
D
engue virus (DENV) is not endemic in the conti-
nental United States (1); most cases occur among
travelers to DENV-endemic areas (2). In Florida, USA,
DENV infections are primarily reported among trav-
elers (https://ndc.services.cdc.gov/case-denitions/
dengue-virus-infections-2015); however, locally ac-
quired cases and limited outbreaks have been report-
ed in Monroe County in 2009–2010 (n = 88), Martin
County in 2013 (n = 24), and Monroe County in 2020
(n = 72) (3–5). During 2009–2021, an annual median of
83 (range 19–413) travel-associated DENV infections
and 7 (range 0–77) locally acquired cases were report-
ed in Florida; all DENV types (DENV-1−4) occurred
among both travel-associated and locally acquired
cases (6). Previous work demonstrated the DENV
vectors Aedes aegypti and A. albopictus mosquitoes are
present across Florida (7).
In early 2022, the Florida Department of Health
(FDOH) identied an increase in travel-associated
DENV infections, primarily among travelers return-
ing from Cuba. In July 2022, a DENV-3 outbreak was
reported in Cuba (8); DENV-3 case increases were
also documented in other countries in the Americas
(9,10). On July 18, Miami-Dade County health of-
cials issued a mosquito-borne illness advisory after
the rst locally acquired DENV infection in 2022 was
conrmed in a Florida resident (11). We document
the DENV-3 outbreak in Florida by describing the
epidemiologic features of reported cases, analyzing
DENV-3 genomic sequences, and reconstructing pos-
sible transmission trees.
The Study
FDOH routinely conducts active case-nding activi-
ties for DENV and conducts IgM and reverse tran-
scription PCR testing for conrmation and DENV
serotype identication. Suspected case-patients are
interviewed to identify risk factors, possible mos-
quito exposure locations, and additional suspected
cases (3). Ethics approval was not required because
this work was part of standard public health outbreak
surveillance and response.
During May 1, 2022–April 30, 2023 (52 weeks),
1,037 DENV infections were reported, 966 (93%)
were travel-associated and 71 (7%) locally acquired.
DENV-3 was the most frequently identied serotype
Introduction and Spread of
Dengue Virus 3, Florida, USA,
May 2022–April 2023
Forrest K. Jones,
1
Andrea M. Morrison,
1
Gilberto A. Santiago, Kristyna Rysava, Rebecca A. Zimler,
Lea A. Heberlein, Edgar Kopp, Florida Department of Health Bureau of Public Health Laboratory Team,
2
Katharine E. Saunders, Samantha Baudin, Edhelene Rico, Álvaro Mejía-Echeverri, Emma Taylor-Salmon,
Verity Hill, Mallery I. Breban, Chantal B.F. Vogels, Nathan D. Grubaugh, Lauren M. Paul, Scott F. Michael,
Michael A. Johansson, Laura E. Adams, Jorge Munoz-Jordan, Gabriela Paz-Bailey, Danielle R. Stanek
376 Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 30, No. 2, February 2024
DISPATCHES
Author aliations: Centers for Disease Control and Prevention,
San Juan, Puerto Rico, USA (F.K. Jones, G.A. Santiago,
M.A. Johansson, L.E. Adams, J. Munoz-Jordan, G. Paz-Bailey,
K. Rysava); Centers for Disease Control and Prevention, Atlanta,
Georgia, USA (F.K. Jones, K.E. Saunders); Florida Department
of Health, Tallahassee, Florida, USA (A.M. Morrison, R.A. Zimler,
L.A. Heberlein, E. Kopp,
K.E. Saunders, S. Baudin, E. Rico,
Á. Mejía-Echeverri, D.R. Stanek); Yale School of Medicine,
New Haven, Connecticut, USA (E. Taylor-Salmon); Yale School of
Public Health, New Haven (E. Taylor-Salmon, V. Hill, M.I. Breban,
C.B.F. Vogels, N.D. Grubaugh); Florida Gulf Coast University,
Fort Myers, Florida, USA (L.M. Paul, S.F. Michael)
DOI: https://doi.org/10.3201/eid3002.231615
1
These authors shared rst authorship.
