
SMRT
®
Tools
reference guide
(v11.0)
Research use only. Not for use in diagnostic procedures.
P/N 102-278-500 Version 01 (April 2022)
© 2022, PacBio. All rights reserved.
Information in this document is subject to change without notice. PacBio assumes no responsibility for any errors or
omissions in this document.
Certain notices, terms, conditions and/or use restrictions may pertain to your use of PacBio products and/or third
party products. Refer to the applicable PacBio terms and conditions of sale and to the applicable license terms at
http://www.pacificbiosciences.com/licenses.html.
Trademarks:
Pacific Biosciences, the PacBio logo, PacBio, Circulomics, Omnione, SMRT, SMRTbell, Iso-Seq, Sequel, Nanobind,
and SBB are trademarks of Pacific Biosciences of California Inc. (PacBio). All other trademarks are the sole property
of their respective owners.
See https://github.com/broadinstitute/cromwell/blob/develop/LICENSE.txt for Cromwell redistribution information.
PacBio
1305 O’Brien Drive
Menlo Park, CA 94025
www.pacb.com

Page 1
Introduction
This document describes the command-line tools included with SMRT
®
Link v11.0. These tools are for use by bioinformaticians working with
secondary analysis results.
• The command-line tools are located in the
$SMRT_ROOT/smrtlink/
smrtcmds/bin
subdirectory.
Installation
The command-line tools are installed as an integral component of the
SMRT Link software. For installation details, see SMRT Link software
installation guide (v11.0).
• Note: SMRT Link v11.0 is for use with Sequel
®
II systems and Sequel IIe
systems only.
• To install only the command-line tools, use the
--smrttools-only
option with the installation command, whether for a new installation or
an upgrade. Examples:
smrtlink-*.run --rootdir smrtlink --smrttools-only
smrtlink-*.run --rootdir smrtlink --smrttools-only --upgrade
Supported chemistry
SMRT Link v11.0 supports all chemistry versions for Sequel II systems.
PacBio command-line tools
Following is information on the PacBio-supplied command-line tools
included in the installation. For information on third-party tools installed, see
“Appendix B - Third party command-line tools” on page 113.
Tool Description
bam2fasta/
bam2fastq
Converts PacBio
®
BAM files into gzipped FASTA and FASTQ files.
See “bam2fasta/bam2fastq” on page 3.
bamsieve Generates a subset of a BAM or PacBio Data Set file based on either a list of hole
numbers, or a percentage of reads to be randomly selected.
See “bamsieve” on page 4.
ccs Calculates consensus sequences from multiple “passes” around a circularized
single DNA molecule (SMRTbell
®
template). See “ccs” on page 7.
dataset Creates, opens, manipulates and writes Data Set XML files.
See “dataset” on page 16.
Demultiplex
Barcodes
Identifies barcode sequences in PacBio single-molecule sequencing data.
See “Demultiplex Barcodes” on page 23.
SMRT
®
Tools reference guide (v11.0)
Page 2
export-datasets Takes one or more PacBio dataset XML files and packages all contents into a
single ZIP archive. See “export-datasets” on page 35.
export-job Takes one SMRT Link Analysis job and packages all contents into a single ZIP
archive. See “export-job” on page 36.
gcpp Variant-calling tool which provides several variant-calling algorithms for PacBio
sequencing data. See “gcpp” on page 38.
Genome Assembly Generates de novo assemblies using HiFi reads.
See “Genome Assembly” on page 41.
HiFiViral SARS-
CoV-2 Analysis
Analyzes multiplexed viral surveillance samples for SARS-CoV-2.
See “HiFiViral SARS-CoV-2 Analysis” on page 49.
ipdSummary Detects DNA base-modifications from kinetic signatures.
See “ipdSummary” on page 52.
isoseq3 Characterizes full-length transcripts and generates full-length transcript isoforms,
eliminating the need for computational reconstruction. See “isoseq3” on page 56.
juliet A general-purpose minor variant caller that identifies and phases minor single
nucleotide substitution variants in complex populations. See “juliet” on page 60.
Microbial
Genome Analysis
Generates de novo assemblies of small prokaryotic genomes between 1.9-10 Mb
and companion plasmids between 2 – 220 kb and performs base modification
detection, using HiFi reads. See “Microbial Genome Analysis” on page 68.
motifMaker Identifies motifs associated with DNA modifications in prokaryotic genomes.
