas in which leishmaniasis was previ-
ously nonendemic (1). Apart from cutaneous, mucocuta-
neous/mucosal, and visceral leishmaniases, HIV-1–associ-
ated leishmaniasis acquired in southern Europe and in
other parts of the world have been observed in increasing
numbers (1,2).
Leishmaniasis is not notifiable in Germany. In
September 2000, a national advice and reference center
was created at the Institute of Tropical Medicine in Berlin;
the aim of the center was to monitor the frequency, origin,
and type of leishmaniases seen in Germany; to advise
physicians; and to improve information for travelers to dis-
ease-endemic areas. The healthcare professionals were
informed about the reference center by the Robert-Koch-
Institute, the center for surveillance of infectious diseases
in Germany, as well as through the Journal of the German
Medical Association, which is received by every registered
physician (3). Leishmaniasis was diagnosed if parasites
were detected in smears, culture, histologic sections, by
polymerase chain reaction (PCR) of lesion biopsy speci-
mens, bone marrow, or peripheral blood. For detection of
Leishmania-specific antigen the small subunit (ssu rRNA),
the internal transcribed spacer (ITS-1) region of the ribo-
somal RNA genes, or both, were amplified by PCR (4).
Leishmania complexes and species were determined by
digestion of the ribosomal ITS-1 PCR product with restric-
tion enzymes (4).
Within 2 years, 70 cases of leishmaniases (43 cutaneous
or mucocutaneous/mucosal; 27 visceral) were reported.
For 58 case-patients (35 cutaneous or mucocutaneous/
mucosal; 23 visceral), data were available on the age, sex,
residence, travel destination, possible exposure location,
reason for travel, duration of stay, duration and type of
symptoms, concomitant diseases or therapies, type of diag-
nosis, and treatment received.
Cutaneous and Mucosal Leishmaniasis
Of the 35 patients with cutaneous or mucocutaneous/
mucosal leishmaniasis, 30 were German tourists (Table 1).
The male-to-female ratio was 1.5:1. Ten had contracted
cutaneous or mucocutaneous/mucosal leishmaniasis in
Europe, 11 in Central and South America, 6 in Asia, and 3
in Africa. Two persons had been infected during work
stays of 1 to 4 months in French Guyana, one each in Peru
and Libya, and one patient had immigrated from
Afghanistan.
The median duration of lesions until the diagnosis of
leishmaniasis was made was 4 months (range 3 weeks to 2
years). Sixteen patients had more than one lesion (median
2, range 1–6 lesions). Seventeen lesions were located in
the face, including mouth and nose, 28 on the upper
extremities and 21 on the lower extremities. Lesions were
ulcerated in 39 cases, papular-nodular in 24, and plaque-
like in 3. Parasites were detected in 13 of 20 smears, in 9
of 10 cultures, and in 14 of 16 histologic sections; by using
PCR, Leishmania-specific DNA was detected in 16 of 16
biopsy specimens.
Patient 1 had lesions in the mouth caused by L. infan-
tum. She was under continuous immunosuppressive treat-
ment for severe bronchial asthma. Patient 2 had received
methotrexate and steroids for treatment of systemic col-
lagenosis for several weeks. Both patients were tested for
leishmanial infection in the blood; in both patients, the
Leishmania-specific PCR of the buffy coat of the blood
was positive. Patient 22 had mucocutaneous leishmaniasis
of the nasal septum. She had been treated for a skin lesion
caused by L. braziliensis 3 years earlier.
Visceral Leishmaniasis
A total of 18 of the 23 visceral leishmaniasis patients
were German tourists; 3 were immigrants from Angola,
Iran, and Togo; and 2 were visitors from Italy and Portugal
(Table 2). The male-to-female ratio was 6.7:1.
The median time between symptom onset and the cor-
rect diagnosis was 4 months (range 1–16 months). All
case-patients had fever, 17 (74%) had splenomegaly, 11
(48%) hepatomegaly, 20 (87%) anemia, 17 (74%) leukope-
nia, and 8 (35%) thrombocytopenia.
Bone marrow smears indicated Leishmania in 18 of 20,
bone marrow culture in 6 of 7, bone marrow histologic
sections in 7 of 8, PCR of the bone marrow in 8 of 9, and
PCR of the buffy coat of the blood in 7 of 7 cases.
Additionally, antibodies were detected in medium to high
concentration by an immunofluorescence test, enzyme-
872 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
*Charité, Humboldt University Berlin, Berlin, Germany; and †Free
University of Berlin, Berlin, Germany

linked immunosorbent assay (ELISA), or both, in 14 of 15
cases. Species was identified in 7 of 18 visceral cases con-
tracted in southern Europe and indicated Leishmania
belonging to the L. donovani complex, which implicated
infection with L. infantum.
Six cases of visceral leishmaniasis occurred in children
2 months of age to 11 years of age. Four German tourists
and two immigrants had long-known HIV infection (medi-
an duration 3 years, range 8 months–6 years). All HIV–co-
infected patients had CD4-cell counts below 200/µL
(median 108, range 23–185 CD4 cells/µL) when the diag-
nosis of visceral leishmaniasis was made. Of the remaining
11 patients, 1 had a thymoma with impaired T-helper-1 cell
function, 2 had received intermittent immunosuppressive
therapy (methotrexate and steroids) for rheumatologic dis-
ease, and 2 patients had their spleens removed. Three
patients were in an impaired general condition because of
combinations of diabetes, hypertonus, hypercholes-
terolemia, and emphysema. In the remaining three patients
(53–68 years of age), apart from hypertonus in one, no
impairing condition was detected.
Discussion
Information on single cases and a small case series of
imported leishmaniases in Germany is available, but sys-
tematic reporting on frequency, type, and origin of leish-
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 873
DISPATCHES
Table 1. Characteristics of patients with cutaneous and mucosal leishmaniasis, Germany
No.
Exposure
Status
Sex
Age (y)
Leishmania species
Treatment
Outcome
Europe
1
France
Tourist
F
64
L. donovani complex
No treatment
No cure
2
Italy
Tourist
F
24
L. donovani complex
Liposomal amphotericin B
Improved
3
Malta
Tourist
M
22
L. donovani complex
Perilesional pentavalent antimonials
Cured
4
Malta
Tourist
M
39
L. donovani complex
Perilesional pentavalent antimonials
Cured
5
Malta
Tourist
F
57
n.d.
Perilesional pentavalent antimonials
Cured
6
Malta
Tourist
M
62
n.d.
Perilesional pentavalent antimonials
Cured
7
Spain (Majorca)
Tourist
F
5
n.d.
Pentamidine isethionate
Cured
8
Spain (Majorca)
Tourist
M
13
L. donovani complex
Perilesional pentavalent antimonials
Cured
9
Spain
Tourist
F
31
L. donovani complex
No treatment
Unknown
10
Spain
Tourist
M
33
L. donovani complex
Antibiotic
Cured
Americas
11
Belize
Tourist
M
36
L. braziliensis complex
Liposomal amphotericin B
Unknown
12
Belize
Tourist
F
32
n.d.
IFN-gamma
Cured
13
Bolivia
Tourist
M
35
L. braziliensis complex
Liposomal amphotericin B
Unknown
14
Brazil
Tourist
M
25
n.d.
Systemic pentavalent antimonials
Cured
15
Brazil
Tourist
M
39
L. braziliensis complex
Liposomal amphotericin B
Unknown
16
Brazil
Tourist
M
33
L. braziliensis complex
Liposomal amphotericin B
Cured
17
Ecuador
Tourist
F
29
L. braziliensis complex
Liposomal amphotericin B
Cured
18
Ecuador
Tourist
M
36
n.d.
Liposomal amphotericin B
Cured
19
French Guyana
Work stay
M
28
L. braziliensis complex
Liposomal amphotericin B
Cured
20
French Guyana
Work stay
M
22
L. braziliensis complex
Liposomal amphotericin B
Cured
21
Guatemala
Tourist
M
31
L. mexicana
Ketoconazole
Cured
22
Peru
Work stay
F
25
L. braziliensis complex
Liposomal amphotericin B
Cured
23
Peru
Tourist
F
33
L. braziliensis complex
Liposomal amphotericin B
Cured
24
Peru
Tourist
M
35
L. braziliensis complex
Liposomal amphotericin B
Cured
Asia
25
Afghanistan
Immigrant
M
21
L. tropica
Aminosidine ointment
No cure
26
Afghanistan
Tourist
F
12
n.d.
Aminosidine ointment
Cured
27
United Arab Emirates
Tourist
F
44
n.d.
Perilesional pentavalent antimonials
Cured
28
Syria
Tourist
M
5
n.d.
Perilesional pentavalent antimonials
Unknown
29
Syria
Tourist
F
3
n.d.
Perilesional pentavalent antimonials
Unknown
30
Turkey
Tourist
F
33
n.d.
Antibiotic
Cured
31
Turkey
Tourist
M
37
L. donovani complex
Liposomal amphotericin B
Improved
Africa
32
Egypt
Tourist
M
25
L. tropica
Aminosidine ointment
No cure
33
Egypt
Tourist
F
27
L. tropica
Aminosidine ointment
Cured
34
Kenya
Tourist
M
50
n.d.
Perilesional pentavalent antimonials
Cured
35
Lybia
Work stay
M
34
n.d.
Aminosidine ointment
Cured
a
n.d., not done; IFN, interferon.
manial infections in Germany did not exist until 2000
(5–7). Our recent surveillance is dependent on passive
consultation and reporting and therefore may have selec-
tion bias because if visceral leishmaniasis, a potentially
fatal disease that requires hospitalization, is suspected,
advice on diagnosis and treatment is sought more often
than for the skin infection. We assume that our system cap-
tures approximately half of the visceral leishmaniasis
cases and approximately one third of the classical cuta-
neous cases imported to Germany.
A total of 47% of all cases, but 78% of the visceral
cases were contracted in the European Mediterranean area
and Portugal, and most of the infections indicated a species
of the L. donovani complex, most probably L. infantum, as
874 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Table 2. Characteristics of patients with visceral leishmaniasis, Germany
No.
Exposure
Status
Sex
Age
Risk factor
Leishmania
species
Treatment
Outcome
1
Italy
Tourist
M
2 y
Child
n.d.
Liposomal amphotericin B
Cured
2
Italy
Tourist
F
5 y
Child
n.d.
Liposomal amphotericin B
Cured
3
Italy
Tourist
M
11 y
Child
n.d.
Liposomal amphotericin B
Cured
4
Spain
Tourist
F
8 mo
Child
n.d.
Liposomal amphotericin B
Cured
5
Spain
Tourist
M
9 mo
Child
L. donovani
complex
Liposomal amphotericin B
Cured
6
Iran
Immigrant
M
7 y
Child
L. donovani
complex
Liposomal amphotericin B
Cured
7
Spain
Tourist
M
43 y
HIV
L. donovani
complex
Liposomal amphotericin B;
maintenance therapy:
HAART plus liposomal
amphotericin B once monthly
No relapse for 6
months
8
Spain (Ibiza)
Tourist
M
48 y
HIV
L. donovani
complex
Liposomal amphotericin B;
maintenance therapy:
HAART plus liposomal
amphotericin B once monthly
No relapse for 8
months
9
Portugal
Visitor
M
29 y
HIV
n.d.
Liposomal amphotericin B;
maintenance therapy:
HAART plus liposomal
amphotericin B once monthly
unknown
10
France
Tourist
M
31y
HIV
L. donovani
complex
Liposomal amphotericin B;
maintenance therapy:
HAART plus liposomal
amphotericin B once monthly
Relapse after 4
months;
retreatment with
liposomal
amphotericin B; No
relapse for 3
months
11
Angola
Immigrant
M
40 y
HIV
L. donovani
complex
Systemic pentavalent
antimonials; maintenance
therapy: HAART
plus pentavalent antimonials
once monthly
unknown
12
Togo
Immigrant
M
37 y
HIV
L. donovani
complex
Liposomal amphotericin B
Cured
13
Italy (Sicily)
Visitor
M
31 y
Thymoma
n.d.
Liposomal amphotericin B
Cured
14
Italy (Ischia)
Tourist
M
67 y
Methotrexate/steroids
L. donovani
complex
Liposomal amphotericin B
Cured
15
Italy (Sicily)
Tourist
M
68 y
Methotrexate
n.d.
Liposomal amphotericin B
Cured
16
Italy (Ischia)
Tourist
M
70 y
Splenectomy
n.d.
Liposomal amphotericin B
Cured
17
Spain
Tourist
M
51 y
Splenectomy
L. donovani
complex
Liposomal amphotericin B
Cured
18
Greece
(Korfu)
Tourist
M
66 y
Diabetes mellitus
Hypertonus
n.d.
Liposomal amphotericin B
Cured
19
Spain
Tourist
M
52 y
Hypertonus
Hypercholesterolemia
n.d.
Liposomal amphotericin B
Cured
20
Greece
(Korfu)
Tourist
M
45 y
Diabetes mellitus
Emphysema
L. donovani
complex
Systemic pentavalent
antimonials
Cured
21
Tunisia
Tourist
M
53 y
Hypertonus
L. donovani
complex
Liposomal amphotericin B
Cured
22
Malta
Tourist
F
55 y
-
n.d.
Liposomal amphotericin B
Cured
23
China
Tourist
M
67 y
-
n.d.
Liposomal amphotericin B
Cured
a
n.d., not done; HAART, highly active anti-retroviral therapy.

the probable causative agent. Thirteen infections (22%)
were acquired on the Mediterranean islands of Ibiza,
Ischia, Majorca, Malta, Korfu, or Sicily.
This distribution reflects the fact, that the
Mediterranean countries, Spain, Italy, and the
Mediterranean islands, in particular, are the favorite vaca-
tion areas for Germans. Annually, Germans take 18 million
vacations to the European Mediterranean area (including 8
million to Spain and 6 million to Italy) with a median dura-
tion of 2 weeks. Sixty percent of travel to Italy and 90% of
travel to Spain are to Leishmania-endemic areas.
While leishmaniasis has always been endemic in the
Mediterranean countries, the maximum northern latitude
for sandfly survival is speculated to move further to the
North, beyond Germany (1) because of global warming. If
this scenario is correct, the imported cases may serve as a
potential substrate for the sandfly vector. Dogs that are
imported as pets from the disease-endemic areas of south-
western Europe or that contract the infection when accom-
panying their owners for vacation are another potential
substrate (8).
Infections with L. infantum in a child, as well as in a
horse who had never left Germany, have recently been
described and have led to speculations about an autochtho-
nous focus (9,10). Also recently, the first sandfly species,
Phlebotomus mascittii Grassi, 1908, was detected in south-
ern Germany, although its potential as a vector of
Leishmania remains to be demonstrated (11).
As expected, visceral leishmaniasis is often manifested
in persons with impaired immunocompetence because of
young age, HIV infection, immunosuppressive therapy and,
in our analysis, in older persons with concomitant diseases.
Notably, 12 (67%) of 18 of the visceral cases contracted
in the European Mediterranean area were in adults, thus
confirming a change in age groups affected. Formerly, vis-
ceral leishmaniasis was known mainly as a disease of chil-
dren (1,2). This change may partly be explained by the
increased proportion of Leishmania and HIV–co-infected
persons and partly by increased travel activities of otherwise
immunocompromised persons, including elderly persons.
Furthermore, even in patients with cutaneous leishma-
niasis, dissemination of parasites has to be excluded in
case of impaired immunocompetence (e.g., immunosup-
pressive treatment). In these cases, Leishmania-specific
PCR of the buffy coat of the peripheral blood is a sensitive
method for detecting parasite spread beyond the skin.
Parents of small children and persons with reduced
immunocompetence should be informed about their
increased susceptibility to infection with Leishmania when
traveling to disease-endemic areas. Measures to reduce the
exposure to sandflies, such as clothes, repellents, and mos-
quito nets as well as collars impregnated with repellents
for accompanying dogs, should be recommended.
Acknowledgments
We thank colleagues at German hospitals and health institu-
tions who discussed their patients with us or kindly contributed
to the data collected on leishmaniases.
Dr. Harms is a senior lecturer and researcher in tropical
medicine and international health at the Institute of Tropical
Medicine and at the Medical Faculty Charité, Humboldt
University Berlin, Germany. Her primary research interests are
the interaction of HIV/AIDS and parasitic infections, leishmani-
asis in particular.
References
1. Desjeux P. The increase in risk factors for leishmaniasis worldwide.
Trans R Soc Trop Med Hyg 2001;95:239–43.
2. World Health Organization. Leishmania/HIV co-infection: south-
western Europe, 1990–1998 (WHO/LEISH/2000.42). Geneva: The
Organization; 2000.
3. Harms G, Bienzle U. Leishmaniosen-importierte Krankheiten. Dtsch
Arztebl 2000;31/32:1589–92.
4. Schönian G, Schnur L, Fahri M. Genetic heterogeneity in the species
Leishmania tropica revealed by different PCR-based methods. Trans
R Soc Trop Med Hyg 2001;95:217–24.
5. Hohenschild S, Feldmeier H. Imported kala azar in children and
adults—comparison of medical history, clinical and diagnostic find-
ings. J Trop Pediatr 1995;41:378–9.
6. Harms G, Zenk J, Martin S, Kokozidou M, Püschel W, Bienzle U, et
al. Localized lymphadenopathy due to leishmanial infection.
Infection 2001;29:355–6.
7. Hölzer E, Kupferschmidt HG. Cutaneous leishmaniasis in East
German citizens. Z Arztl Fortbild (Jena) 1986;80:381–3.
8. Gothe R, Nolte I, Kraft W. Leishmaniasis in dogs in Germany: epi-
demiological case analysis and alternatives to conventional causal
therapy. Tierarztl Prax 1997;25:68–73.
9. Bogdan C, Schönian G, Banuls AL, Hide M, Pratlong F, Lorenz E, et
al. Visceral leishmaniasis in a German child who had never entered a
known endemic area: case report and review of the literature. Clin
Infect Dis 2001;32:302–6.
10. Koehler K, Stechele M, Hetzel U, Domingo M, Schönian G, Zahner
H, et al. Cutaneous leishmaniasis in a horse in southern Germany
caused by Leishmania infantum. Vet Parasitol 2002;109:9–17.
11. Naucke TJ, Pesson B. Presence of Phlebotomus (Transphlebotomus)
mascittii Grassi, 1908 (Diptera: Psychodidae) in Germany. Parasitol
Res 2000;86:335–6.
Address for correspondence: Gundel Harms, Institute of Tropical
Medicine Berlin, Spandauer Damm 130, 14050 Berlin, Germany; fax:
+49-30-30116-888; email: [email protected]
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 875
DISPATCHES