2
Team members are listed at the end of this article.
During May 2022–April 2023, dengue virus serotype 3
was identied among 601 travel-associated and 61 lo-
cally acquired dengue cases in Florida, USA. All 203
sequenced genomes belonged to the same genotype
III lineage and revealed potential transmission chains in
which most locally acquired cases occurred shortly after
introduction, with little sustained transmission.
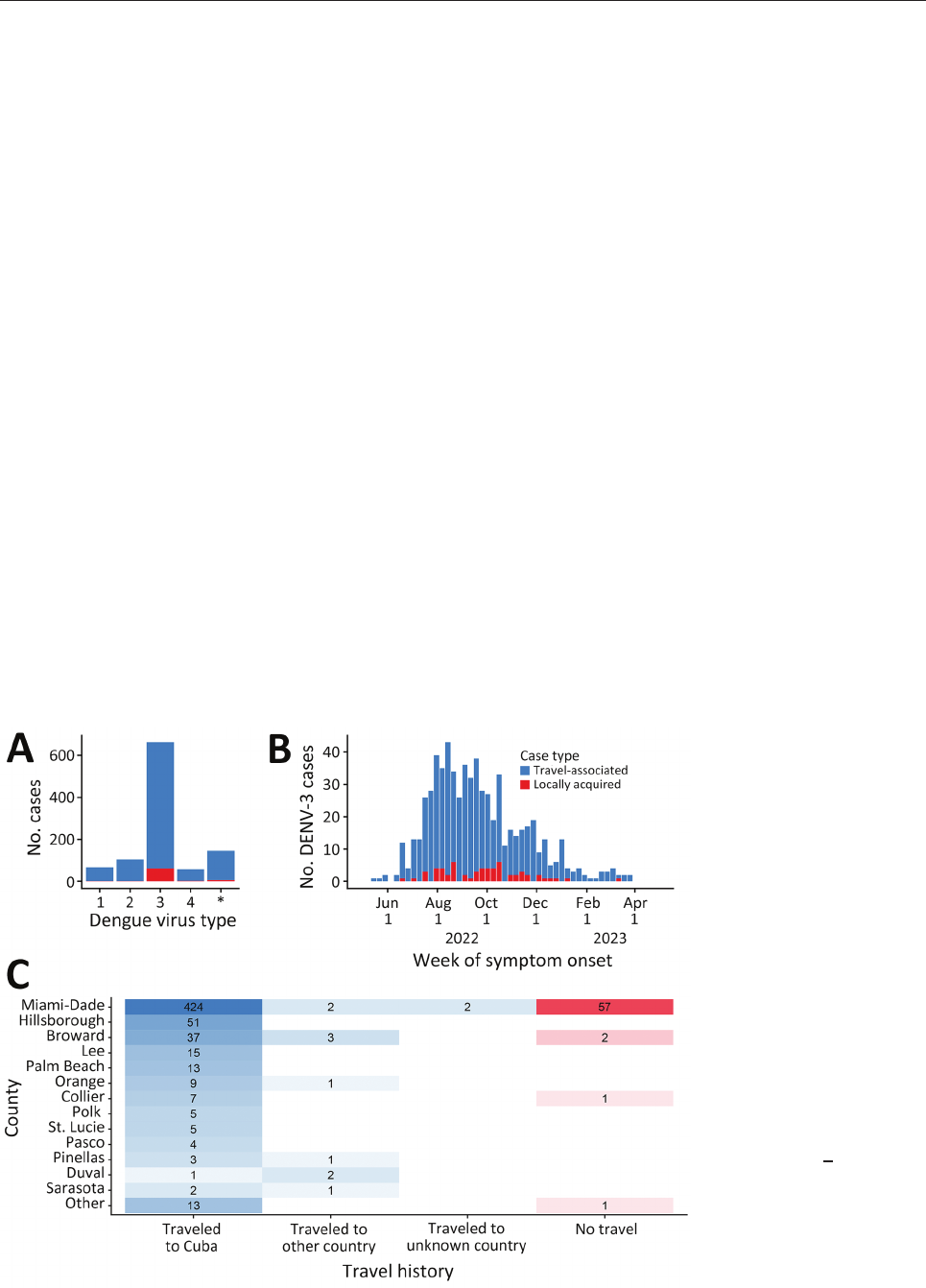
Dengue Virus 3, Florida, USA, May 2022–April 2023
(64%, n = 662), followed by DENV-2 (10%, n = 104),
DENV-1 (7%, n = 68), and DENV-4 (5%, n = 57); in 146
(14%) cases, multiple serotypes or no serotype was
identied (Figure 1, panel A). Among DENV-3 cases,
601 (91%) were travel-associated and 61 (9%) were lo-
cally acquired cases (Figure 1, panel B). Most DENV-3
case-patients identied as White (n = 609; 92%) and
Hispanic or Latino (n = 642, 97%).