Probable Dengue
Virus Infection
among Italian
Troops, East
Timor, 1999–2000
Mario Stefano Peragallo,* Loredana Nicoletti,†
Florigio Lista,* and ‡Raffaele D’Amelio
for The East Timor Dengue Study Group
1
To investigate the attack rate and risk factors for prob-
able dengue fever, a cross-sectional study was conducted
of an Italian military unit after its deployment to East Timor.
Probable dengue was contracted by 16 (6.6%) of 241
army troops and caused half of all medical evacuations
(12/24); no cases were detected among navy and air force
personnel.
D
engue fever (DF), caused by dengue virus (DENV)
serotypes 1 to 4, is an emerging public health problem
in many tropical countries (1). Dengue hemorrhagic fever
(DHF) and dengue shock syndrome (DSS), the severe
manifestations of DENV infection, were first recognized
in the 1950s in Southeast Asia and are today a leading
cause of childhood illness and death in many tropical
countries. More recently, DHF and DSS have emerged in
Central and South America and in the Pacific region (2,3).
DF is also recognized as an emerging health problem for
international travelers (4,5) and for troops deployed to
tropical countries (6,7).
In 1999, following a United Nations Security Council
recommendation, the International Force for East Timor
(INTERFET) was formed to restore peace on the island. In
November 1999, INTERFET troops totaled 11,000 from
17 countries. The Italian Armed Forces contributed 640
soldiers.
DF is endemic in East Timor. The peak transmission
periods for DF are July–August and December–January,
corresponding to the rainy months (8). In 1998, at least
11% of hospital inpatient deaths in East Timor were attrib-
uted to DHF (9). In October 1999, a localized outbreak of
DF in a western district was attributed to serotype 3 (9) and
serotype 2 was isolated in December 1999 (10). Serotypes
2 and 3 were also responsible for DF cases among
Australian troops returning from East Timor in January–
February 2000 (11).
During deployment, a high attack rate of febrile illness
consistent with DF was reported among Italian troops. A
seroepidemiologic survey was therefore conducted in
February 2000 among soldiers returning home, in an
attempt to determine the cause of this outbreak and to
define infection rates and risk factors for infection.
The Study
All Italian troops eligible for deployment are routinely
vaccinated against diphtheria/tetanus, tetravalent meningo-
coccal meningitis, measles/mumps/rubella, hepatitis A and
B, polio (with inactivated virus), typhoid fever (orally), and
yellow fever (YF). In this situation, troops were also vacci-
nated against Japanese encephalitis (JE) (Nakajama strain, 3
doses on days 0, 7, and 14) just before landing in East Timor.
DF prevention consisted of the use of personal protec-
tion measures against mosquitoes (repellents applied to the
skin; permethrin-treated bed nets and uniforms) along with
environmental mosquito control. Adulticide spraying was
conducted weekly by pesticide-dispersal units but only
within the campsite and in its nearest surroundings, which
were also inspected daily to reduce or eliminate breeding
sites of vectors.
Italian troops were deployed in East Timor from late
September 1999 to mid-February 2000, and all 640 partic-
ipating military personnel were eligible for inclusion in the
study. Army soldiers were permanently based on the
ground and operated in Dili and surrounding areas, while
air force and navy personnel had only logistical tasks and
their presence in Dili was episodic, since they were main-
ly aboard ship or based in Darwin (Australia).
A seroepidemiologic survey was conducted February
15–28, 2000, among troops returning to Italy after their 3-
month period of duty in East Timor. After informed con-
sent was obtained, peripheral blood specimens were drawn
and a written questionnaire administered. The question-
naire asked for personal health data, including all symp-
toms experienced during deployment and information
about compliance with personal protection measures.
Immunization status and clinical data concerning febrile
illness cases consistent with DF were obtained from stan-
dardized records kept by medical personnel. Soldiers and
navy/air force personnel were studied according to their
serologic status and disease status during deployment.
876 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
*Centro Studi e Ricerche Sanità e Veterinaria Esercito, Rome,
Italy; †Istituto Superiore di Sanità, Rome, Italy; and ‡Stato
Maggiore della Difesa and Università “La Sapienza,” Seconda
Facoltà di Medicina, Rome, Italy
1
The East Timor Dengue Study Group: Roberto Biselli, Glauco
Calì, Maria Rosaria Capobianchi, Maria Grazia Ciufolini, Raffaele
D’Amelio, Paolo Di Zenzo, Giovanni Fascia, Cristiano Fiorentini,
Alberto Germani, Michele Giattino, Giuseppe Ippolito, Florigio
Lista, Fabio Magurano, Antonella Marchi, Loredana Nicoletti,
Mario Stefano Peragallo, Alessandro Polidori, Giuseppe Sarnicola,
and Antonio Stella.

All specimens were screened for antibodies to dengue
virus serotype 2 (DEN-2), yellow fever virus (YVF), and
West Nile virus (WNV) by hemagglutination-inhibition
test (HI). All serum specimens positive for DEN-2 were
tested by neutralization test (NT) for DEN-2. Additionally,
serum samples from participants who had experienced an
acute clinical syndrome suggestive of DF were directly
tested by NT for antibodies to DEN-2. Serum specimens
negative for DEN-2 were then tested for neutralizing anti-
bodies to dengue virus serotypes 1, 3, and 4 (DEN-1,
DEN-3 and DEN-4).
The HI test was performed by the method of Clarke and
Casals (12) and NT as 90% plaque reduction neutralization
test (PRNT) on Vero cells. Briefly, serum specimens
(twofold dilutions) and virus (10
2
PFU) were incubated
overnight at 4°C, injected onto monolayers of Vero cells,
and overlaid with 1% Tragacanth gum (Sigma-Aldrich
S.r.I., Milan, Italy). Seven days postinfection, cells were
washed with saline and stained with 1% crystal violet in
20% ethanol (DEN-2 and DEN-3) or by immunodetection
assay (DEN-1 and DEN-4) as described (13). Vero cells
were propagated in minimum essential medium with
Earle’s salts (EMEM), supplemented with nonessential
amino acids, 10% fetal calf serum, 100 IE/mL of penicillin
G, and 100 IE/mL of streptomycin.
The following viruses were used in the study: DEN-1
(Hawaii), DEN-2 (NGB), DEN-3 (H87), DEN-4 (H241),
YF (Asibi), and WN (Bratislava). Viruses were injected
into suckling mice by the intracerebral route. For NT, viral
stocks were prepared as 10% brain suspension in Hank’s
saline+7.5% bovine serum albumin (Sigma-Aldrich). For
HI, test antigens were prepared by sucrose-acetone extrac-
tion from mouse infected brains (12). Monoclonal antibod-
ies specific for DEN-1 or broadly reactive with flavivirus-
es were purchased from ATCC (ATCC HB112, ATCC
HB47) and used as mouse ascitic fluid after injection into
adult BALB/c mice.
Undetermined febrile illness was defined as an acute
clinical syndrome with temperature >38.5°C, unrelated to
diarrhea, malaria, or other identified infections. Suspected
dengue (14) was defined as an undetermined febrile illness
of 2–7 days’ duration, associated with two or more of the
following manifestations: headache, retroorbital pain,
myalgia, arthralgia, cutaneous rash. Antibody levels
>
1:1,280 dilutions by HI (1,15) for DEN-2 and >1:20 dilu-
tions by NT to at least one of the four DENV serotypes
were considered supportive serologic evidence of a recent
dengue infection. Probable dengue (1,14) was defined as a
case compatible with the clinical description of suspected
DF and serologic findings supportive of a recent dengue
infection.
The prevalence of undetermined febrile illness, sus-
pected dengue, and probable dengue was compared by chi-
square test among army and navy/air force personnel.
Since navy and air force personnel had a limited exposure
to the environment of East Timor, risk factors for probable
dengue were studied only in the army contingent. A uni-
variate analysis was first performed by Fisher exact test;
each risk variable was crossed with the prevalence of prob-
able dengue. Significance was tested at a level of α=0.05.
A multiple logistic regression model was used to deter-
mine the relationship between the outcome of probable
dengue and a set of explanatory variables, and test the sig-
nificance of each variable while simultaneously account-
ing for demographic and risk factors. The following vari-
ables were included in the model: age, rank, previous
deployments in dengue-endemic areas, YF/JE vaccination,
night guards, skin repellents/permethrin-treated uni-
forms/bed nets use, and operational versus logistic tasks.
To identify a subset of variables significantly related to
probable DF, the stepwise procedure was performed with
the likelihood ratio test, by using at each step the p value
of 0.05 as entry criterion and the p value of 0.10 as
removal criterion. Univariate statistical analysis was per-
formed with EpiInfo 6.04d software (Centers for Diseases
Control and Prevention, Atlanta, GA, January, 2001) and
multivariate analysis by SPSS 11.0 software (SPSS Inc.,
Chicago, IL).
Conclusions
Of 640 eligible participants (280 army, 93 air force, and
267 navy), 595 (93%) were included in the study: 241
army, 88 air force, and 266 navy personnel (Table 1).
Serum specimens and questionnaires were obtained within
2 weeks after the troops’ return, in late February 2000.
Some (14.5%) of the troops had previously been
deployed to DF-endemic areas, primarily Somalia and
Mozambique in 1992–1994. According to their immuniza-
tion status versus YF and JE viruses, 100 (41.5%) of the
241 army soldiers had received vaccinations against YFV
and JEV, 119 (49.4%) had been vaccinated against JEV
only, 2 (0.8%) against YF only, and 20 (8.3%) had not been
vaccinated.
Undetermined febrile illness was more frequently
reported (p<0.01) among army soldiers than among navy
and air force personnel: 85 (35.3%) of 241 versus 13
(3.7%) of 354 , respectively. All participants with suspect-
ed dengue (n=30), with serologic results supportive of a
recent dengue infection (n=27), and with a probable case
of dengue n=16), belonged to the army group (Table 2).
The 16 participants with probable dengue showed also
a significant increase (p<0.01) in HI antibody titers to YFV
(>1:1,280 in 15/16 infected soldiers vs. 14/225 uninfected
soldiers) and WNV (>1:1,280 in 10/16 vs. 6/225). The
average interval between the onset of clinical manifesta-
tions suggestive of DF and the date when blood samples
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 877
DISPATCHES

were taken was 36±25 days. All 16 case-patients with
probable DF had a fever >38.5°C; a saddle-back fever pat-
tern was recorded for 5 (31.3%). Other reported symptoms
included myalgia and rash in 13 (81.3%); headache in 11
(68.8%); retroorbital pain in 9 (56.3%), and adenopathy in
3 (18.8%). No patients had DHF/DSS.
The mean duration of probable DF cases was 7±3 days.
Moreover, 12 of the 16 patients with probable DF were
evacuated because of their clinical status. Univariate
analysis of risk factors for probable DF suggested a possi-
ble protective effect of JEV vaccination and personal pro-
tection measures (Table 3). However, logistic regression
analysis identified only a subset of variables significantly
related to probable dengue, whose risk was higher among
soldiers on duty in operational rather than logistic units,
and lower among participants with regular use of bed nets
(Table 4).
Since most of soldiers had been previously vaccinated
with a flavivirus vaccine (YFV, JEV, or both), their
immune response to an eventual dengue infection was
expected to be a secondary (anamnestic) response, with
high-titer antibodies cross-reacting with several DENV
serotypes, as well as other flaviviruses (15). Thus, in spite
of the lack of paired serum specimens, high antibody titers
to DEN-2 by HI (>1:1,280) (1,16) and to any of the four
dengue virus serotypes by NT (>1:20), after an average of
36 days from the onset of clinical manifestations compati-
ble with dengue infection, may be considered supportive
serology of a recent flavivirus infection, likely acquired
during deployment.
Overall, 6.6% of army soldiers contracted probable
dengue. No cases of probable DF were detected in the low-
exposure group of navy and air force personnel. The high
attack rate of probable dengue among the army contingent
may be due to several reasons. First, DF and DHF/DSS are
epidemic throughout Southeast Asia (3), including
Indonesia (17); in particular, the incidence of DF marked-
ly increased in East Timor in 1998–1999 (18). Secondly,
the multinational deployment to East Timor took place
during the rainy season (December–January), when the
risk of infection is high.
Approximately 60% of troops with supportive serolog-
ic evidence of a recent dengue infection showed the clini-
cal manifestations of classic DF, 20% had milder symp-
toms, and 20% were asymptomatic. This finding agrees
with the U.S. troops’ experience in Somalia in 1993, where
>85% of all DENV infections were symptomatic (6). In
contrast, the overall ratio of inapparent to clinical DENV
infections is quite high in persons living in disease-endem-
ic areas, as in Indonesia, where it has been reported to be
as high as 9.3 (17).
Performing duties outside the camp was associated
with a significantly higher risk of infection, probably
because vector control activities were regularly carried out
within the compound. Regular use of bed nets was the only
personal protection measure that significantly decreased
the risk of contracting probable dengue. This finding is not
new (6) and may have been because some of the troops
were frequently on duty at night and thus slept during the
day when the biting activity of dengue vectors is highest.
Otherwise, the regular use of repellents (applied to the
skin) and permethrin-treated uniforms seemed to decrease
the risk for dengue infection, but the differences between
those who did not follow these practices and those who did
were not significant statistically.
DF is therefore an emerging problem for troops
deployed to dengue-endemic areas, mainly because of the
lack of effective preventive measures, the high attack rate,
878 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Table 1. Characteristics of Italian military personnel, East Timor, 1999–2000
Feature
Army
Navy
Air Force
Total
No. participants
241
266
88
595
Mean age (years ± sd)
27 ± 7
28 ± 7
35 ± 7
–
Time of deployment
22/Sep/99–16/Feb/00
21/Oct/99–19/Feb/00
19/Sep/99–17/Feb/00
–
Mean duration of deployment (days ± sd)
100 ± 25
109 ± 14
41 ± 23
–
Person months
803
968
102
1,873
Presence in East Timor for >90 days (no. soldiers)
241
0
0
241
Episodical presence in East Timor (no. soldiers)
0
266
88
354
Table 2. Clinical and serologic findings of recent dengue infection among Italian troops
Serologic findings of recent dengue infection
a
Clinical assessment
No. supportive (%)
No. not supportive (%)
Total no. (%)
Undetermined febrile illness
a
6 (22.2)
49 (22.9)
55 (22.8)
Suspected dengue
b
16 (59.3)
14 (6.5)
30 (12.4)
Asymptomatic
5 (18.5)
151 (70.6)
156 (64.7)
Total
27 (100)
214 (100)
241 (100)
a
All military personnel with supportive serologic findings belonged to the army contingent (N=241). Probable dengue cases are represented by the 16 soldiers with
clinical manifestations compatible with DF (suspected dengue) and serologic findings supportive of a recent dengue infection.
b
Cases are defined in the section “Materials and Methods.”

the high symptomatic/inapparent infection ratio, and the
long period of being unfit for duty after the acute phase of
the disease. DF may thus seriously disrupt the readiness of
a military unit. Moreover, previously infected soldiers
redeployed to disease-endemic areas may be at increased
risk for DHF/DSS complications. Persons previously
infected by a DENV serotype may be at higher risk of
developing DHF/DSS, if they are subsequently infected by
a different serotype. Such risks should be taken into
account while planning international peace-keeping opera-
tions, and the risk of DHF among previously dengue-
infected military personnel should be evaluated.
Cross-reaction by antiflavivirus antibodies induced by
JEV vaccine may otherwise afford some cross-protection
against DF. JEV vaccine (Nakajama strain) seems to
decrease the attack rate of DHF and reduce the severity of
cases for a short time (19). More recently, researchers have
noted that prior vaccination of hamsters with a live, atten-
uated JEV vaccine strain (not licensed for human use) and
a St. Louis encephalitis virus wild strain seems to reduce
the severity of a subsequent WNV infection (20). Our data
suggest that prior vaccination with the commercially avail-
able JEV inactivated vaccine for human use (Nakajama
strain) may have some protective effect against subsequent
probable DF. The decrease was, however, not significant,
according to the multiple logistic regression model we
used.
Our data suggest that effectiveness of routine protective
measures against vector mosquitoes is far from satisfacto-
ry. A tetravalent dengue vaccine is needed to effectively
reduce the risk for DF and DHF/DSS among troops
deployed to tropical areas as well as to protect long-term
international travelers to dengue-endemic countries.
Acknowledgements
We thank David Vaughn, Ashley Croft, and Tom Jefferson
for critical review of the manuscript and Antonino Bella and
Fortunato “Paolo” D’Ancona for statistical analysis.
Dr. Peragallo is a researcher at the Centro Studi e Ricerche
di Sanità e Veterinaria of the Italian Army. His main research top-
ics are the epidemiology and control of infectious diseases, par-
ticularly in tropical settings.
References
1. Gibbons RV, Vaughn DW. Dengue: an escalating problem. BMJ
2002;324:1563–6.
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 879
DISPATCHES
Table 3. Risk factors associated with probable dengue (univariate analysis)
Demographic and risk factors
No. cases of probable DF/no. soldiers exposed (%)
OR 95% CI
p value
b
Age, <26 y
Yes
No
10/124 (8.1)
6/117 (5.1)
1.62
(0.51 to 5.61)
0.26
Lower rank (enlisted men vs. NCOs/officers)
Yes
No
9/145 (6.2)
7/96 (7.3)
0.84
(0.27 to 2.76)
0.47
Previous deployments in dengue-endemic areas
Yes
No
3/44 (6.8)
13/197 (6.6)
1.04
(0.18 to 4.01)
0.59
YFV vaccination
Yes
No
5/102 (4.9)
11/139 (7.9)
0.60
(0.16 to 1.95)
0.26
JEV vaccination
Yes
No
9/219 (4.1)
7/22 (31.8)
0.09
(0.03 to 0.34)
<0.01
Night guard at least once a week
Yes
No
6/142 (4.2)
10/99 (10.1)
0.39
(0.11 to 1.25)
0.06
Skin repellents, regular use (at least once a day)
Yes
No
9/209 (4.3)
7/32 (21.9)
0.16
(0.05 to 0.56)
<0.01
Use of permethrin-treated uniform
Yes
No
9/186 (4.8)
7/55 (12.7)
0.35
(0.11 to 1.17)
0.05
Bed nets, regular use (every night)
Yes
No
9/223 (4.0)
7/18 (38.9)
0.07
(0.02 to 0.26)
<0.01
On duty in operational vs. logistic units
Yes
No
15/179 (8.4)
1/62 (1.6)
5.55
(0.82 to 238.67)
0.05
a
DF, dengue fever; OR, odds ratio; CI, confidence interval; NCOs, noncommissioned officers; YFV, yellow fever virus; JEV, Japane
se encephalitis virus.
b
Fisher exact test.
Table 4. Risk factors associated with probable dengue by
multivariate analysis
a
Risk factors
OR estimate
p value
On duty in operational vs. logistic units
11.29
<0.05
Bed nets, regular vs. nonregular use
0.04
<0.01
a
OR, odds ratio.

2. Gubler DJ and Clark GG. Dengue/dengue hemorragic fever: the
emergence of a global health problem. Emerg Infect Dis
1995;1:55–7.
3. World Health Organization. Dengue/dengue haemorragic fever: situ-
ation in 2000. Wkly Epidemiol Rec 2000;75:193–200.
4. Potasman I, Srugo I, Schwartz E. Dengue seroconversion among
Israeli travelers to tropical countries. Emerg Infect Dis 1999;5:824–7.
5. Jelinek T. Dengue fever in international travelers. Clin Infect Dis
2000;31:144–7.
6. Sharp TW, Wallace MR, Hayes CG, Sanchez JL, DeFraites RF, Arthur
RR, et al. Dengue fever in U.S. troops during operation Restore
Hope, Somalia, 1992–1993. Am J Trop Med Hyg 1995;53:89–94.
7. Trofa AF, DeFraites RF, Smoak BL, Kanesa-thasan N, King AD,
Burrous JM, et al. Dengue fever in US military personnel in Haiti.
JAMA 1997; 277:1546–8.
8. World Health Organization, Dili Office, East Timor. East Timor
health sector situation report: January-June 2000. Available from:
URL: http://www.who.int/eha/emergenc/etimor/14082000.htm
9. World Health Organization, Dili Office, East Timor. Weekly Report
45. 1999. Available from: URL: http://www.who.int/eha/emergenc/
etimor/191199.htm
10. World Health Organization, Dili Office, East Timor. Weekly Report
50–52 1999 & 01 2000. Available from: URL: http://www.who.int/
eha/emergenc/etimor/141200.htm
11. Hills S, Piispanen J, Foley P, Smith G, Humphreys J, Simpson J, et al.
Public health implications of dengue in personnel returning from East
Timor. Communicable Disease Intelligence [serial online]
2000;24:365–8. Available from: URL: http:// http://www.health.
gov.au/pubhlth/cdi/cdi2000.htm#december
12. Clarke DH, Casals J. Techniques for the hemagglutination and
hemagglutination-inhibition with arthropod-borne viruses. Am J Trop
Med Hyg 1958;7:561–77.
13. Desprès P, Frenkiel MP and Deubel V. Differences between cell
membrane fusion activities of two Dengue type-1 isolates reflect
modification of viral structure. Virology 1993;196:209–19.
14. World Health Organization. Recommended surveillance standards.
2
nd
edition. Geneva: The Organization; 1999. p. 39.
15. World Health Organization. Laboratory diagnosis. In: Dengue
haemorragic fever. Diagnosis, treatment, prevention and control. 2nd
edition. Geneva: The Organization, 1997. p. 34–47.
16. Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol
Rev 1998;11:480–96.
17. Corwin AL, Larasati RP, Bangs MJ, Wuryadi S, Arjoso S, Sukri N, et
al. Epidemic dengue transmission in southern Sumatra, Indonesia.
Trans R Soc Trop Med Hyg 2001;95:257–65.
18. Krause V. Increase in dengue fever notifications in visitors to East
Timor. Northern Territory Disease Control Bulletin [serial online]
2000;7:6-7. Available from: URL: http:// http://www.nt.gov.au/
health/cdc/bulletin/june_2000.pdf
19. Hoke CH, Nisalak A, Sangawhipa N, Jatanasen S, Laorakapongse T,
Innis BL, et al. Protection against Japanese encephalitis by inactivat-
ed vaccine. N Engl J Med 1988;319:608–14.
20. Tesh RB, Travassos da Rosa APA, Guzman H, Araujo TP, Xiao SY.
Immunization with heterologous Flaviviruses protective against fatal
West Nile encephalitis. Emerg Infect Dis 2002;8:245–51.
Address for correspondence: Mario Stefano Peragallo, Capo Primo
Reparto, Centro Studi e Ricerche di Sanità e Veterinaria dell’Esercito, Via
S. Stefano Rotondo 4, 00184 Rome, Italy; fax: ++39 06 7009005; email:
880 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
The print journal is available at no charge to public health professionals
YES, I would like to receive Emerging Infectious Diseases.
Please print your name and business
address in the box and return by fax to
404-371-5449 or mail to
EID Editor
CDC/NCID/MS D61
1600 Clifton Road, NE
Atlanta, GA 30333
Moving? Please give us your new address (in the box) and print the number of your old
mailing label here_________________________________________
Full text free online at
www.cdc.gov/eid

HIV Epidemic
among Young Thai
Men, 1991–2000
Kalyanee Torugsa,* Scott Anderson,†
Nucharee Thongsen,* Narongrid Sirisopana,*
Achara Jugsudee,‡ Pitak Junlananto,‡
Sorachai Nitayaphan,*
Suebpong Sangkharomya,*
and Arthur E. Brown*
Characterization of the HIV epidemic in Thailand has
benefited from the systematic testing of young men upon
entry into the military. These data, which have shown that
public health measures can reverse an HIV epidemic, have
been reanalyzed with current geographic information sys-
tems methods. The resulting maps are, thus far, the best
means of visualizing the geography of the dynamic HIV epi-
demic in Thailand.
I
n few nations is the HIV epidemic characterized as well
as it is in Thailand. As elsewhere, sentinel surveillance
of high-risk populations is performed and more general
populations are partially assessed by HIV testing of blood
donors and pregnant women. In addition, Thailand has
benefited from another measure to assess the HIV epidem-
ic in the general population. Since very early in the epi-
demic, young men have been tested for HIV at the time
they entered into military service. Military medicine and
public health leaders recognized that, in addition to using
this information to benefit individual men, maintaining
this information in a confidential database linked to area of
residence would allow monitoring of national demograph-
ic trends in the epidemic. Data generated in this way have
provided key evidence that public health policy has
reversed the trend of increasing HIV infections (1), pre-
venting an estimated 200,000 HIV infections by 2000 (2).
The HIV prevalence data on young Thai men is provid-
ed to public health policy makers on a regular basis. It has
also been published in the scientific literature (3,4) with
data presented that uses provinces as the scale of analysis.
More recently, this unique dataset has been analyzed by
using geographic information systems (GIS). GIS methods
allow enhanced visualization of the trends, both geograph-
ic and chronologic, within this well-documented HIV epi-
demic. Using a GIS approach, we present the Royal Thai
Army dataset, expanded to include the decade of
1991–2000 and data precision refined to the district level.
Methods
The methods of recruiting young men into the Thai mil-
itary have been described previously (3,4). Approximately
50,000–60,000 men, mostly 21 years of age, are selected
each year by lottery in their home province (the province
in which they are listed on the house registration). The sys-
tem produces a representative national sampling of Thai
men. Because of their young age, HIV prevalence in these
annual recruit classes may serve as a proxy for HIV inci-
dence. Induction into the military occurs in May (M) and
November (N) of each year. At the time when blood is col-
lected, recruits provide information about the location of
their main residence (including province and district) dur-
ing the previous 2 years (3). Although the actual locations
where infections occurred are unknown, residential data
enables analysis of the association between HIV preva-
lence and this key geographic marker.
To refine the HIV prevalence analyses, geographic
localization uses districts as the first administrative subunit
of provinces. When data were analyzed by using annual
classes grouped at the district level, calculations of the per-
centage testing positive for HIV were statistically unreli-
able because the number of men tested in some rural dis-
tricts was so small. Therefore, we merged data in two ways
to decrease variability attributable to the small sample size
of the prevalence figures: classes were combined across
time, and districts were combined across space. Sixteen
classes of men recruited from 1991 to 2000 were combined
temporally into four larger classes, each representing dis-
crete 2-year periods: 1) N91, M92, N92, M93; 2) N94,
M95, N95, M96; 3) N96, M97, N97, M98; and 4) N98,
M99, N99, M00. Data on classes recruited from the M91,
N93, M94, and N00 lotteries were not available for analy-
sis (M91, before full implementation; N93 and M94, pro-
tocol under revision; N00, completed after dataset closed).
However, even after combining classes into these 2-year
periods, a number of districts still had numbers too low for
statistical reliability.
We also merged some districts with neighboring dis-
tricts so that each had a minimum denominator of 20 in the
HIV prevalence calculation. Numbers of >
20 persons pro-
vide minimal, but acceptable, reliability in the percentage-
positive calculations. Districts with <20 were combined
with other districts according to the following protocol,
following a sequence of priorities: we combined districts if
they were in the same province, had historic connections
(formerly part of single larger district), had similarly small
numbers tested, had similar demographics, and had similar
topography or other geographic features.
For the GIS analysis, data tables provided in Excel files
(Microsoft Corp., Redmond, WA) by the Armed Forces
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 881
DISPATCHES
*Armed Forces Research Institute of Medical Sciences, Bangkok,
Thailand; †State University of New York at Cortland, Cortland,
New York, USA; and ‡Army Institute of Pathology, Bangkok,
Thailand
Research Institute of Medical Sciences were joined to dis-
trict-level GIS maps obtained from the Thai
Environmental Institute and National Statistical Office. We
used Arcview 3.2a software (ESRI, Redlands, CA) to cre-
ate dot density and choropleth maps. (A choropleth map
uses shades or colors to demonstrate the geographic distri-
bution of a range of values.)
Results and Discussion
Figure 1 provides visual evidence of the geographic
distribution of the epidemic. Each of the 10,043 dots rep-
resents a recruit who tested positive for HIV infection
(positive for HIV-1 by enzyme immunoassay and Western
blot) among the 442,923 recruits tested. Arcview software
uses an algorithm that allows the random placement of
dots within a geographic area (in this case a district). By
creating these dots at the district level but presenting them
at the province level, we show the vivid geographic pattern
of infection while illustrating the human impact of the epi-
demic. The dot distribution does not distinguish between
population density and HIV prevalence. Nevertheless,
these absolute numbers provide important information
regarding the potential HIV-related impact on the health-
care system and various other social structures.
The evolution of the HIV epidemic over the course of
the decade is seen in the four maps shown in Figure 2.
These maps use gradients of color shading to distinguish
the percentage of men who tested positive for HIV during
each 2-year period. The initial high prevalence of HIV in
men from the upper north region is illustrated, as is the
decline in prevalence over time (which likely occurred as
a result of public health interventions). The relative per-
sistence of HIV prevalence in some districts in the far
south of the country is also shown, suggesting that locale-
specific features of the epidemic need to be well under-
stood in that area, potentially leading to changes in public
health interventions.
Previous analyses conducted at the province level iden-
tified regional variation quite clearly, but at a less detailed
level (3,4). The maps from our study indicate considerable
variation even to the district level of analysis, which sug-
gests important local factors may be at work in determin-
ing rates of transmission. The HIV epidemic in Thailand
seems to have largely missed some districts and devastat-
ed others, even within the same province. Chiang Mai and
Chiang Rai Provinces are examples of this phenomenon.
These maps also show that, although the growth of the epi-
demic has slowed in most parts of the country, the epidem-
ic has not decreased in some districts in south Thailand.
Choropleth maps best demonstrate the changing preva-
lence of HIV among recruits from each district. The dot
map best demonstrates the intensity of the epidemic with-
in and across provinces. Both types of imagery are
required to describe and understand the epidemic and to
suggest locales in which public health initiatives are need-
ed. In areas where the intensity of the epidemic is highest,
health and social services may require additional resources
to serve persons with the disease. In areas where the preva-
lence rates are highest or are increasing, socially and cul-
turally appropriate interventions may be needed to
strengthen and focus HIV programs.
These GIS maps are the most accurate maps available
to date and are built upon the unique HIV prevalence
datasets collected over a decade by the Royal Thai Army.
GIS mapping at this descriptive level enhances visualiza-
tion of these data (5,6). More sophisticated methods of
multivariate GIS visualization could enhance this analysis
even more but would require the Royal Thai Army to
assess recruits at induction on other variables, especially
risk factors, or to use recruits’ home addresses rather than
districts or provinces of residence. Nonetheless, changes in
time and place of chronic viral infection can be added to
882 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
DISPATCHES
Figure 1. Dot density map of young men who tested positive for
HIV at time of entry into the Royal Thai Army, Thailand, November
1991–May 2000. Each dot represents one man. Location of dots
based on recruit’s residence during the previous 2 years. Data on
recruits entering in November 1993 and May 1994 are not avail-
able.

the recognized areas of GIS application in epidemiology,
transmission of vector-borne and water-borne diseases,
and environmental health (7). The maps also illustrate the
point that political borders may have no relationship to epi-
demiologic boundaries. Differences in HIV prevalence
often occur, not only between regions and provinces but
also within provinces and across provincial borders. When
an epidemic can be visualized at this level of detail and
accuracy, researchers can formulate and address questions
regarding the bases of these differences. At the same time,
policy makers can better direct intervention strategies and
more finely assess the outcomes of these interventions.
Coordination of data collection and joint GIS analysis by
neighboring countries would enhance regional understand-
ing of the emergence and spread of HIV epidemics and
monitoring of control programs.
Dr. Torugsa is a research scientist at the Armed Forces
Research Institute of Medical Sciences (AFRIMS) in Bangkok,
Thailand. Her research interests are in infectious disease epi-
demiology, and her current focus is the establishment of a disease
surveillance and outbreak response system in the Royal Thai
Army Medical Department.
References
1. Phoolcharoen W. HIV/AIDS prevention in Thailand: success and
challenges. Science 1996:280;1873–4.
2. World Bank. Thailand’s response to AIDS: building on success, con-
fronting the future. p. 1, Nov 3, 2000 [cited 2002 Jun 21]. Available
from: URL: http://www.worldbankor.th/social/index.html
3. Sirisopana N, Torugsa K, Mason CJ, Markowitz LE, Jugsudee A,
Supapongse T, et al. Correlates of HIV-1 seropositivity among young
men in Thailand. J Acquir Immune Defic Syndr Hum Retrovirol
1996:11;492–8.
4. Mason CJ, Kitsiripornchai S, Markowitz LE, Chanbancherd P,
Supapongse T, Jugsudee A, et al. Nationwide surveillance of HIV-1
prevalence and subtype in young Thai men. J Acquir Immune Defic
Syndr Hum Retrovirol 1998:19:165–73.
5. Albert DP, Gesler WM, Levergood B. Spacial analysis, GIS, and
remote sensing applications in the health sciences. Chelsea (MI): Ann
Arbor Press; 2000.
6. Gatrell AC, Loytonen M. GIS and health. GISDATA VI. London:
Taylor & Francis; 1998.
7. Clarke KC, McLafferty SL, Tempalski BJ. On epidemiology and geo-
graphic information systems: a review and discussion of future direc-
tions. Emerg Infect Dis 1996:2;85–92.
Address for correspondence: Arthur E. Brown, Department of
Retrovirology, AFRIMS, APO AP 96546; fax: 66-02-644-4824; email:
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 883
DISPATCHES
Figure 2. Choropleth maps of HIV prevalence in four classes of young men at time of entry into the Royal Thai Army, Thailand,
1991–2000. Location determined by residence during the previous 2 years. Prevalence is stratified by color and localized to district or
group of districts so that calculations are based on >20 men.