Among 601 travel-associated DENV-3 cases,
the median age was 52 (interquartile range 41–61)
years; 51% of patients were male and 49% female.
Most (98%, n = 589) case-patients with travel-associ-
ated DENV-3 had recently traveled from Cuba; they
were reported in 21/67 Florida counties (Figure 1,
panel C). Miami-Dade County had the most travel-
associated DENV-3 cases (71%, n = 428). Among 61
locally acquired DENV-3 cases, the median age was
54 (interquartile range 36–58) years; 67% of patients
were male and 33% female, and nearly all (93%, n =
57) were reported in Miami-Dade County. The 485
DENV-3 case-patients in Miami-Dade County were
identied in 60/82 postal (ZIP) codes.
We performed genomic characterization of
DENV-3 by sequencing the complete genomes of
203 cases at the Centers for Disease Control and Pre-
vention (San Juan, Puerto Rico, USA), Yale School
of Public Health (New Haven, CT, USA), and
FDOH (Appendix 1, https://wwwnc.cdc.gov/EID/
article/30/2/23-1615-App1.pdf) (12). Sequencing
was prioritized and successful for 34 locally acquired
cases, as well as case-patients with recent travel his-
tory to Cuba (n = 168) or Guyana (n = 1). To assess the
representativeness of DENV sequences, we evaluated
symptom onset dates and counties of residence for
cases selected for sequencing and all cases detected
(Appendix 1 Figure 1). We conducted maximum-
likelihood phylogenetic analysis to infer the genetic
relatedness of DENV-3 to contemporary circulation
globally. Global context was provided with a subsam-
ple of 146 publicly available genomes that represent
relevant genotypes.
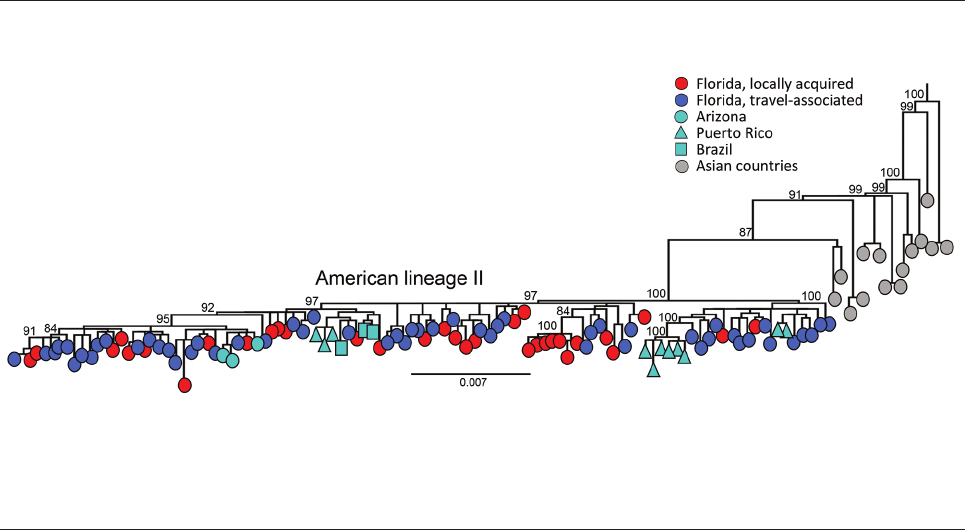
The DENV-3 genomes identied in Florida are
classied as genotype III and cluster within the novel
American II lineage (9). We observed a close relation-
ship with DENV-3 genomes recently identied in
Arizona, Puerto Rico, and Brazil, indicating that the
lineage is spreading across the Americas (Figure 2).