884 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
Taenia solium
Cysticercosis, Irian
Jaya, Indonesia
To the Editor: Cysticercosis, a tis-
sue infection caused by accidental
ingestion of eggs released from
humans harboring the pork tapeworm,
Taenia solium (TsCysti), is one of the
most serious reemerging parasitic dis-
eases worldwide (1). Taeniasis is an
intestinal infection caused by the
adult stage of the large tapeworm.
Carriers of T. solium acquire infection
through eating undercooked pork con-
taminated with cysticerci (larvae).
Although most Indonesian people are
Muslim and do not eat pork, infection
with T. solium has occurred in some
areas or islands where most local peo-
ple are Christian or Hindi.
The area most affected by this
infection is Irian Jaya, Indonesia, the
western half of New Guinea Island
(2–4). In field surveys conducted in
2000 and 2001, we found that 5
(8.6%) of 58 local people and 7 (11%)
of 64 local dogs living approximately
1 km from the local capital city,
Wamena, in Jayawijaya District, har-
bored adult tapeworms and cysticerci
of T. solium, respectively (5,6). We
have further seroepidemiologic data
from 1996 and molecular confirma-
tion of subcutaneous nodules (SCN)
as cysticerci of the T. solium Asian
genotype. We believe this organism is
an emergent problem in Irian Jaya.
We previously reported that
TsCysti was highly endemic in
Jayawijaya District, Irian Jaya (2–6).
A total of 96 local people >
18 years of
age from Assologaima, Jayawijaya
District, were chosen at random and
examined by serologic testing and by
administering questionnaires in
February 1996 after the local and
Indonesian governments gave their
ethical approval. The 96 persons were
divided into three groups on the basis
of a history of epileptic seizures (ES,
n=17), physical examination of SCN
by palpation (n=32), or good health
(including no ES or SCN; n=47). A
total of 14 subcutaneous nodules
removed from 14 men in both ES and
SCN groups were confirmed to be
cysticerci of T. solium by morpholog-
ic observation and to be T. solium
Asian genotype by mitochondrial
DNA analysis with cytochrome c oxi-
dase subunit 1 gene (3,7). For serolog-
ic analysis, we conducted an enzyme-
linked immunosorbent assay (ELISA)
that used glycoproteins purified from
cyst fluid of T. solium cysticerci by
preparative isoelectric focusing (frac-
tions of pH 9.1) (8) in 2001.
On the basis of serologic results,
12 (70.6%) of 17, 20 (62.5%) of 32,
and 12 (25.5%) of 47 of ES, SCN, and
healthy groups, respectively, were
infected with the larval stage of T.
solium. Serologically positive rates
increased to 83.3% (10/12) of people
with subcutaneous nodules in the ES
group. A follow-up study of seroposi-
tive persons in the healthy group in
1997 showed that five of eight per-
sons had ES (two persons), headache
(one person), or SCNs in upper arm
(two persons). Seropositive persons in
all three groups (ES, SCN, and health)
were considered to be infected with
TsCysti. Persons of the SCN and
healthy groups who showed optical
density values higher than the cut-off
value were considered to have asymp-
tomatic TsCysti cases.
The local persons we examined
ranged from 18 to 29 years of age
(n=30), 30–44 years of age (n=36),
and >45 years of age (n=30).
Seropositive persons (n=12) from the
ES group (n=17) were 18 to 29 years
of age (40.0%, 2/5), 30–44 years
(71.4%, 5/7), and >
45 years (100%,
5/5). The prevalence of TsCysti did
not vary statistically by sex (males
53.6% [37/69] versus females 33.3%
[9/27], Pearson’s chi-square test,
p=0.074).
That 14 persons confirmed to have
subcutaneous cysticerci of T. solium
were seropositive strongly suggests
that the serologic test (ELISA) is
highly reliable for detecting TsCysti
in patients, whether their infection is
symptomatic or asymptomatic. In
contrast, one of the following scenar-
ios was expected for cases in three of
five persons in the ES group who did
not have SCN and were seronegative:
1) the case was not due to TsCysti, 2)
the case was caused by TsCysti but
without antibody response, rather
common in cases of a solitary cyst, or
3) the case was caused by TsCysti
with calcified cysts and without anti-
body response. Twelve (approximate-
ly 40%) of seronegative persons from
the SCN group (n=32) were expected
to have cases of TsCysti without anti-
body response or to have calcified
cysts without antibody response.
Cases without antibody response
would be most expected because of
the heavily contaminated environ-
ment in Irian Jaya (3–6). However,
further evaluation with computed
tomography or magnetic resonance
imaging scans is necessary. Based on
serologic results and mitochondrial
DNA confirmation of T. solium Asian
genotype (3,7), we concluded that
47.9% (46/96) of local people exam-
ined at random, 53.6% of men (37/69)
and 33.3% of women (9/27) >
18 years
of age had TsCysti.
An additional 30 local people in non
–TsCysti-endemic Merauke District
underwent serologic testing. One
woman had an exceptionally high
antibody titer. She was a transmigrant
from another island (South Sulawesi
Province). Although Paniai, Jayawijaya,
and Manokwari Districts are contami-
nated with T. solium taeniasis and cys-
ticercosis (2–4), no additional critical
evidence exists to show that Merauke
District has already been contaminat-
ed with this parasite.
Taeniasis and cysticercosis may
have been accidentally introduced
into Irian Jaya in 1969 when the coun-
try was governed by Indonesia, since
the governing body came from Bali,
the only area in Indonesia where

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 885
LETTERS
TsCysti was exclusively endemic (2).
The contaminated areas in Irian Jaya
have increased from the central area
(Paniai), to the east (Jayawijaya) (3),
and then to the west (Manokwari),
where 54 TsCysti cases have been
reported (Papua Province Health
Office Services, 1997, unpub. data).
We wanted to know if taeniasis/cys-
ticercosis had been introduced into
the eastern half of New Guinea Island,
called Papua New Guinea (PNG) (9).
We had already serologically con-
firmed that 16 (3.0%) of 541 local res-
idents and Irianese refugees in Alice
River villages along the border in
PNG had asymptomatic TsCysti (Ito
et al., unpub. data). Follow-up sur-
veys will be crucial in several other
districts including Merauke District in
Irian Jaya, PNG, and other islands
such as Timor Island, where most of
the population is Christian and many
suspected cases have recently been
reported by the District Health Office
Services (10). Schoolchildren should
also be checked so that cases can be
detected and treated early. Sustainable
education of the local community in
Irian Jaya, Indonesia, and Papua New
Guinea is also necessary.
Financial support was provided by a
grant-in-aid from the Nissan Foundation
and the Ministry of Education, Japan
(09044279, 10557029, 11694259,
1255702414, and 14256001) (A.I.) and by
the Japan Health Science Foundation
(S.S.M.).
Toni Wandra,* Akira Ito,†
Hiroshi Yamasaki,†
Thomas Suroso,*
and Sri S. Margono‡
*Ministry of Health, Jakarta, Indonesia;
†Asahikawa Medical College, Asahikawa,
Japan; and ‡University of Indonesia,
Jakarta, Indonesia
References
1. Schantz PM, Wilkins PP, Tsang VWC.
Immigrants, imaging and immunoblot: the
emergence of neurocysticercosis as a sig-
nificant public health problem. In: Scheld
WM, Craig WA, Hughes GM, editors.
Emerging infections 2. Washington, DC:
ASM Press; 1998. p. 213–42.
2. Simanjuntak GM, Margono SS, Okamoto
M, Ito A. Taeniasis/cysticercosis in
Indonesia as an emerging disease. Parasitol
Today 1997;13:321–3.
3. Wandra T, Subahar R, Simanjuntak GM,
Margono SS, Suroso T, Okamoto M, et al.
Resurgence of cases of epileptic seizures
and burns associated with cysticercosis in
Assologaima, Jayawijaya, Irian Jaya,
Indonesia, 1991–95. Trans R Soc Trop Med
Hyg 2000; 94:46–50.
4. Subahar R, Hamid A, Purba W, Wandra T,
Karma C, Sako Y, et al. Taenia solium
infection in Irian Jaya (West Papua),
Indonesia: a pilot serological survey of
human and porcine cysticercosis in
Jayawijaya District. Trans R Soc Trop Med
Hyg 2001;95:388–90.
5. Ito A, Putra MI, Subahar R, Sato MO,
Okamoto M, Sako Y, et al. Dogs as alterna-
tive intermediate hosts of Taenia solium in
Papua (Irian Jaya), Indonesia confirmed by
highly specific ELISA and immunoblot
using native and recombinant antigens and
mitochondrial DNA analysis. J Helminthol
2002;76:311–4.
6. Margono SS, Ito A, Sato MO, Okamoto M,
Subahar R, Yamasaki H, et al. Taenia soli-
um taeniasis/cysticercosis in Papua,
Indonesia in 2001: detection of human
worm carriers. J Helminthol 2003;
77:39–42.
7. Nakao M, Okamoto M, Sako Y, Yamasaki
H, Nakaya K, Ito A. A phylogenetic hypoth-
esis for the distribution of two genotypes of
the pig tapeworm Taenia solium world-
wide. Parasitology 2002;124:657–62.
8. Ito A, Plancarte A, Ma L, Kong Y, Flisser
A, Cho SY, et al. Novel antigens for neuro-
cysticercosis: simple method for prepara-
tion and evaluation for serodiagnosis. Am J
Trop Med Hyg 1998;59:291–4.
9. McManus DP. Improved diagnosis as an aid
to better surveillance of Taenia solium cys-
ticercosis, a potential health threat to Papua
New Guinea. Papua New Guinea Medical
Journal 1995;38:287–94.
10. Suroso T. Petunjuk pemberantasan taenia-
sis/sistiserkosis di Indonesia. Depkes RI,
Ditjen. Jakarta: PPM & PL; 2000. p. 1–30
(in Indonesian).
Address for correspondence: Akira Ito,
Department of Parasitology, Asahikawa
Medical College, Midorigaoka-Higashi 2-1-1-
1, Asahikawa 078-8510, Japan; fax: +81-
(0)166-68-2429; email: akiraito@asahikawa-
med.ac.jp
Recombinant
Vaccine–Derived
Poliovirus in
Madagascar
To the Editor: Between October
2001 and April 2002, five cases of
acute flaccid paralysis associated with
vaccine-derived poliovirus (VDPV)
type 2 isolates were reported in the
southern province of the Republic of
Madagascar. The first patient, an 11-
year-old child from the urban district
of Toliara, first experienced paralysis
on October 29, 2001. Three other chil-
dren, 6, 9, and 14 months of age from
Ebakika village, in a rural district of
Taolagnaro (250 miles east of Toliara),
showed signs of poliomyelitis
between March 21 and March 26,
2002. The last case-patient, a 20-
month-old child from Ambanihazo
village (6 miles north of Ebakika),
came into contact with one of the three
case-patients in Ebakika in March
2002, and symptoms developed on
April 12, 2002 (1). None of the
patients had been fully vaccinated
against poliomyelitis.
Nine type 2 poliovirus (PV) strains
were isolated. A restriction fragment
length polymorphism (RFLP) assay,
with three different genomic regions
amplified by reverse transcription–
polymerase chain reaction (RT-PCR)
and four different restriction enzymes
(HinfI, DpnII, RsaI, and DdeI) were
used to characterize the PV isolates at
the molecular level (2). The RFLP
profiles of all of the isolates in the two
capsid protein regions were identical
to that of the type 2 strain of the oral
polio vaccine (OPV) in the VP1-2A
region (nucleotides 2,872 to 3,647)
but slightly different in the VP3-VP1
region (nucleotides 1,915 to 2,883).
The observed differences allowed us

886 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
to distinguish two groups (isolates
from Toliara and isolates from
Taolagnaro) and two subgroups (iso-
lates from March and isolates from
April). The RFLP profiles of isolates
in the noncapsid region, at the 3′-ter-
minal end of the genome (polymerase
3D and 3′ noncoding regions:
nucleotides 6,535 to 7,439) also con-
firmed the presence of two separate
groups. These last profiles were com-
pletely different from those of the
three reference vaccine strains, sug-
gesting recombination with other
enteroviruses.
Partial genomic sequencing con-
firmed these observations. The entire
VP1 region (903 nucleotides) of the
type 2 PV strains from Toliara and
Taolagnaro differed from the type 2
OPV strain by 1% and 2.5%
nucleotides, respectively. This differ-
ence may indicate that the two strains
had been multiplying or circulating
for approximately 1 and 2.5 years,
respectively. Taolagnaro strains are
closely related to each other (<1%
nucleotide difference) but appear to be
very different from Toliara strains
(2.9% nucleotide difference), indicat-
ing the existence of two genetic line-
ages. The sequencing of the noncapsid
region (440 nucleotides corresponding
to nucleotide positions 6,705 to 7,144
of the Sabin 2 genome) confirms the
existence of two lineages derived
from different recombination events
with two nonidentified enteroviruses
of the phylogenetic cluster C. This
cluster, based on sequence similarity,
includes some coxsackieviruses and
all PV strains (3).
We tried to identify the donor
strains for sequences in the 3’ terminal
end of these recombinant strains by
aligning the nonidentified sequences
with homologous enterovirus
sequences available in a nucleotide
sequence database (FASTA, version
3.3 applied to GenBank) (4). The
highest percentages of nucleotide
sequence identity were those with
PVs and with most other cluster C
enteroviruses available in the data-
base (87% to 91% nucleotide identi-
ties). No wild PV strains have been
isolated in Madagascar since 1997
despite surveillance and investigation
of viral causes of acute flaccid paraly-
sis cases (5). Thus, that the detected
VDPVs were the product of recombi-
nation between OPV strains and two
nonpolio enteroviruses is more likely
than that they were the product of
OPV strains and two different unde-
tected wild PV strains. However, we
cannot exclude the possibility that
wild PVs were imported or circulating
silently for a while.
In response to the outbreak, the
local health authorities conducted
house-to-house vaccination with
OPV. Further field investigations
were carried out to determine the
extent to which VDPV had spread and
to search actively for other cases.
Data analysis is in progress.
As with the other epidemics in
Egypt and Hispaniola, VDPV circu-
lated in a province of Madagascar
with low OPV coverage (6,7).
Because a high OPV coverage rate
helps prevent the circulation of both
VDPVs and wild PVs, obtaining and
maintaining high rates of immuniza-
tion coverage are essential (8).
Moreover, two recombinant VDPV
lineages in Madagascar indicate that
recombination is frequent between
OPV and cluster C enteroviruses.
Similar recombinant VDPVs have
been implicated in the epidemics in
Hispaniola and in the Philippines
(6,9). Determining whether the neu-
rovirulence and transmissibility of
these VDPVs could be the result of
The print journal is available at no charge to public health professionals
YES, I would like to receive Emerging Infectious Diseases.
Please print your name and business
address in the box and return by fax to
404-371-5449 or mail to
EID Editor
CDC/NCID/MS D61
1600 Clifton Road, NE
Atlanta, GA 30333
Moving? Please give us your new address (in the box) and print the number of your old
mailing label here_________________________________________
Full text free online at
www.cdc.gov/eid

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 887
LETTERS
the recombination with nonpolio
enteroviruses is important. These
VDPVs have major implications for
the cessation of immunization with
OPV after certification that wild PV
has been eradicated.
Acknowledgments
We thank R. Crainic for support
and encouragement, O. Kew for pro-
viding unpublished results; and E. de
Gourville, O. Tomori, D. Wood, and
F. Colbere-Garapin for their interest
and advice.
Dominique Rousset,*
Mala Rakoto-Andrianarivelo,*
Richter Razafindratsimandresy,*
Bakolalao Randriamanalina,†
Sophie Guillot,‡ Jean Balanant,‡
Philippe Mauclère,*
and Francis Delpeyroux‡
*Institut Pasteur de Madagascar,
Antananarivo, Republic of Madagascar;
†Ministry of Health, Antananarivo, Republic
of Madagascar; and ‡Institut Pasteur,
Paris, France
References
1. Centers for Disease Control and
Prevention. Poliomyelitis—Madagascar
2002. MMWR Morb Mortal Wkly Rep
2002;51:622.
2. Guillot S, Caro V, Cuervo N, Korotkova E,
Combiescu M, Persu A, et al.] Natural
genetic exchanges between vaccine and
wild poliovirus strains in humans. J Virol
2000;74:8434–43.
3. Pringle CR. Virus taxonomy at the XIth
International Congress of Virology, Sydney,
Australia, 1999. Arch Virol
1999;144:2065–-70.
4. Pearson WR, Lipman DJ. Improved tools
for biological sequence comparison. Proc
Natl Acad Sci U S A 1988;85:2444–8.
5. World Health Organization. Vaccines,
immunization, and biologicals. Accessed
May 13, 2003. Available from: URL:
http://www.who.int/vaccines/casecount/afp
extractnew.cfm
6. Centers for Disease Control and
Prevention. Circulation of a type 2 vaccine-
derived poliovirus—Egypt, 1982–1993.
MMWR Morb Mortal Wkly Rep 2001;
50:41–2.
7. Kew O, Morris-Glasgow V, Landaverde M,
Burns C, Shaw J, Garib Z, et al. Outbreak
of poliomyelitis in Hispaniola associated
with circulating type 1 vaccine-derived
poliovirus. Science 2002;296:356–9.
8. Wood DJ, Sutter RW, Dowdle WR.
Stopping poliovirus vaccination after eradi-
cation: issues and challenges. Bull World
Health Organ 2000;78:347–57.
9. Centers for Disease Control and
Prevention. Acute flaccid paralysis associ-
ated with circulating vaccine-derived
poliovirus—Philippines, 2001. MMWR
Morb Mortal Wkly Rep 2001;50:874–5.
Address for correspondence: Mala Rakoto-
Andrianarivelo, Unité de Virologie, Institut
Pasteur de Madagascar, BP 1274, Antananarivo
101, Madagascar; fax: (261.20) 22.415.34;
email: mala@pasteur.mg
West Nile Virus
Infection in
Crocodiles
To the Editor: Recently West Nile
virus (WNV) infection has been
reported in three alligators (Alligator
sp.) from central Florida (1) and one
captive crocodile monitor (Varanus
salvadori) with neurologic signs from
the District of Columbia and
Maryland area (2). These first reports
of the virus in American reptiles high-
light the possible role of this group of
vertebrates in the WNV life cycle. To
our knowledge, WNV in a reptile was
reported only once before in a sero-
survey conducted in Israel from 1965
to 1966, in which 22 reptiles and 96
amphibians were tested for hemagglu-
tination-inhibiting antibodies against
several viruses, including WNV; one
turtle (Clemmys caspica) was
seropositive (3). Experimental infec-
tion of the lake frog (Rana ridibunda)
with a Russian strain of WNV result-
ed in high levels of viremia (4). At
present, the role of reptiles and
amphibians in the life cycle and epi-
demiology of WNV is not known.
We report, for the first time, WNV
infection in crocodiles (Crocodylus
niloticus). To assess the potential role
of crocodiles in the life cycle of WNV
in Israel, serum specimens were col-
lected from 20 healthy crocodiles on a
commercial farm in the Negev Desert,
in southern Israel (31°14′N, 34°19′E).
The crocodiles came from two sepa-
rate breeding farms (32°03′N,
35°26′E and 30°18′N, 35°07′E) in the
Syrian-African Rift Valley, which is
on the main route of bird migration
from Africa to Europe. Five males and
15 females, 1–2.5 years of age, were
examined. Blood was withdrawn from
the crocodiles’ ventral caudal vein,
separated by centrifugation, and kept
at –20°C until analyzed. Neutralizing
antibody titers were determined
against WN-goose-98 (5) and
attempts to isolate the virus were per-
formed by using Vero cell culture (6)
and by using direct reverse transcrip-
tion–polymerase chain reaction (RT-
PCR) on the serum specimens. To
eliminate the possibilities of nonspe-
cific reaction, all serum samples were
concurrently tested for the only other
flavivirus known to be present in
Israel; Israeli turkey meningo-
encephalitis virus (ITV) (7). Because
ITV does not produce cytopathic
effects (CPE) in Vero cells, virus neu-
tralization was conducted on BHK
cells for both WNV and ITV by using
WN-goose-98 and ITV (vaccine
strain). In this case, the virus stocks
(10
-4.2
50% tissue culture infective
dose) were diluted 1:400, and virus
neutralization titers were checked 3
days later.
Viral RNA was extracted from
serum samples with the QIAamp
RNA blood kit (QIAGEN, Valencia,
CA) , according to the manufacturer’s
protocol and resuspended in 30 µl of
RNase-free water. The primer pair
WN240-Kun848 (respective genome
positions 5′: 848 and 1,645) was used
to synthesize an 800-bp product in the
E gene region (8,9). The resulting
DNA fragment was visualized on

888 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
1.5% agarose gel stained with ethidi-
um bromide.
The seroprevalence rate in the first
set of virus neutralization assays in
Vero cells was 14/20 (70%, with titers
ranging from 1:20 to 1:320 [3x1:20,
3x1:40, 3x1:80, 2x1:160, 3x1:320]).
No differences were discernible in
either the seroprevalence rate or in the
average titers of crocodiles from two
different breeding farms. In BHK
cells, a similar seroprevalence rate
was observed, with titers ranging from
1:40 to 1:1,280 (3x1:40, 2x1:80,
1x1:160, 4x1:320, 3x1:640,
1x1:1280). All serum samples, except
one, were <1:10 against ITV virus,
which had a titer of 1:640 against
WNV and 1:10 against ITV. Viremia
was not detected in any of the 20 sam-
ples in Vero cell culture or by RT-PCR.
These results demonstrate a high
rate of infection with WNV in croco-
diles in Israel. The crocodiles may
have been exposed to the virus during
the summer at their present location,
since no difference in prevalence was
seen between the two groups (which
differed only in the farm of origin)
and since the younger crocodiles had
been hatched in the spring of 2002.
Furthermore, a cross-reaction with the
other prevalent flavivirus in Israel,
ITV, was ruled out. Preliminary
results from an equine seroprevalence
study (involving 800 horses over a 3-
year period) of virus neutralization
antibodies to WNV collected during
fall 2002, indicate that most horses
sampled in Israel’s Arava Valley (a
desert in the Syrian-African Rift near
the Jordanian and Egyptian borders)
and the Gulf of Aqaba/Eilat (30°59′N,
35°18′E to 29°34′N, 34°57′E) (85%,
79/90) were positive (A. Steinman
and S. Tal, unpub. data,). WNV was
also isolated from mosquitoes in the
same region (10). The seroprevalence
of WNV antibodies among horses and
local birds from the Negev Desert is
not known nor is the time when the
horses acquired WNV infection.
However, the isolation of WNV from
mosquitoes (10) and the presence of
antibodies to WNV in young croco-
diles demonstrate arboviral activity in
this region in the summer of 2002,
although clinical cases were few. That
virus was not isolated from crocodiles
in late November (past outbreaks of
WNV in Israel mainly occurred
between August and October) (6,11).
WNV has been endemic in Israel
since the early 1950s (12). More
recently, in the summer of 2000, an
extensive outbreak occurred, affecting
hundreds of people (11), dozens of
horses (6), and several flocks of geese
(5). However, no deaths of crocodiles
were reported. This contrasts with the
report from Florida (1), where WNV
was isolated from dead alligators, and
where hundreds of cases of sudden
death had been reported in previous
years; these deaths are now suspected
to result, at least in part, from WNV
disease.
The role of various reptile species
in the epidemiology of other
arboviruses such as western equine
encephalitis, eastern equine
encephalitis, and Venezuelan equine
encephalitis is well documented
(13–15). At present, the role of rep-
tiles and amphibians in the life cycle
and epidemiology of WNV is not
known, and further research is neces-
sary.
Acknowledgments
We thank Kubbi Ofer for assistance
in the collection of serum samples from
the crocodiles.
Amir Steinman,* Caroline Banet-
Noach,† Shlomit Tal,‡ Ohad Levi,*
Lubov Simanov,† Shimon Perk,†
Mertyn Malkinson,†
and Nahum Shpigel*
*Hebrew University of Jerusalem Koret
School of Veterinary Medicine, Rehovot,
Israel; †Kimron Veterinary Institute, Bet
Dagan, Israel; and ‡Tel Aviv University
Sackler Faculty of Medicine, Tel Aviv, Israel
References
1. ProMED-mail. Florida: West Nile virus
identified in alligators for the first time.
ProMED-mail 2002; 14 Nov:
20021114.5797. Available from: URL:
http://www.promedmail.org
2. Travis D, McNamara T, Glaser A,
Campbell R. A national surveillance system
for WNV in zoological institutions.
Available from: URL: http://www.cdc.gov/
ncidod/dvbid/westnile/conf/ppt/1a-travis.
ppt
3. Nir Y, Lasowski Y, Avivi A, Goldwasser R.
Survey for antibodies to arboviruses in the
serum of various animals in Israel during
1965–1966. Am J Trop Med Hyg
1969;18:416–22.
4. Kostiukov MA, Gordeeva ZE, Bulychev
VP, Nemova NV, Daniiarov OA. The lake
frog (Rana ridibunda)—one of the food
hosts of blood-sucking mosquitoes in
Tadzhikistan—a reservoir of the West Nile
Dejà Vu
"I must have made this molecule before;
It is familiar to the core!"
Said a yeast cell emerging from mitosis,
With no experience yet in synthesis.
Then, guided by a transmigrant human gene,
It assembled that "alien" protein.
Boghos L. Artinian

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 889
LETTERS
fever virus. Med Parazitol (Mosk)
1985;3:49–50.
5. Malkinson M, Banet C, Weisman Y,
Pokamunski S, King R, Drouet MT, et al.
Introduction of West Nile virus in the
Middle East by migrating white storks.
Emerg Infect Dis 2002;8:392–7.
6. Steinman A, Banet C, Sutton GA, Yadin H,
Hadar S, Brill A. Clinical description of
equine West Nile encephalomyelitis during
the outbreak of 2000 in Israel. Vet Rec
2002;151:47–9.
7. Ianconescu M. Turkey meningoen-
cephalitis: a general review. Avian Dis
1976;20:135–8.
8. Berthet FX, Zeller HG, Drouet MT, Rauzier
J, Digoutte JP, Deubel V. Extensive
nucleotide changes and deletions within the
envelope glycoprotein gene of Euro-
African West Nile viruses. J Gen Virol
1997;78:2293–7.
9. Savage HM, Ceianu C, Nicolescu G,
Karabatsos N, Lanciotti RS, Vladimirescu
A, et al. Entomologic and avian investiga-
tions of an epidemic of West Nile fever in
Romania in 1996, with serologic and
molecular characterization of a virus isolate
from mosquitoes. Am J Trop Med Hyg
1999;61:600–11.
10. Ministry of the Environment, State of
Israel. Available from: URL: http://
www.sviva.gov.il
11. Weinberger M, Pitlik SD, Gandacu D, Lang
R, Nassar F, Ben David D, et al. West Nile
fever outbreak, Israel, 2000: epidemiologic
aspects. Emerg Infect Dis 2001;7:686–91.
12. Bernkopf H, Levine S, Nerson R. Isolation
of West Nile virus in Israel. J Infect Dis
1953;93:207–18.
13. Bowen G.S. Prolonged western equine
encephalitis viremia in the Texas tortoise
(Gopherus berlandieri). Am J Trop Med
Hyg 1977;26:171–5.
14. Thomas LA, Eklund CM, Rush WA.
Susceptibility of garter snakes
(Thamnophis spp.) to western equine
encephalomyelitis virus. Proc Soc Exper
Biol Med 1958;99:698–700.
15. Walder R, Suarez OM, Calisher CH.
Arbovirus studies in the Guajira region of
Venezuela: activities of Eastern equine
encephalitis and Venezuelan equine
encephalitis viruses during an interepizoot-
ic period. Am J Trop Med Hyg
1984;33:699–707.
Address for correspondence: Amir Steinman,
Koret School of Veterinary Medicine, Hebrew
University of Jerusalem, P.O.B. 12, Rehovot
76100, Israel; fax: 972-3-9688539; email:
Rickettsia
aeschlimannii in
Spain: Molecular
Evidence in
Hyalomma
marginatum and
Five Other Tick
Species that Feed
on Humans
To the Editor: Rickettsia aeschli-
mannii is a pathogenic spotted fever
group rickettsia first isolated from
Hyalomma marginatum ticks collect-
ed in Morocco in 1997 (1). Later
found in H. marginatum ticks from
Zimbabwe, Niger, Mali, and Portugal
(2), R. aeschlimannii has also been
found in a Rhipicephalus appendicu-
latus tick attached to the right thigh of
a patient in South Africa (3). These
data suggest a broad geographic dis-
tribution for R. aeschlimannii and the
possibility that tick species other than
H. marginatum may also be suitable
vectors for this rickettsia.
The pathogenicity of Rickettsia
aeschlimannii in humans has been
demonstrated by Raoult et al. (4) in a
French patient who became ill after
returning from a trip to Morocco. The
patient exhibited symptoms similar to
those of Mediterranean spotted fever
(MSF) produced by R. conorii, with a
tache noire–like eschar on his ankle,
fever (39.5°C), and a generalized
maculopapular skin rash. The second
documented and most recent case of
human infection by R. aeschlimannii
occurred in a South African man who
was bitten by R. appendiculatus; an
eschar also developed around the tick
attachment site on this patient (3). He
was aware of the risk for tick-trans-
mitted disease, so he removed the tick
and administered doxycycline; he did
not develop additional symptoms.
Over the past 6 years, throughout
the region of Castilla y León, north-
western Spain, we have collected and
identified 3,059 ticks belonging to 15
species (unpub. data) that were
attached to persons living in this terri-
tory. We have systematically analyzed
the ticks by polymerase chain reaction
(PCR) to detect those infected with
Borrelia burgdorferi, Anaplasma
phagocytophila, and Rickettsia spp.
This procedure enabled us to identify,
for the first time in Spain, R. aeschli-
mannii in 35 tick specimens belong-
ing to H. marginatum and to another
five species.
During the 6-year study, ticks
found on patients who sought medical
advice in the hospitals and healthcare
centers of Castilla y León were
removed and referred to our laborato-
ry for identification and analysis. Each
tick was first disinfected by immersion
in 70% alcohol, rinsed in sterile water,
and dried on sterile filter paper. We
then extracted DNA in 5% Chelex-
100, according to the method of
Guttman et al. (5). In searching for
Rickettsia spp., we proceeded as
described by La Scola and Raoult (6):
All DNA samples were first tested for
a fragment of the rickettsial gltA gene
(7), and then, in the gltA-positive sam-
ples, a fragment of the rickettsial
ompA gene (8) was amplified,
sequenced, and compared with gene
databases for identification. The gltA
amplicon was sequenced only when
the ompA was not successfully ampli-
fied. To prevent DNA contamination
and the carryover of amplified prod-
ucts, we used sterile tools at all times
and carried out each step of the analy-
sis (extracting DNA, preparing the
reaction mixture, and amplifying and
analyzing the PCR product) in separat-
ed work areas. Two negative controls
(Milli-Q water and DNA from labora-
tory-reared noninfected ticks) were
included in each amplification trial.
These controls were never amplified.
We obtained 21 ompA amplicons
(629–632 bp) from 21 ticks. One

890 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
amplicon, from a Haemaphysalis
punctata tick, had 100% sequence
identity with the ompA of R. aeschli-
mannii (GenBank accession no.
U43800). The nucleotide sequences
of the remaining 20 ompA amplicons
shared >99% similarity with the
ompA of R. aeschlimannii. These 20
amplicons were obtained from nine
Hyalomma marginatum, five
Rhipicephalus bursa, three R. turani-
cus, one R. sanguineus, and two
Ixodes ricinus ticks.
In an additional 14 ticks (10 H.
marginatum, 2 R. bursa, 1 R. san-
guineus, and 1 I. ricinus) we
sequenced 14 gltA amplicons (382
bp), which were 100% identical to the
gltA of R. aeschlimannii (GenBank
accession no. U59722). No other tick-
borne pathogens were detected in the
35 R. aeschlimannii–containing ticks.
R. aeschlimannii has never been
detected in Spain; therefore, our study
constitutes its first citation in this
country. Because R. aeschlimannii
had been already detected in ticks in
Portugal, we believe that its presence
in Spain was expected and our finding
is not surprising. However, the high
number of tick species in which we
found this rickettsia was unexpected:
six species belonging to four genera.
For these six species, the ratio
between the specimens infected and
the specimens analyzed (as well as the
infection rates) were as follows: I.
ricinus (3/1,320; 0.23%), H. mar-
ginatum (19/324; 5.86%), H. punctata
(1/106; 0.94%), R. bursa (7/425,
1.64%), R. sanguineus (2/102;
1.96%), and R. turanicus (3/330;
0.91%). Although H. marginatum was
the fourth most anthropophilic species
in our study, this species simultane-
ously showed the highest number of
infected specimens and the highest
infection rate, making H. marginatum
the main vector of R. aeschlimannii in
our region. The next most important
vectors are Rhipicephalus spp., and in
particular R. bursa.
The 35 R. aeschlimannii–positive
ticks were removed in the first 6 to 12
hours after attachment, before they
could have ingested any blood, thus
indicating that they were previously
infected with the bacterium. Persons
bitten by these specimens had never
had symptoms of spotted fever, and
they remained asymptomatic after the
bite, suggesting that, as expected
because of the rapid removal of the
ticks, they did not acquire the infection.
Although MSF is endemic in
Castilla y León (9), we only found
one tick infected with R. conorii
(0.03%) among the 3,059 analyzed
(unpub. data), whereas R. aeschliman-
nii was much more prevalent in these
same ticks (1.14%). Hence, in accor-
dance with what was proposed by
Raoult et al. (4) for MSF cases in
Morocco, we suspect that many cases
of MSF in Castilla y León may really
have been due to R. aeschlimannii.
Our findings show that R. aeschli-
mannii is present in Castilla y León,
the largest region in Spain, in six tick
species that frequently feed on
humans. Our observations expand the
geographic distribution of this bac-
terium and the range of its potential
tick vectors.
Acknowledgments
We thank Rufino Álamo-Sanz for his
invaluable contributions and N. Skinner
for revising the English version of the
manuscript.
This work was supported by the
Consejería de Sanidad y Bienestar Social
of the Junta de Castilla y León (Spain).
Pedro Fernández-Soto,*
Antonio Encinas-Grandes,*
and Ricardo Pérez-Sánchez†
*Universidad de Salamanca, Salamanca,
Spain; and †Instituto de Recursos
Naturales y Agrobiología, Consejo Superior
de Investigaciones Científicas, Salamanca,
Spain
References
1. Beati L, Meskini M, Thiers B, Raoult D.
Rickettsia aeschlimannii sp. nov., a new
spotted fever group rickettsia associated
with Hyalomma marginatum ticks. Int J
Syst Bacteriol 1997;47:548–54.
2. Parola P, Raoult D. Ticks and tickborne
bacterial diseases in humans: an emerging
infectious threat. Clin Infect Dis
2001;32:897–928.
3. Pretorius A, Birtles RJ. Rickettsia aeschli-
mannii: a new pathogenic spotted fever
group rickettsia, South Africa. Emerg Infect
Dis 2002;8:874.
4. Raoult D, Fournier PE, Abboud P, Caron F.
First documented human Rickettsia aeschli-
mannii infection. Emerg Infect Dis
2002;8:748–9.
5. Guttman D S, Wang PW, Wang IN, Bosler
EM, Luft BJ, Dykhuizen DE. Multiple
infections of Ixodes scapularis ticks by
Borrelia burgdorferi as revealed by single-
strand conformation polymorphism analy-
sis. J Clin Microbiol 1996;34:652–6.
6. La Scola B, Raoult D. Laboratory diagnosis
of rickettsioses: current approaches to diag-
nosis of old and new rickettsial diseases. J
Clin Microbiol 1997;35: 2715–27.
7. Regnery RL, Spruil CL, Plikaytis BD.
Genotypic identification of Rickettsiae and
estimation of intraspecies sequence diver-
gence for portions of two rickettsial genes.
J Bacteriol 1991;173:1576–89.
8. Roux V, Furnier PE, Raoult D.
Differentiation of spotted fever group rick-
ettsiae by sequencing and analysis of
restriction fragment length polymorphism
of PCR-amplified DNA of the gene encod-
ing the protein rOmpA. J Clin Microbiol
1996;34:2058–65.
9. Boletines Epidemiológicos de Castilla y
León 1997–2002. (Spain): 2000;16:9–12.
Address for correspondence: Ricardo Pérez-
Sánchez, Departamento de Patología Animal,
Instituto de Recursos Naturales y Agrobiología,
Consejo Superior de Investigaciones
Científicas, C/ Cordel de Merinas, 40-52,
37007 Salamanca, Spain; fax: +34 923 219609;
email: [email protected]
The opinions expressed by authors con-
tributing to this journal do not necessari-
ly reflect the opinions of the Centers for
Disease Control and Prevention or the
institutions with which the authors are
affiliated.