However, the limited sampling of the new American
II lineage prevented us from inferring a potential time
of emergence in Florida. The short branch lengths and
similarity between locally acquired and travel-associ-
ated cases in the phylogenetic tree demonstrate low
genomic diversity during the sampling period, where
genomes from locally acquired cases cluster random-
ly with travel-associated cases. The tree topology sug-
gests frequent importation events occurred during
Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 30, No. 2, February 2024 377
Figure 1. DENV serotype
distribution and DENV-3 case
distribution by week of symptom
onset, county of reporting, and
origin of travel, Florida, USA,
May 1, 2022–April 30, 2023.
A) Number of dengue cases
by each virus serotype. Cases
with an unknown dengue virus
type (asterisk) only had a
positive serologic test or multiple
serotypes identied. B) Epidemic
curve of reported cases of
DENV-3, showing 601 travel-
associated cases and 61 locally
acquired cases. C) Heat map
indicating number of DENV-3
cases by county and by travel
history. Other countries were
Bangladesh, Colombia, Guyana,
India, Jamaica, Mexico, Pakistan,
and Sri Lanka. The names of
counties reporting >3 DENV-3
cases are shown and sorted
by the total number of cases
reported. DENV, dengue virus;
DENV-3, DENV serotype 3.
DISPATCHES
the sampling period and indicate frequent movement
of DENV between Cuba and Florida without estab-
lishing sustained local transmission in Florida.
To model a possible transmission tree, we adapted
a graph-based model using genomic sequences and
symptom onset dates from 31 locally acquired and
144 travel-associated cases (Appendix 1) (13,14). To ac-
count for infections in transmission chains that went
undetected between reported cases, we included a sur-
veillance reporting probability (i.e., the probability an
infection was detected as a case) and performed sensi-
tivity analyses assuming different reporting probabili-
ties of 1%, 5%, 10%, and 15%. Assuming a 5% reporting
probability, we identied 22 travel-associated cases
(15%) with most compatible linkages leading to the 31
locally acquired cases (Appendix 1 Figure 2). Overall,
122 (85%) travel-associated cases had no likely link-
age to locally acquired cases, 17 (11%) were linked to 1
case, 2 (1%) were linked to 2 cases, 2 (1%) were linked
to 3 cases, and 1 (1%) was linked to 4 cases.
Conclusions
We documented an unprecedented number of travel-
associated and locally acquired DENV-3 cases in
Florida during May 2022–April 2023; circulation of
the DENV-3 genotype III was recently identied in
the Americas. Our investigation illustrates that local
transmission and spread in Florida was limited, de-
spite multiple introductions from outside the coun-
try. Sequencing and phylogenetic analysis revealed
that cases were from the same DENV-3 genotype III
lineage and were highly related to one another and to
cases identied in Puerto Rico, Arizona, and Brazil.
Assessment of possible linkages between sequenced
cases indicated that local transmission during this
outbreak was limited; most travel-associated cases
did not lead to further transmission.