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 891
LETTERS
Hantaviruses in
São Paulo State,
Brazil
To the Editor: Hantavirus pul-
monary syndrome (HPS) is an emerg-
ing health problem in Brazil. This
syndrome was first reported in 1993
in three persons living in a rural area
of Juquitiba County; two of them died
of acute respiratory failure (1).
Although Juquitiba County is part of
the metropolitan area of greater São
Paulo City, patients lived in a recently
deforested region. From 1993 through
2002, approximately 200 HPS cases
were reported in Brazil, with a 40%
case-fatality ratio (Ministry of Health
of Brazil, Report on Hantavirus cases
1993–2002, unpub. data).
The wild rodent Bolomys laziurus
is believed to be the most important
hantavirus reservoir in the State of
São Paulo, based on high levels of
specific antibodies observed in serum
from captured specimens (L.E.
Pereira, Adolpho Lutz Institute, pers.
comm., 2001). The economy of the
inland region of Ribeirão Preto in the
State of São Paulo, with its 3.5 million
inhabitants, is based on the sugar cane
agroindustry. The region has been
almost completely deforested, with
important consequences to the envi-
ronment and wild rodent ecology.
Twenty HPS cases were reported in
Ribeirão Preto in the last 5 years, with
a 60% case-fatality ratio. Review of
medical records showed that a pro-
dromic fever occurred in all 14 case-
patients studied; dyspnea, cough,
hypotension, and tachycardia
occurred in about two thirds of
patients; and hemorrhagic phenomena
(hematuria, melena, and hypermenor-
rhea) in about one third.
Thrombocytopenia was observed in
all the patients, elevated hematocrit in
about three fourths, and leukocytosis
with neutrophilia and a left shift in the
differential count in about two thirds.
Serum creatinine levels were also
increased (average level 2 mg/dL).
Chest radiographs showed diffuse
alveolar flocculant infiltrates in most
cases (2,3). Laboratory diagnosis of
HPS was made by serologic testing
(enzyme-linked immunosorbent assay
[ELISA]) in 18 cases and by reverse
transcription–polymerase chain reac-
tion (RT-PCR) in 11 cases; for 7 cases,
both techniques were used. We per-
formed a nucleotide sequence analy-
sis of the N gene of hantavirus
(residues 236–477) obtained from the
blood of 11 of the 20 patients. This
analysis showed that the infections
were caused by Araraquara virus, a
previously known hantavirus that had
been detected by RT-PCR in the
serum of an HPS patient living in a
nearby county (4). Thus, Araraquara
virus is the causative agent of a severe
form of HPS, with a high death rate.
This high death rate could also be
related to the lack of adequate initial
therapy provided by clinicians who
probably did not immediately suspect
HPS and may have not recommended
hospitalization in intensive-care units.
In addition, some hospitalized
patients were in shock when first seen
and were rehydrated with massive
quantities of fluids, which may have
aggravated pulmonary edema and
contributed to death.
The occurrence of 10% of the
Brazilian HPS reported cases in
Ribeirão Preto indicates that this
region is suitable for studying the epi-
demiology of hantavirus infections. A
serologic survey conducted in the
region in 1999, which included 567
primary-care patients from Ribeirão
Preto, Guariba, and Jardinópolis
Counties, found that 7 (1.23%) of
them had immunoglobulin (Ig) G
antibodies to Sin Nombre virus by
ELISA and that 5 of those lived in
Jardinópolis (population 30,000), a
county where a fatal case of HPS
occurred in 1999 (5). Thus,
Jardinópolis County was chosen for a
population-based survey. In May
2001, we obtained personal informa-
tion and collected fingerprick blood
samples from 818 participants, 15–70
years of age, living in urban and rural
areas of the county. IgG antibodies to
the N recombinant protein of Andes
virus were detected by ELISA in the
blood samples of 14.3% of the partic-
ipants (5). Even though all HPS cases
in Ribeirão Preto were associated
with rural activity and rodent expo-
sure, these serologic data suggest that
hantavirus infections are common in
Jardinópolis County, independent of
sex, profession, or history of contact
with rodents. None of the 14.3% par-
ticipants with IgG antibodies to han-
tavirus had a history of HPS-like dis-
ease, and the ELISA test showed
cross-reactions with most of the South
American hantaviruses, including
Araraquara. Persons living in the
urban area had higher levels of anti-
bodies to hantavirus than those from
rural areas. In Ribeirão Preto, the
physical boundaries of cities have
expanded to incorporate other areas,
encroaching upon rural areas with
many popular subsidized housing
complexes. Work-related and recre-
ational rural activities in that region
are also frequent, which makes it dif-
ficult to interpret these data. These
results suggest that in this region of
southeast Brazil, hantaviruses may be
causing undiagnosed asymptomatic or
clinically minor infections in addition
to typical HPS. This finding envokes
important questions. Is more than one
hantavirus circulating in this region,
causing mostly benign infections? Is
Araraquara virus widespread, causing
mostly inapparent infections and only
rarely causing HPS? Would HPS be
associated with some predisposing
condition in the infected person? If
more than one hantavirus is circulat-
ing in the region, could urban rodents
be reservoirs?
Further studies are necessary to
better understand the epidemiology
and clinical signs and symptoms of
hantavirus infection in the region of
Ribeirão Preto. Such studies should

892 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
emphasize determining the reservoirs,
the modes of virus transmission to
people, and the possible distinct clini-
cal forms of hantavirus infections.
Acknowledgments
We thank the employees of the
Health Bureau of the County of
Jardinópolis for the dedicated collabora-
tion on the serologic survey and Paula
Padula for supplying antigen for enzyme-
linked immunoabsorbent assays.
This work was supported by the State
of São Paulo Research Foundation
(FAPESP).
Luiz T.M. Figueiredo,*
Marcos L. Moreli,*
Gelse M. Campos,*
and Ricardo L.M. Sousa*
*University of São Paulo, São Paulo, Brazil
References
1. Iversson LB. Hantavirus pulmonary
syndrome in the rural area of Juquitiba, São
Paulo Metropolitan Area, Brazil. Rev Inst
Med Trop Sao Paulo 1997;39:237–8.
2. Figueiredo LTM, Moreli ML, Kashima S,
Almeida VSO, Félix PR, Bruno JC, et al.
Hantavirus pulmonary syndrome (HPS) in
Guariba, SP, Brazil. Report of 2 cases. Rev
Inst Med Trop Sao Paulo 1999;41:131–7.
3. Figueiredo LTM, Campos GM, Rodrigues
FB. Síndrome pulmonar e cardiovascular
por hantavirus: aspectos epidemiológicos,
clínicos, do diagnóstico laboratorial e do
tratamento. [in Portuguese]. Rev Soc Bras
Med Trop 2001;34:15–27.
4. Johnson AM, de Souza LT, Ferreira IB,
Pereira LE, Ksiazek TG, Rollin PE, et al.
Genetic investigation of novel hantaviruses
causing fatal HPS in Brazil. J Med Virol
1999;59:527–35.
5. Holmes RR, Boccanera R, Figueiredo
LTM, Mançano SR, Pane C.
Seroprevalence of human hantavirus
infection in Ribeirão Preto Region of São
Paulo State, Brazil. Emerg Infect Dis
2000;6:560–1.
Address for correspondence: Luiz Tadeu
Moraes Figueiredo, Faculdade de Medicina de
Ribeirão Preto–USP, Av. Bandeirantes 3900,
Ribeirão Preto, SP, 14049-900, Brazil; fax: 55
16 6336695; email: [email protected]
Israeli Spotted
Fever Rickettsia
in Sicilian
Rhipicephalus
sanguineus Ticks
To the Editor: Mediterranean
spotted fever (MSF) is endemic in
Italy, where it is a reportable disease.
From 1992 to 1998, the Italian
Ministry of Health was notified of
approximately 8,500 cases of human
rickettsioses presumed to be MSF.
MSF occurs more commonly in some
central (Lazio) and southern
(Sardinia, Sicily, and Calabria)
regions (1,2); in 1998, an average of
8.8 cases occurred for every 100,000
persons in Sicily, compared with the
national average of 1.6 cases per
100,000 persons. Rickettsia conorii
has been thought to be the only path-
ogenic Rickettsia of the spotted fever
group in Sicily (3,4) or the western
Mediterranean area. Recently, three
different spotted fever group rickettsi-
ae, including R. helvetica, were
detected in Ixodes ricinus ticks from
central and northern Italy. This find-
ing suggests that bacteria other than
R. conorii are involved in rickettsial
diseases in Italy (5).
To investigate whether unusual
tick-transmitted rickettsiae are also
present in Sicily, we used molecular-
sequence–based identification tech-
niques to study two strains isolated
from the hemolymph of
Rhipicephalus sanguineus ticks col-
lected in 1990 in western Sicily. These
isolates had been previously identi-
fied by serologic tests as belonging to
the spotted fever group rickettsiae. We
obtained bacterial DNA and per-
formed polymerase chain reaction
(PCR) for ompA gene and restriction
analysis under conditions previously
described by Roux et al. (6). Our
observation of a peculiar PstI profile
allowed a presumptive identification
of one of the two tick isolates as
belonging to the Israeli spotted fever
rickettsiae, while the other showed a
restriction profile corresponding to
that of R. conorii strain Seven. To
confirm the identification of the
Israeli spotted fever Rickettsia isolate,
we sequenced the PCR-amplified
fragment of ompA gene (MWG-
Biotech AG, Ebersberg, Germany)
and aligned sequence data with
homologous sequences of reference
strains of the spotted fever group rick-
ettsiae retrieved from the GenBank
database. Sequence analysis showed
100% similarity with the homologous
sequence of Israeli spotted fever
Rickettsia reference strain ISTT
CDC1 (GenBank accession no.
U43797). The Israeli spotted fever
Rickettsia belongs to the R. conorii
complex (7,8) and was first isolated in
1974 from ticks and humans. Initially,
Israeli spotted fever rickettsiae distri-
bution appeared to be restricted to
Israel (9), but more recently the
organism has also been isolated from
patients with MSF in Portugal (10).
Our finding of Israeli spotted fever
Rickettsia infection in a R. sanguineus
tick, the main vector for MSF in
Sicily, also suggests that the geo-
graphic distribution of Israeli spotted
fever might be wider than previously
thought, including not only Israel and
the Iberian Peninsula but also Italy.
Molecular analysis of spotted
fever group Rickettsia isolates from
Sicilian MSF patients is under way to
verify this hypothesis. Because initial
signs and symptoms of Israeli spotted
fever are particularly uncharacteristic,
awareness of the presence of Israeli
spotted fever Rickettsia in our geo-
graphic area may hasten provision of
the appropriate treatment. The
Sicilian ompA gene sequence
described in this study has been

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 893
LETTERS
deposited in the GenBank database
(accession no. AY197565).
Giovanni M. Giammanco,*
Serafino Mansueto,*
Pietro Ammatuna,*
and Giustina Vitale*
*Università di Palermo, Palermo, Italy
References
1. Scaffidi V. Current endemic expansion of
boutonneuse fever in Italy. Minerva Med
1981;72:2063–70.
2. Tringali G, Intonazzo V, Perna AM,
Mansueto S, Vitale G, Walker DH.
Epidemiology of Boutonneuse fever in
western Sicily. Distribution and prevalence
of spotted fever group rickettsial infection
in dog ticks (Rhipicephalus sanguineus).
Am J Epidemiol 1986;123:721–7.
3. Mansueto S, Vitale G, Lavagnino A, Di
Rosa S, Merulla R. Rickettsiae of the spot-
ted fever group in dog fleas
(Ctenocephalides spp.) in western Sicily.
Ann Trop Med Parasitol 1989;83:325.
4. Vitale G, Di Stefano R, Damiani G,
Mansueto S. Characterization of Sicilian
strains of spotted fever group rickettsiae by
using monoclonal antibodies. J Clin
Microbiol 1989;27:1081–5.
5. Beninati T, Lo N, Noda H, Esposito F,
Rizzoli A, Favia G, et al. First detection of
spotted fever group Rickettsiae in Ixodes
ricinus from Italy. Emerg Infect Dis
2002;8:983–6.
6. Roux V, Fournier PE, Raoult D.
Differentiation of spotted fever group
Rickettsiae by sequencing and analysis of
restriction fragment length polymorphism
of PCR-amplified DNA of the gene
encoding the protein rOmpA. J Clin
Microbiol 1996;34:2058–65.
7. Regnery RL, Spruill CL, Plikaytis BD.
Genotypic identification of Rickettsiae and
estimation of interspecies sequence diver-
gence for portions of two rickettsial genes.
J Bacteriol 1991;173:1576–89.
8. Fournier PE, Roux V, Raoult D.
Phylogenetic analysis of spotted fever
group Rickettsiae by study of the outer sur-
face protein rOmpA. Int J Syst Bacteriol
1998;48:839–49.
9. Roux V, Raoult D. Phylogenetic analysis
and taxonomic relationships among the
genus Rickettsia. In: Raoult D, Brouqui P,
editors. Rickettsiae and rickettsial diseases
at the turn of the third millennium. Paris:
Elsevier; 1999. p. 52–66.
10. Bacellar F, Beati L, Franca A, Pocas J,
Regnery R, Filipe A. Israeli spotted fever
rickettsia (Rickettsia conorii complex)
associated with human disease. Emerg
Infect Dis 1999;5:835–6.
Address for correspondence: Giovanni M.
Giammanco, Department of Hygiene and
Microbiology, Università di Palermo, via del
Vespro 133, I-90127, Palermo, Italy; fax: +39
0916553676; email: [email protected]
Co-feeding
Transmission and
Its Contribution to
the Perpetuation of
the Lyme Disease
Spirochete
Borrelia afzelii
In Reply: Richter et al. (1) have
asked an important question: To what
extent does the transmission of non-
systemic infections of the Lyme borre-
liosis spirochete (Borrelia afzelii)
between co-feeding nymphal and lar-
val Ixodes ricinus ticks apply to natu-
ral tick infestations on wild rodents?
The authors conclude that the trans-
mission of infections 3 days after inoc-
ulation by tick bite is >100 times less
efficient than the transmission of infec-
tions that have lasted at least 14 days.
That answer depends on a critical cal-
culation based on experimental results
combined with field observations.
Unfortunately, this calculation is incor-
rect by a factor of approximately 20.
When hairless laboratory mice
were restrained within wire mesh
tubes and larvae were allowed to
attach at random over their bodies,
13.6% of these larvae became infect-
ed with B. afzelii if they fed 3 days
after the attachment of a single infect-
ed nymph (i.e., transmission probabil-
ity of 0.136, as used below). By con-
trast, 85.4% of larvae that fed 14 days
after the nymph became infected (1).
At three sites in Germany and France,
over the period April–October in each
of the years from 1993 through 1995,
17.6% of mice (Apodemus flavicollis
and A. sylvaticus) and voles
(Clethrionomys glareolus) fed larval
and nymphal ticks together, while
1.5% fed nymphs alone. Of these
nymphs, 26.4% were infected with B.
burgdorferi s.l. before attachment.
The probability of a larva’s acquiring
an infection equals the product of 1)
the probability of transmission from
host to larva and 2) the probability of
the host’s being infected, while the
larva feeds, via an infected nymphal
tick bite. For a short-lived (3-day)
infection, the probability is 0.136 ×
0.176 × 0.264 = 0.0063; for longer-
lived (14-day) infections, the proba-
bility is 0.854 × (0.176 + 0.015) ×
0.264 = 0.0431. The ratio is therefore
1:6.8. Richter et al. erroneously con-
cluded that the ratio was 1:116
because they did not take into account
the probability of wild rodents’
acquiring a long-lived, ”systemic”
infection; the authors assumed the
probability was 1. A greater propor-
tion of garden dormice (Eliomys
quercinus) carried ticks and so would
yield much higher transmission prob-
abilities but in almost the same ratio,
1:6.4.
In fact, how much of the increase
from 13.6% transmission at day 3 to
85.4% at day 14 was due to the devel-
opment of systemic infections (i.e.,
disseminated to parts of the hosts’
bodies >2 cm from the infected tick
bite) is not clear because the feeding
sites of the larvae attached ad libitum
on the hairless mice were not report-
ed. In the original discovery of co-
feeding transmission of B. burgdor-
feri s.l. (2), the infection prevalence in
larvae feeding close to infected
nymphs increased from 33% on day 2
to 96% on day 11 and 100% on day 14
(3; see Figure 2 therein) in the demon-
strated absence of a systemic infec-
tion. Mice skin and ticks feeding at
distant sites remained uninfected.
Only after day 14 had a systemic
894 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
infection developed (2). Because
spirochetes are not transmitted to the
host until at least 17.6 h after an
infected nymph starts feeding (4–6)
and then disseminate only slowly
from the feeding site (7), co-feeding
in space rather than in time is the cru-
cial feature in Lyme borreliosis (2,8)
(so-called “extended co-feeding”[3]).
Larvae that attach to hosts simultane-
ously with infected nymphs rarely
acquire spirochetes (1,9), wherever
they attach. This pattern is distinct
from the more immediate and short-
lived co-feeding transmission of tick-
borne viruses (10–12). In both cases,
however, the key feature is a nonsys-
temic infection.
Despite the uncertainties in Richter
et al.’s study, their corrected ratio is
very similar to that (1:5.7) calculated
(3) with a “synthetic model . . . based
on major assumed parameters” (sic)
(1). In that model we assumed that
50% of larvae were likely to be feed-
ing within 1 to 2 cm of any infected
nymph, the distance over which co-
feeding ticks can pick up nonsystemic
infections (1,2), because in the wild
very few rodents carry nymphs in the
absence of larvae (1,13), and >95% of
all immature stage ticks feed in aggre-
gations, mostly on the ears and also
around the eyes or on the snouts of
mice and voles. Considerable risks
exist in using laboratory experimental
results to quantify the epidemiologic
importance of nonsystemic infections
in the wild because of differences
between host species, unnatural spa-
tial distributions of introduced ticks
on hosts, and the subtleties of natural
tick-host relationships. Coincident
aggregated distributions of larvae and
nymphs among their rodent hosts,
whereby the same individual hosts
carry the largest numbers of both
stages, increase the number of larvae
co-feeding with any infected nymph,
and so augment the potential amplifi-
cation of infection prevalence in ticks
(13). Nevertheless, in the case of
rodents, nonsystemic infections are
soon rendered redundant by the much
longer lived systemic infections. In
contrast, in the case of host species in
which systemic infections do not
develop, the transmission of nonsys-
temic infections between co-feeding
ticks is the only way in which infec-
tion prevalence can be amplified in
feeding ticks. Field data suggest that
this route of transmission occurs on
wild Sika deer (Cervus nippon) (14).
Natural experimental systems have
confirmed that on sheep this transmis-
sion pathway exists and is sufficient
alone to maintain enzootic cycles of
Lyme borreliosis (8).
Sarah Randolph* and Lise Gern†
*University of Oxford, Oxford, United
Kingdom, and †Université de Neuchâtel,
Neuchâtel, Switzerland
References
1. Richter D, Allgöwer R, Matuschka F-R.
Co-feeding transmission and its contribu-
tion to the perpetuation of the Lyme disease
spirochete Borrelia afzelii. Emerg Infect
Dis 2002;8:1421–5.
2. Gern L, Rais O. Efficient transmission of
Borrelia burgdorferi between cofeeding
Ixodes ricinus ticks (Acari: Ixodidae). J
Med Entomol 1996;33:189–92.
3. Randolph SE, Gern L, Nuttall PA. Co-feed-
ing ticks: epidemiological significance for
tick-borne pathogen transmission. Parasitol
Today 1996;12:472–9.
4. Kahl O, Janetzki-Mittmann C, Gray JS,
Jonas R, Stein J, de Boer R, et al. Risk of
infection with Borrelia burgdorferi sensu
lato for a host in relation to the duration of
nymphal Ixodes ricinus feeding and the
method of tick removal. Zentbl Bakt
1998;287:41–52.
5. Crippa M, Rais O, Gern L. Investigations
on the mode and dynamics of transmission
and infectivity of Borrelia burgdorferi ss
and Borrelia afzelii in Ixodes ricinus ticks.
Vector-borne and Zoonotic Diseases
2002;2:3–9.
6. Piesman J. Dispersal of the Lyme disease
spirochete Borrelia burgdorferi to salivary
glands of feeding nymphal Ixodes scapu-
laris (Acari: Ixodidae). J Med Entomol
1995;32:519–21.
7. Shih C-M, Pollack RJ, Telford SR,
Spielman A. Delayed dissemination of
Lyme disease spirochetes from the site of
deposition in the skin of mice. J Infect Dis
1992;166:827–31.
8. Ogden NH, Nuttall PA, Randolph SE.
Natural Lyme disease cycle maintained via
sheep by co-feeding ticks. Parasitology
1997;115:591–9.
9. Piesman J, Happ CM. The efficacy of co-
feeding as a means of maintaining Borrelia
burgdorferi: a North American model sys-
tem. J Vector Ecol 2001;26:216–20.
10. Jones LD, Davies CR, Steele GM, Nuttall
PA. A novel mode of arbovirus transmis-
sion involving a nonviraemic host. Science
1987;237:775–7.
11. Alekseev AN, Chunikhin SP. Exchange of
tick-borne encephalitis virus between
Ixodidae simultaneously feeding on the ani-
mals with sub-threshold levels of viraemia.
Med Parazitol Parazit Bolezni
1990;2:48–50.
12. Labuda M, Jones LD, Williams T,
Danielova V, Nuttall PA. Efficient transmis-
sion of tick-borne encephalitis virus
between cofeeding ticks. J Med Entomol
1993;30:295–9.
13. Randolph SE, Miklisová D, Lysy J, Rogers
DJ, Labuda M. Incidence from coinci-
dence: patterns of tick infestations on
rodents facilitate transmission of tick-borne
encephalitis virus. Parasitology 1999;
118:177–86.
14. Kimura K, Isogal E, Isogal H, Kamewaka
Y, Nishikawa T, Ishii N, et al Detection of
Lyme disease spirochetes in the skin of nat-
urally infected wild Sika deer (Cervus nip-
pon yesoensis) by PCR. Appl Environ
Microbiol 1995;61:1641–2.
Address for correspondence: Sarah Randolph,
University of Oxford, South Parks Road,
Oxford OX1 3PS, UK; fax: +44 1865 310447;
email: sarah.randolph@zoology.ox.ac.uk