DENV activity in Cuba and Florida are linked
given their proximity and the extensive travel be-
tween them. Our results are similar to ndings in
Florida in 2019 (5), where many DENV case-patients
reported recent travel to Cuba, leading to an in-
crease in locally acquired cases. An elevated number
of locally acquired DENV cases in Florida might be
expected after a high number of introductions, but
our analysis suggests that DENV introductions did
not result in sustained local transmission beyond
small-scale outbreaks. Factors potentially reducing
transmission include living conditions (e.g., use of
air conditioning and screens), rapid case notica-
tion that enabled vector interventions (e.g., spray-
ing insecticide, conducting surveillance, community
education, and removing standing water), or limited
availability of mosquito breeding sites (15).
The relatively low genetic diversity in this da-
taset limited our ability to estimate the timing of ini-
tial DENV-3 introductions and fully reconstruct local
spread. We did not use case locations to determine the
compatibility of transmission links. DENV case detec-
tion continued through 2023 in Florida; efforts to un-
derstand those transmission dynamics are ongoing.
In summary, we used epidemiologic surveil-
lance and genomic sequencing to identify a newly
emerging lineage of DENV-3 genotype III that
caused an unusually large number of travel-as-
sociated and locally acquired DENV infections in
378 Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 30, No. 2, February 2024
Figure 2. Evolutionary analysis of dengue virus serotype 3 sampled in Florida, USA, May 1, 2022–April 30, 2023. Maximum-likelihood
phylogenetic tree was generated from a subset of 203 complete genomes from Florida (34 local cases, 168 cases in persons with recent
travel history to Cuba, and 1 traveler case from Guyana) and 146 complete genomes publicly available (1985–2022) from GenBank
representing genotype III, American lineage II. A subset of the sequences was used because of the low diversity in the population
sample, which was limiting the phylogenetic signal and hampering the statistical analyses that supported the tree accuracy and certainty
in major nodes. Sampling locations are coded by shape and color. Scale bar represents nucleotide substitutions per site.

Dengue Virus 3, Florida, USA, May 2022–April 2023
Florida, particularly in Miami-Dade County. Our
analysis suggests that locally acquired cases were
driven by large numbers of case-patients with recent
travel to Cuba and that DENV persistence in Florida
was limited. Close monitoring of DENV activity in-
ternationally, as well as increasing healthcare pro-
vider awareness about DENV identication and test-
ing, can strengthen preparedness and response to
future introductions in non–DENV-endemic areas.
Florida Department of Health Bureau of Public Health
Laboratory Team: Sylvia Bunch, Natalia Cano, Amanda
Davis, Yibo Dong, Rayah Jaber, Timothy Locksmith,
Charles Panzera, Brittany Rowlette, Sarah Schmedes,
Julieta Vergara.
Acknowledgments
We thank Joshua Wong for help with initial discussions
on analysis plans. We also gratefully acknowledge the
Arizona Department of Health Services and the Maricopa
County Department of Public Health for contributing
specimens for the phylogenetic analysis.
Code presented in this study is available at
https://github.com/fjones2222/denv-3-orida-2022/.
Research reported in this publication was supported by
the National Institute of Allergy and Infectious Diseases
of the National Institutes of Health under award number
DP2AI176740 (NDG), and by CTSA Grant Number UL1
TR001863 from the National Center for Advancing
Translational Science (NCATS), a component of the
National Institutes of Health (CBFV), and by the
National Institute of General Medical Sciences of the
National Institutes of Health under award number
R21GM142011 (SFM).
About the Author
Dr. Jones is an Epidemic Intelligence Service ofcer
stationed at the Centers for Disease Control and
Prevention Dengue Branch (Division of Vector-Borne
Diseases, National Center for Emerging and Zoonotic
Infectious Diseases) in San Juan, Puerto Rico.
His research interest is surveillance and modeling
of infectious diseases.
References
1. Centers for Disease Control and Prevention. Dengue [cited
2023 Aug 19]. https://www.cdc.gov/dengue/index.html
2. Wong JM, Rivera A, Volkman HR, Torres-Velasquez B,
Rodriguez DM, Paz-Bailey G, et al. Travel-associated dengue
cases—United States, 2010–2021. MMWR Morb Mortal
Wkly Rep. 2023;72:821–6. https://doi.org/10.15585/
mmwr.mm7230a3
3. Rowe D, McDermott C, Veliz Y, Kerr A, Whiteside M,
Coss M, et al.; Florida Department of Health Dengue
Investigation Team. Dengue outbreak response during
COVID-19 pandemic, Key Largo, Florida, USA, 2020.
Emerg Infect Dis. 2023;29:1643–7. https://doi.org/10.3201/
eid2908.221856
4. Graham AS, Pruszynski CA, Hribar LJ, DeMay DJ,
Tambasco AN, Hartley AE, et al. Mosquito-associated
dengue virus, Key West, Florida, USA, 2010. Emerg Infect
Dis. 2011;17:2074–5. https://doi.org/10.3201/eid1711.110419
5. Sharp TM, Morris S, Morrison A, de Lima Corvino D,
Santiago GA, Shieh WJ, et al.; 2019 Florida Dengue
Investigation Team. Fatal dengue acquired in Florida.
N Engl J Med. 2021;384:2257–9. https://doi.org/10.1056/
NEJMc2023298
6. Centers for Disease Control and Prevention. Historic data
(2010–2022) [cited 2023 Aug 19]. https://www.cdc.gov/
dengue/statistics-maps/historic-data.html
7. Parker C, Ramirez D, Connelly CR. State-wide survey of
Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in
Florida. J Vector Ecol. 2019;44:210–5. https://doi.org/
10.1111/jvec.12351
8. U.S. Embassy in Cuba. Health alert for U.S. citizens in Cuba
on dengue fever [cited 2023 Aug 19]. https://cu.usembassy.
gov/health-alert-for-u-s-citizens-in-cuba-on-dengue-fever
9. Naveca FG, Santiago GA, Maito RM, Ribeiro Meneses CA,
do Nascimento VA, de Souza VC, et al. Reemergence of
dengue virus serotype 3, Brazil, 2023. Emerg Infect Dis.
2023;29:1482–4. https://doi.org/10.3201/eid2907.230595
10. Kretschmer M, Collins J, Dale AP, Garrett B, Koski L,
Zabel K, et al. Notes from the eld: rst evidence of
locally acquired dengue virus infection—Maricopa County,
Arizona, November 2022. MMWR Morb Mortal Wkly Rep.
2023;72:290–1. https://doi.org/10.15585/mmwr.mm7211a5
11. Florida Health, Miami-Dade County. Health ofcials issue
mosquito borne illness advisory following conrmation of
one dengue case [cited 2023 Aug 19]. https://miamidade.
oridahealth.gov/newsroom/2022/07/2022-07-18-
mosquito-borne-illness-advisory.html
12. Vogels C. DengueSeq: A pan-serotype whole genome
amplicon sequencing protocol for dengue virus v1
[cited 2023 Sep 27]. https://www.protocols.io/view/
dengueseq-a-pan-serotype-whole-genome-amplicon
-seq-kqdg39xxeg25/v2
13. Cori A, Nouvellet P, Garske T, Bourhy H, Nakouné E,
Jombart T. A graph-based evidence synthesis approach to
detecting outbreak clusters: an application to dog rabies.
PLOS Comput Biol. 2018;14:e1006554. https://doi.org/
10.1371/journal.pcbi.1006554
14. Hampson K, Dushoff J, Cleaveland S, Haydon DT, Kaare M,
Packer C, et al. Transmission dynamics and prospects for the
elimination of canine rabies. PLoS Biol. 2009;7:e53.
https://doi.org/10.1371/journal.pbio.1000053
15. Reiter P, Lathrop S, Bunning M, Biggerstaff B, Singer D,
Tiwari T, et al. Texas lifestyle limits transmission of dengue
virus. Emerg Infect Dis. 2003;9:86–9. https://doi.org/
10.3201/eid0901.020220
Address for correspondence: Forrest Kirby Jones, Centers for
Disease Control and Prevention, 1324 Calle Cañada, San Juan,
00920, Puerto Rico, USA; email: [email protected]
Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 30, No. 2, February 2024 379