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 895
LETTERS
Co-feeding
Transmission and
Its Contribution to
the Perpetuation of
the Lyme Disease
Spirochete
Borrelia afzelii
In Reply to Randolph and Gern:
Although transmission between co-
feeding vector ticks may perpetuate
particular tick-borne viruses, this
mode of transmission plays no role in
the epizootiology of Lyme disease
spirochetes (1,2). In their letter,
Randolph and Gern defend their sug-
gestion that tick-borne pathogens per-
petuate effectively by direct passage
from one feeding tick to another by
criticizing our analysis (3). These
researchers mainly address our com-
parison of the transmission efficiency
between simultaneously feeding ticks
with that between ticks feeding
sequentially on a persistently infected
rodent. Our experiments demonstrate
that approximately six times as many
larvae (85.4%) acquire Borrelia
afzelii spirochetes from a systemical-
ly infected mouse than from a mouse
on which an infected nymph is feed-
ing simultaneously (13.6%) (1). In
nature, however, larval ticks rarely
co-feed with nymphs on mice or
voles; only approximately one fifth
(18.8%) of these hosts harbor both
subadult stages simultaneously. And
of the nymphs, only approximately
one quarter (26.4%) are infected by
Lyme disease spirochetes. As a result,
the natural transmission efficiency
between simultaneously feeding ticks
would be only one twentieth (5%) of
that observed in the laboratory.
Multiplying the experimentally
observed efficiency of co-feeding
transmission (13.6%) by the likeli-
hood of larval and nymphal ticks co-
infesting small rodents, as well as by
the prevalence of infected nymphal
ticks, reduces the efficiency of co-
feeding transmission in nature to
<1%. Although Randolph and Gern
commit several minor mathematical
errors, their calculations support our
argument that few larval vector ticks
would acquire spirochetal infection
directly from an infected nymph (3).
Randolph and Gern err, however,
by applying the same mathematical
modifications to the transmission effi-
ciency by which larvae acquire spiro-
chetes from a persistently infected
host (3). Whereas the efficiency of co-
feeding transmission observed in the
laboratory must be modified to pay
tribute to the rare event of larvae co-
feeding with an infected nymph in
nature, the efficiency by which larvae
acquire infection from a persistently
infected host is independent of such
limiting parameters. Because a com-
petent rodent host remains infectious
to larval ticks throughout its life, the
proportion of hosts infested by partic-
ular subadult stages of the vector is
irrelevant. Thus, the transmission effi-
ciency on a persistently infected host
is unchanged in the laboratory and the
field. Almost 85.4% of larvae feeding
on mice or voles in nature would,
therefore, acquire spirochetal infec-
tion—far more than by co-feeding.
We are correct in stating that natural

896 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
LETTERS
transmission by sequentially feeding
ticks is more efficient than transmis-
sion between co-feeding ticks.
Randolph and Gern suggest that
we could have recorded the distance
between the feeding ticks to clarify
whether the increase from a 13.6%
transmission efficiency between co-
feeding ticks to a transmission effi-
ciency of 85.4% from a persistently
infected host is associated with the
development of a systemic infection.
Our experimental observation (Table
1 in our article [1]), as well as a study
on the movement of spirochetes
through their host’s skin (4), conclu-
sively demonstrates that the increase
in transmission efficiency is due to the
progressive dissemination of spiro-
chetes from the site of inoculation.
The likelihood of a larva’s acquiring
spirochetes from any site of its host’s
skin increases with the passage of
time since the infected nymph
attached. To compare the two modes
of transmission in terms of efficiency
(Table 2 in our article [1]), we permit-
ted the larvae to attach randomly to
their rodent hosts, just as they would
do in nature.
In the epizootiology of Lyme dis-
ease spirochetes, “simultaneous”
transmission between co-feeding ticks
(<1%) is some two orders of magni-
tude less efficient than sequential
transmission between ticks feeding on
persistently infected reservoir rodents
(85.4%). The two studies that relied
on natural infestation densities and
refrained from using artificial feeding
capsules conclusively demonstrated
that transmission of Lyme disease
spirochetes between ticks feeding
simultaneously and in close proximity
contributes little to the perpetuation of
this pathogen, either in North America
or in Europe (1,2). We are correct in
concluding that Lyme disease spiro-
chetes are maintained in nature main-
ly by sequential attachment of ticks to
persistently infected reservoir hosts.
Dania Richter,* Rainer Allgöwer,*
and Franz-Rainer Matuschka*
*Humboldt-Universität zu Berlin, Berlin,
Germany
References
1. Richter D, Allgöwer R, Matuschka F-R.
Co-feeding transmission and its contribu-
tion to the perpetuation of the Lyme disease
spirochete Borrelia afzelii. Emerg Infect
Dis 2002;8:1421–5.
2. Piesman J, Happ CM. The efficacy of co-
feeding as a means of maintaining Borrelia
burgdorferi: a North American model sys-
tem. J Vector Ecol 2001;26:216–20.
3. Randolph S, Gern L. Reply to Richter et al:
Co-feeding transmission and its contribu-
tion to the perpetuation of the Lyme disease
spirochete Borrelia afzelii. Emerg Infect
Dis 2003;9:893–4.
4. Shih CM, Telford SR, Pollack RJ, Spielman
A. Rapid dissemination by the agent of
Lyme disease in hosts that permit fuminat-
ing infection. Infect Immun
1993;61:2396–9.
Address for correspondence: Dania Richter,
Abteilung Parasitologie, Institut für Pathologie,
Charité, Humboldt-Universität zu Berlin,
Malteserstraße 74-100, 12249 Berlin,
Germany; fax: 49 30 776 2085; email:

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 897
NEWS & NOTES
Conference Summary
West Nile Virus
Southeast
Conference
1
On January 14–15, 2003, more
than 60 scientists, public health offi-
cials, and clinicians from throughout
the southeastern United States gath-
ered in Atlanta to present data from
the 2001 and 2002 West Nile virus
(WNV) epidemics. The aim of the
daylong conference, hosted by the
Southeastern Center for Emerging
Biological Threats at Emory
University, was to assemble a diverse
perspective on WNV by sharing
knowledge, identifying key questions
for research about the disease and pre-
vention, and fostering collaborations
between epidemiologists, veterinari-
ans, laboratorians, and clinicians.
In the overview presentation, the
experiences of state health officials,
clinical spectrum and pathogenesis,
laboratory diagnostics, veterinary
issues, surveillance, and vector control
were discussed. The 2002 epidemic
produced the largest annual number of
cases to date, and four novel modes of
transmission were discovered: 1)
transplanted organs or tissue, includ-
ing blood; 2) breast milk from an
infected mother; 3) percutaneous
exposure to infected tissue or serum
among laboratory and hospital work-
ers; and 4) transplacental exposure to
fetuses in utero, resulting in birth
defects. In addition, WNV-associated
acute flaccid paralysis was discussed;
the paralysis is caused by localized
infection of the anterior horn cells of
the spinal cord, resulting in signs and
symptoms similar to poliomyelitis.
State epidemiologists or their rep-
resentatives presented information
about the state epidemics in the south-
eastern United States. The experi-
ences in four states (Florida, Georgia,
Louisiana, and Mississippi) were
common in their themes of expansion
of WNV epidemics, concentrated
nature of outbreaks, importance of
protection from mosquito bites, limi-
tations of diagnostic methods, and
dynamics of WNV spread in the
United States from 2001 to 2002.
Significant differences also emerged
regarding the observed benefits of
mosquito control activities and the
value of animal surveillance as an
early detection system. Also, while
the states unanimously agreed that
collaboration among local, state, and
federal public health agencies, aca-
demic research institutions, and other
nongovernmental organizations is
critical to responding effectively to
WNV, the degree to which such col-
laborations actually occurred and the
existence of previously established
relationships varied.
Several clinicians discussed the
pathogenesis and clinical aspects of
the disease. The pathogenesis of
WNV was reviewed; clinicians sug-
gested that infections of the central
nervous system can demonstrate any
or all of three distinct characteristics:
neuroinvasiveness (the ability to enter
the nervous system), neurotropism
(the ability to infect neural cells), and
neurovirulence (the ability to cause
neurologic disease). WNV possesses
all three. The virus can enter the nerv-
ous system, as shown by the
encephalitis that occurs in approxi-
mately 1 in 150 infected persons; it
has been shown to infect neurons;
approximately 10% of patients in
which the virus has invaded the nerv-
ous system eventually die. One pre-
senter suggested that the virus had
changed in the last 60 years, evolving
into a more virulent strain. The neuro-
logic manifestations of the disease
were described, including weakness
and flaccid paralysis (which can occur
even without fever or meningoen-
cephalitis).
WNV infection in patients with
HIV was also described. Two HIV-
infected patients, with CD4 cell
counts below 200 cells/µL were iden-
tified with WNV by the presence of
immunoglobulin M antibodies to
WNV. The first was a 50-year-old
homeless man co-infected with tuber-
culosis. Despite treatment including
intravenous acyclovir, the patient con-
tinued to deteriorate, and died 18 days
after admission. An autopsy revealed
meningitis. The second HIV-positive
patient diagnosed with WNV was a
48-year-old man who arrived at the
emergency room with fever,
headache, and confusion; he reported
feeling “slow,” and indeed was slow
to respond to questions. This patient
improved rapidly and was discharged
after 3 days.
Information about intracellular
host-virus interactions was summa-
rized. Studies in mice have identified
a genetic allele that apparently con-
fers resistance to flaviviruses.
Although humans do not have a direct
genetic homologue, studies to identify
genetic differences to explain differ-
ent clinical outcomes should be pur-
sued. Results of a case-control study
suggested that the greatest increases
in risk were related to environmental
factors favoring mosquito popula-
1
Presentations: James J. Sejvar, Centers for Disease Control and Prevention (CDC); Mary Currier, Mississippi Department of Health;
Raoult Ratard, Louisiana Department of Health & Human Services; Lisa Conti, Florida Department of Health; Susan Lance-Parker,
Georgia Department of Human Resources; Richard T. Johnson, Johns Hopkins University School of Medicine; A. Arturo Leis, University
of Mississippi Medical Center; Margo Brinton, Georgia State University; Sally Slavinski, Mississippi State Health Department; Carlos del
Rio, Emory University; Sharif R. Zaki, CDC; Lillian Stark, Florida Department of Health; Elizabeth Howerth, University of Georgia; Brigid
Elchos, Mississippi Department of Health; Jonathan Day, University of Florida; Tom Bevan, Georgia Institute of Technology; Mike
Bunning, CDC; and David Stallknecht, University of Georgia.

898 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
NEWS & NOTES
Conference Summary
West Nile Virus and
Wildlife Health
The West Nile Virus and Wildlife
Health Workshop, hosted by the
Smithsonian Institution, National
Audubon Society, U.S. Geological
Survey, and U.S. Department of
Agriculture, was held February 5–7,
2003, at the Smithsonian Environ-
mental Research Center in Edgewater,
Maryland. The event was attended by
more than 100 scientists, who heard
29 speakers and participated in strate-
gy discussions during the 2-day meet-
ing. The main focus of the conference
was the present and future impact of
West Nile virus on wildlife popula-
tions. Talks and discussions empha-
sized how basic research, public
health, and land management can con-
tribute to our understanding of the dis-
ease’s impact and spread. A primary
objective of this meeting was to devel-
op future research priorities from both
basic and applied perspectives.
The conference centered around
four main themes: 1) host, vector, and
pathogen interactions (disease ecolo-
gy); 2) vertebrate behavior and ecolo-
gy; 3) vector behavior and ecology;
and 4) modeling and spatial statistics.
We describe some of the findings
from the meeting. For an in-depth
summary of this meeting, please visit
the conference website for meeting
abstracts and a downloadable confer-
ence white paper (available from:
URL: www.serc.si.edu/migratory-
birds/migratorybirds_index.htm).
West Nile virus (WNV) has spread
rapidly across North America since its
probable introduction to the New
York City area in 1999 (D.J. Gubler,
Centers for Disease Control and
Prevention, Fort Collins, CO). By
December 2002, the Canadian
provinces of Saskatchewan, Quebec,
Ontario, Nova Scotia, and Manitoba
reported dead birds that tested posi-
tive for WNV. By winter 2002, only
four states in the continental United
States remained free of confirmed
WNV infection; the virus was expect-
ed to reach the West Coast later in the
year. WNV has also found its way into
tropical regions. One case in a person
was reported in 2001 from the
Cayman Islands. Additionally, resi-
dent birds from Jamaica (January
2002) and the Dominican Republic
tions. These findings in turn raised
questions about factors involved in
human susceptibility, risks of pesti-
cide exposure, efficacy of mosquito
control, the value of sentinel animals
in surveillance, and the roles played
by various species in virus transmis-
sion and amplification.
The ability to diagnose WNV in
the laboratory emphasized the role of
pathology, including histopathology,
electron microscopy, immunohisto-
chemistry, polymerase chain reaction,
and virus isolation. Few of these tests
are entirely sensitive or specific. The
challenges involved in the pathologic
diagnosis in animals because of the
large diversity of infected species
were discussed. The single most
urgent concern, repeatedly empha-
sized by state public health officials
and clinicians, is the need for a faster,
simpler diagnostic test for WNV that
would ease the amount of work by
public health laboratories, assist
physicians in correctly diagnosing
infected patients, and improve surveil-
lance by identifying subclinical cases.
The problems of vector control
and the best application of these meth-
ods were also emphasized. The cost of
these programs and proof of effective-
ness will require careful research,
including the identification of specific
mosquito vectors and assessment of
the long-term safety of pesticides and
personal repellant applications. The
most important principle in attempt-
ing emergency vector control is to
consider it early, before the epidemic
has evolved.
Data from in vitro studies evaluat-
ed the interaction of viral vectors and
amplification hosts. These studies
might elucidate the importance of
birds, horses, and household pets in
maintenance of epidemics. Because
wild birds play a key role in the
spread of WNV, the exact nature of
that role must be clarified to predict
the development and expansion of
future epidemics.
The variation in changing epi-
demiology of the states’ experiences
to date with WNV, even within the
southeastern United States, clearly
demonstrates that research needs to be
replicated in numerous localities;
what succeeds in one state may not
prove successful in another. Whether
epidemics will continue to expand in
size and geographic distribution or
whether a more sporadic pattern of
occurrence will emerge is still
unclear. Controlling WNV in the
southeastern United States will take a
concerted, cohesive effort. The con-
tinued collaboration of the diverse
scientists in this meeting will aid in
this effort. Presentations from this
conference are available on the Web
site for Southeastern Center for
Emerging Biological Threats (avail-
able from: URL: www.secenterbio-
threats.org).
David Rimland,* Jeffrey Koplan,†
David S. Stephens*†‡
*Emory University School of Medicine,
Atlanta, Georgia, USA; †Southeastern
Center for Emerging Biologic Threats,
Atlanta, Georgia, USA; and ‡Centers for
Disease Control and Prevention, Atlanta,
Georgia, USA
Address for correspondence: David Stephens,
Southeastern Center for Emerging Biologic
Threats, 1440 Clifton Road NE, Suite 410,
Atlanta, GA 30322, USA; fax: 404-639-3059;
email: dstep01@emory.edu
Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 899
NEWS & NOTES
(November 2002) have shown WNV
antibodies. Recent reports note that
the virus has also reached Mexico’s
Yucatan peninsula. Since 1999, WNV
has killed thousands of birds and
other wildlife, and the impact on
regional wildlife populations is
unclear.
A primary theme of the meeting
was that we still have much to learn
about how WNV is dispersed, trans-
mitted, and amplified by competent
vectors and still relatively unknown
reservoir hosts. At the time of the con-
ference, WNV had been detected in
37 mosquito species, 157 bird species,
horses, 16 other mammals, and alliga-
tors (D.J. Gubler, Centers for Disease
Control and Prevention, Fort Collins,
CO). The Culex genus, particularly
Culex pipiens in the northern United
States and C. quinquefasciatus in the
southern United States, appears to be
the most important mosquito group
for the avian vector amplification
cycle. However, opportunistic mos-
quito species are probably important
bridge vectors to humans, horses, and
other deadend hosts. While the avian
amplification cycle appears to the
most dominant, other cycles may also
be occurring at the same time (i.e., in
mammal and ticks). Reptiles, amphib-
ians, and associated mosquito vectors
may also play important roles (M.J.
Turell, The United States Army
Medical Research Institute for
Infectious Diseases, Fort Detrick,
MD, and E. Jacobson, University of
Florida, Gainesville, FL). Although
mosquitoes appear to be the main vec-
tor, other ectoparasites such as ticks,
louse flies, and fleas should also be
examined as potential vectors.
Several scientists reported that
transmission of WNV is more compli-
cated than previously thought. The
presence of WNV in avian reproduc-
tive organs suggests that vertical
transmission may be a possibility
(T.S. McNamara, Wildlife Conser-
vation Society, Bronx, NY). WNV in
the kidneys leads to cloacal excretion,
which may lead to cloacal-oral mouth
infection. Bird-to-bird transmission
has been demonstrated in the labora-
tory and may be an important infec-
tion route among social birds like the
American crow (R.G. McLean, U.S.
Department of Agriculture, Fort
Collins, CO). Evidence suggests that
ingesting infected vertebrates and
mosquitoes can infect birds.
The impact of WNV on animal
populations is another unknown area.
Data from individually marked popu-
lations of crows in New York State
and Oklahoma (K.J. McGowan,
Cornell University, Ithaca, NY; A.
Clark, State University of New York-
Binghamton, Binghamton, NY; and
C.L. Caffrey, National Audubon
Society, Ivyland, PA) show that these
populations are experiencing impor-
tant declines after the initial WNV
outbreak. Analysis of breeding bird
surveys and annual winter bird cen-
suses (Christmas bird count) from a
wide array of passerine bird species
showed local declines in WNV
“hotspots” but no declines at the
range-wide scale that can be attrib-
uted to WNV (J. Sauer, United States
Geological Survey, Laurel, MD; P.P.
Marra, Smithsonian Environmental
Research Center, Edgewater, MD; and
W. Hochachka, Cornell University,
Ithaca, NY).
Another important issue discussed
at this conference was the secondary
impact that pest management might
have on organisms not pinpointed for
WNV, especially in aquatic environ-
ments. This issue is especially impor-
tant in nature reserves (W.K. Reisen,
University of California, Davis, CA).
Modelers attending the meeting
stressed the importance of standardiz-
ing sampling methods, such as the
dead bird surveillance programs oper-
ated across the nation by many state
health departments. These programs
must consistently and conscientiously
monitor sampling efforts and report
the total sample sizes of dead birds
collected, including the number of
birds that test negative (D.J. Rogers,
Oxford, UK). In addition, a better
understanding of the real-world per-
sistence of WNV antibodies in live
bird surveillance programs would be
useful for virus dispersal models.
Scientists at the meeting felt
strongly that we need to closely mon-
itor how WNV impacts organisms in
tropical regions, including humans
and the many endemic avian species
already threatened or endangered.
Species in Hawaii, many of which are
still endangered after malaria’s centu-
ry-old invasion, should be of special
concern. WNV is not the first and will
not be the last virus to enter our bor-
ders. By developing techniques to sur-
vey, monitor, and control WNV in
wildlife, we prepare ourselves for the
next pathogen species. Our experi-
ences with WNV emphasize the need
to strengthen and integrate animal
monitoring programs with basic
research on population and disease
ecology. A conference white paper,
several review articles, and a list of
research priorities are planned as
products of this meeting.
Peter P. Marra,* Sean M. Griffing,*
and Robert G. McLean†
*Smithsonian Environmental Research
Center, Edgewater, Maryland, USA; and
†National Wildlife Research Center, Fort
Collins, Colorado, USA
Address for correspondence: Peter P. Marra,
Smithsonian Environmental Research Center,
P.O. Box 28, Edgewater, MD 21037, USA; fax:
443-482-2380; email: [email protected]

900 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
NEWS & NOTES
Upcoming Infectious
Disease Conferences
July 7–10, 2003
ASM Conference on Bio-,
Micro- and Nanosystems
American Society for
Microbiology
New York, New York
Contact: ASM Conferences
Phone: 202-942-9261
Fax: 202-942-9340
Email: [email protected]
July 13–16, 2003
The 2nd IAS Conference on
HIV Pathogenesis and Treatment
Paris, France
Contact: JCD Conseil/IAS 2003
Phone: 33 1 40 64 2000
Fax: 33 1 40 64 2050
Website: http://www.ias2003.org
July 16–18, 2003
ISAAR 2003—4th International
Symposium on Antimicrobial
Agents and Resistance
Antimicrobial Treatment in the
21st Century: Current Challenges
and Future Strategies
Seoul, Korea
Contact: Ms. Susan Chung
Phone: 82-2-3410-0327
Fax: 82-2-3410-0023
Email: [email protected]
Website: http://www.ansorp.org/
isaar2003/intro.htm
July 16–19, 2003
11th Annual Conference:
Boards of Health: Stewards
of Public Health
National Association of Local
Boards of Health
Baltimore, MD
Website: http://www.nalboh.org/
confer/confmain.htm

Emerging Infectious Diseases • Vol. 9, No. 7, July 2003 901
As the Gods began one world, and man another,
So the snakecharmer begins a snaky sphere
With moon-eye, mouth pipe.
He pipes. Pipes green. Pipes water
Sylvia Plath, “Snakecharmer”
C
ritics called him “naïve,” the term for painters with no
formal training in art. Henri Rousseau, self-made late-
bloomer from Laval, France, fit the definition. But his
work proved that in art as in all ventures, training, though
valuable, is not the key ingredient—not as key perhaps as
talent, inspiration, or originality. Untrained in art but not
uneducated by the standards of his day, Rousseau was a
teacher and a military man. He was interested in politics
and the realm of ideas. He knew music and poetry and
even tried his hand as playwright. Dubbed “Le Douanier”
(customs officer) after his main occupation outside art, he
struggled in anonymity until near the end of his life, when
he was discovered by Pablo Picasso and others and was
recognized for his powerful individual style (1).
Like other naïve or primitive artists, Rousseau found art
late in life. He took up painting as a hobby and soon retired
from his job in the customs office to devote time to this
new vocation. He copied the masters, struggled to learn
their craft (particularly the academic style of Ingres), and
aspired to paint like them. He exhibited often at the Salon
des Indépendents in Paris, where artists could show their
paintings without selection restrictions, and what he might
have lost to technical clumsiness he seemed to make up in
ingenuous charm. Although his work is difficult to catego-
rize, he seems to have been influenced by his contempo-
rary Paul Gauguin and his followers, the Nabis, who pro-
moted directness of feeling and color harmony (2).
Rousseau eventually found his own formal language and
style, but what elevated his mature work to greatness were
perhaps the very oblivion of convention, the freshness of
approach, and the depth of discovery that comes from a
truly unique perspective.
Rousseau’s exotic compositions owe nothing to tradi-
tional art methods yet defy modern labels. The fantastic
vegetation in his jungle paintings (for which he is best
known) has no equivalent in nature. These exotic land-
scapes, oversized and filled with exuberant color, were
entirely imaginary. Although often inhabited by half-con-
cealed wild beasts and laced with conflict, they exuded an
eerie stillness. The images, smooth, vivid, and clearly
defined, were flat and fluid against dense but dimension-
less greenery, and although unreal and extraordinary, were
rendered in meticulous botanical detail.
The Snake Charmer, on this month’s cover of Emerging
Infectious Diseases, is one of Rousseau’s finest and most
celebrated works. Like his other jungle paintings, it is
filled with lush greenery. Punctuated by an uncoiling rep-
tile at arm’s length, the thick vegetal screen that makes up
most of the landscape is live with tension. The dark, undu-
lating figure of the snake charmer dances ambiguously
amidst a tangle of wildlife. Nature, framed by “a wave of
flickering-grass tongues,” (3) looms in the foreground
immediate and tangible, yet dreamlike and distant as the
moon. In a trance, the animals are guided (it seems as
much by the glossy stream as by the snake charmer’s reed)
into a tight ecological web, where unbeknownst to them,
they share more than the music.
Rousseau’s imagination, like that of many of his con-
temporaries in Paris, succumbed to the allure of exotic
lands, where plants grew larger than life, wild animals held
unknown powers and magnetism, and humans lounged in
“Eden’s navel” (3) amidst all that was lost in the fall from
grace. To explain the products of his inflamed imagination,
Rousseau falsely claimed that he had visited Mexico. But
unlike Gauguin, who went to Tahiti in search of inspira-
tion, Rousseau traveled only vicariously and found his
models in local gardens and the Paris zoo.
Exotic lands have become prosaic to us. What remains
naïve and primitive is our knowledge of the forest’s archi-
tecture and the perils of its convergence with human habi-
tat. But like the uncoiling snake in Rousseau’s painting,
out of the impenetrable jungle comes knowledge about
pestilences, piece by piece: a favorable environment, a sta-
ble population, a reservoir host, the agent. The emergence
of West Nile virus in North America is a case in point. We,
modern snake charmers, must pipe the pieces (bird, horse,
reptile) into a knowable, harmonious fabric.
Polyxeni Potter
1. Shattuck R, Behar H, Hoog M, Lanchner C, Bubin W. Henri
Rousseau. New York: New York Graphic Society; 1985.
2. Janson HW, Janson AF. History of art. New York: Harry N. Abrams
Inc.; 2001.
3. Plath S. Snakecharmer [cited 2003 May 14]. Available from: URL:
http://www.eng.fju.edu.tw/English_Literature/us_poetry/Plath/Plath_
poem.html
ABOUT THE COVER
Henri Rousseau—known as Le Douanier Rousseau (1844–1910) The Snake Charmer (1907). Oil on canvas, 169 cm x 189.5 cm
Musée d’Orsay, Paris, France. Credit: Réunion des Musées Nationaux/Art Resource, NY

902 Emerging Infectious Diseases • Vol. 9, No. 7, July 2003
NEWS & NOTES
A Peer-Reviewed Journal Tracking and Analyzing Disease Trends Vol.9, No.8, August, 2003
Upcoming Issue
For a complete list of articles included in the August issue,
and for articles published online ahead of print publication,
see http://www.cdc.gov/ncidod/eid/upcoming.htm
Look in the August issue for the following topics:
Porcine Reproductive and Respiratory Syndrome Virus: Origin Hypothesis
Detecting Bioterror Attack by Screening Blood Donors: Best-Case Analysis
Survival of Batrachochytrium dendrobatidis in
Water: Quarantine and Disease Control Implications
Legionnaires' Disease Outbreak in Murcia, Spain
Enzootic Transmission of Yellow Fever Virus in Peru
NmcA Carbapenem-hydrolyzing Enzyme in
Enterobacter cloacae in North America
Echovirus 13 Associated with Aseptic Meningitis, Spain
Molecular Characteristics of a Non–Babesia divergens
Organism Causing Zoonotic Babesiosis in Europe
Multidrug-Resistant Tuberculosis in HIV-Negative Patients,
Buenos Aires, Argentina Invasive Group A
Streptococcal Disease: Risk Factors for Adults
Severe Tungiasis in Underprivileged Communities: Case Series from Brazil


S
S
e
e
a
a
r
r
c
c
h
h
p
p
a
a
s
s
t
t
i
i
s
s
s
s
u
u
e
e
s
s
o
o
f
f
E
E
I
I
D
D
a
a
t
t
w
w
w
w
w
w
.
.
c
c
d
d
c
c
.
.
g
g
o
o
v
v
/
/
e
e
i
i
d
d

Instructions to Authors
Manuscript Preparation. For word processing, use MS Word. Begin
each of the following sections on a new page and in this order: title page,
keywords, abstract, text, acknowledgments, biographical sketch, refer-
ences, tables, figure legends, appendixes, and figures. Each figure should
be in a separate file.
Title Page. Give complete information about each author (i.e., full name,
graduate degree(s), affiliation, and the name of the institution in which
the work was done). Clearly identify the corresponding author and pro-
vide that author's mailing address (include phone number, fax number,
and e-mail address). Include separate word counts for abstract and text.
Keywords. Include up to 10 keywords; use terms listed in Medical
Subject Headings Index Medicus.
Text. Double-space everything, including the title page, abstract, refer-
ences, tables, and figure legends. Printed manuscript should be single-
sided, beginning with the title page. Indent paragraphs; leave no extra
space between paragraphs. After a period, leave only one space before
beginning the next sentence. Use 12-point Times New Roman font and
format with ragged right margins (left align). Italicize (rather than under-
line) scientific names when needed.
Biographical Sketch. Include a short biographical sketch of the first
author—both authors if only two. Include affiliations and the author's pri-
mary research interests.
References. Follow Uniform Requirements (www.icmje.org/index.html).
Do not use endnotes for references. Place reference numbers in parenthe-
ses, not superscripts. Number citations in order of appearance (including
in text, figures, and tables). Cite personal communications, unpublished
data, and manuscripts in preparation or submitted for publication in
parentheses in text. Consult List of Journals Indexed in Index Medicus for
accepted journal abbreviations; if a journal is not listed, spell out the jour-
nal title. List the first six authors followed by “et al.” Do not cite refer-
ences in the abstract.
Tables and Figures. Create tables within MS Word’s table tool. Do not
format tables as columns or tabs. Send graphics in native, high-resolution
(200 dpi minimum) .TIF (Tagged Image File), or .EPS (Encapsulated
Postscript) format. Graphics should be in a separate electronic file from
the text file. For graphic files, use Arial font. Convert Macintosh files into
the suggested PC format. Figures, symbols, letters, and numbers should
be large enough to remain legible when reduced. Place figure keys with-
in the figure. For more information see EID Style Guide (http://www.
cdc.gov/ncidod/EID/style_guide.htm).
Manuscript Submission. Include a cover letter indicating the proposed
category of the article (e.g., Research, Dispatch) and verifying that the
final manuscript has been seen and approved by all authors. To submit a
manuscript, access Manuscript Central from the Emerging Infectious
Diseases website (www.cdc.gov/eid).
Manuscript Types
Perspectives. Articles should be under 3,500 words and should include
references, not to exceed 40. Use of subheadings in the main body of the
text is recommended. Photographs and illustrations are encouraged.
Provide a short abstract (150 words) and a brief biographical sketch of
first author. Articles in this section should provide insightful analysis and
commentary about new and reemerging infectious diseases and related
issues. Perspectives may also address factors known to influence the
emergence of diseases, including microbial adaptation and change,
human demographics and behavior, technology and industry, economic
development and land use, international travel and commerce, and the
breakdown of public health measures. If detailed methods are included, a
separate section on experimental procedures should immediately follow
the body of the text.
Synopses. Articles should be under 3,500 words and should include ref-
erences, not to exceed 40. Use of subheadings in the main body of the text
is recommended. Photographs and illustrations are encouraged. Provide a
short abstract (150 words) and a brief biographical sketch of first
author—both authors if only two. This section comprises concise reviews
of infectious diseases or closely related topics. Preference is given to
reviews of new and emerging diseases; however, timely updates of other
diseases or topics are also welcome. If detailed methods are included, a
separate section on experimental procedures should immediately follow
the body of the text.
Research Studies. Articles should be under 3,500 words and should
include references, not to exceed 40. Use of subheadings in the main body
of the text is recommended. Photographs and illustrations are encour-
aged. Provide a short abstract (150 words) and a brief biographical sketch
of first author—both authors if only two. Report laboratory and epidemi-
ologic results within a public health perspective. Although these reports
may be written in the style of traditional research articles, they should
explain the value of the research in public health terms and place the find-
ings in a larger perspective (i.e., "Here is what we found, and here is what
the findings mean").
Policy and Historical Reviews. Articles should be under 3,500 words
and should include references, not to exceed 40. Use of subheadings in
the main body of the text is recommended. Photographs and illustrations
are encouraged. Provide a short abstract (150 words) and a brief biogra-
phical sketch. Articles in this section include public health policy or his-
torical reports that are based on research and analysis of emerging disease
issues.
Dispatches. Articles should be 1,000–1,500 words and need not be divid-
ed into sections. If subheadings are used, they should be general, e.g.,
“The Study” and “Conclusions.” Provide a brief abstract (50 words); ref-
erences (not to exceed 15); figures or illustrations (not to exceed two);
and a brief biographical sketch of first author—both authors if only two.
Dispatches are updates on infectious disease trends and research. The
articles include descriptions of new methods for detecting, characterizing,
or subtyping new or reemerging pathogens. Developments in antimicro-
bial drugs, vaccines, or infectious disease prevention or elimination pro-
grams are appropriate. Case reports are also welcome.
Commentaries. Thoughtful discussions (500–1,000 words) of current top-
ics. Commentaries may contain references but should not include figures
or tables.
Another Dimension. Thoughtful essays, short stories, or poems on philo-
sophical issues related to science, medical practice, and human health.
Topics may include science and the human condition, the unanticipated
side of epidemic investigations, or how people perceive and cope with
infection and illness. This section is intended to evoke compassion for
human suffering and to expand the science reader's literary scope.
Manuscripts are selected for publication as much for their content (the
experiences they describe) as for their literary merit.
Letters. This section includes letters that present preliminary data or
comment on published articles. Letters (500–1,000 words) should not be
divided into sections, nor should they contain figures or tables.
References (not more than 10) may be included.
Book Reviews. Short reviews (250–500 words) of recently published
books on emerging disease issues are welcome. The name of the book,
publisher, and number of pages should be included.
Announcements. We welcome brief announcements (50–150 words) of
timely events of interest to our readers. (Announcements may be posted
on the journal Web page only, depending on the event date.)
Conference Summaries. (500–1,000 words) of emerging infectious dis-
ease conferences may provide references to a full report of conference
activities and should focus on the meeting's content rather than on indi-
vidual conference participants.
Editorial Policy and Call for Articles
Emerging Infectious Diseases is a peer-reviewed journal established expressly to promote the recognition of new and reemerging infectious diseases
around the world and improve the understanding of factors involved in disease emergence, prevention, and elimination.
The journal has an international scope and is intended for professionals in infectious diseases and related sciences. We welcome contributions from
infectious disease specialists in academia, industry, clinical practice, and public health, as well as from specialists in economics, demography, sociol-
ogy, and other disciplines. Inquiries about the suitability of proposed articles may be directed to the Editor at 404-371-5329 (tel), 404-371-5449 (fax),
or [email protected] (e-mail).
Emerging Infectious Diseases is published in English and features the following types of articles: Perspectives, Synopses, Research Studies, Policy
and Historical Reviews, Dispatches, Commentaries, Another Dimension, Letters, Book Reviews, and News and Notes. The purpose and requirements
of each type of article are described in detail below. To expedite publication of information, we post journal articles on the Internet as soon as they are
cleared and edited.
Chinese, French, and Spanish translations of some articles can be accessed through the journal’s home page at http://www.cdc.gov/eid.