See “motifMaker” on page 73.
pbcromwell PacBio’s wrapper for the cromwell scientific workflow engine used to power
SMRT Link. For details on how to use pbcromwell to run workflows, see
“pbcromwell” on page 75.
pbindex Creates an index file that enables random access to PacBio-specific data in BAM
files. See “pbindex” on page 80.
pbmarkdup Marks or removes duplicates reads from CCS reads.
See “pbmarkdup” on page 81.
pbmm2 Aligns PacBio reads to reference sequences; a SMRT wrapper for minimap2.
See “pbmm2” on page 84.
pbservice Performs a variety of useful tasks within SMRT Link. See “pbservice” on page 91.
pbsv Structural variant caller for PacBio reads. See “pbsv” on page 96.
pbvalidate Validates that files produced by PacBio software are compliant with PacBio’s own
internal specifications. See “pbvalidate” on page 100.
primrose Performs motif-calling to detect
5mC CpG sites in HiFi BAM files.
See
“primrose” on page 103 for details.
runqc-reports Generates Run QC reports. See “runqc-reports” on page 104.
summarizeModifi
cations
Generates a GFF summary file from the output of base modification analysis
combined with the coverage summary GFF generated by resequencing pipelines.
See “summarize Modifications” on page 105.
Tool Description
Page 3
bam2fasta/
bam2fastq
The bam2fasta and bam2fastq tools convert PacBio BAM or Data Set files
into gzipped FASTA and FASTQ files, including demultiplexing of barcoded
data.
Usage
Both tools have an identical interface and take BAM and/or Data Set files
as input.
bam2fasta [options] <input>
bam2fastq [options] <input>
Examples
bam2fasta -o projectName m54008_160330_053509.subreads.bam
bam2fastq -o myEcoliRuns m54008_160330_053509.subreads.bam
m54008_160331_235636.subreads.bam
bam2fasta -o myHumanGenomem54012_160401_000001.subreadset.xml
Input files
•One or more *.bam files
•
*.subreadset.xml file (Data Set file)
Output files
• *.fasta.gz
• *.fastq.gz
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
-o,--output Specifies the prefix of the output file names. - implies streaming.
Note: Streaming is not supported with compression or with the split_barcodes
option.
-c Specifies the Gzip compression level. Values are [1,2,3,4,5,6,7,8,9].
(Default = 1)
-u Specifies that the output FASTA/FASTQ files are not compressed. .gz is not
added to the output file names, and -c settings are ignored.
--split-barcodes Splits the output into multiple FASTA/FASTQ files, by barcode pairs.
Note: The bam2fasta/bam2fastq tools inspect the bc tag in the BAM file to
determine the 0-based barcode indices from their respective positions in the
barcode FASTA file.
-p,--seqid-prefix Specifies the prefix for the sequence IDs used in the FAST/FASTQ file headers.

Page 4
bamsieve
The bamsieve tool creates a subset of a BAM or PacBio Data Set file
based on either a list of hole numbers to include or exclude, or a
percentage of reads to be randomly selected, while keeping all subreads
within a read together. Although
bamsieve is BAM-centric, it has some
support for dataset XML and will propagate metadata, as well as scraps
BAM files in the special case of SubreadSets.
bamsieve is useful for
generating minimal test Data Sets containing a handful of reads.
bamsieve operates in two modes: list mode where the ZMWs to keep or
discard are explicitly specified, or percentage/count mode, where a
fraction of the ZMWs is randomly selected.
ZMWs may be listed in one of several ways:
• As a comma-separated list on the command line.
• As a flat text file, one ZMW per line.
• As another PacBio BAM or Data Set of any type.
Usage
bamsieve [-h] [--version] [--log-file LOG_FILE]
[--log-level {DEBUG,INFO,WARNING,ERROR,CRITICAL} | --debug | --quiet
| -v]
[--show-zmws][--include INCLUDE LIST] [--exclude EXCLUDE LIST]
[--percentage PERCENTAGE] [-n COUNT] [-s SEED]
[--ignore-metadata][--barcodes]
input_bam [output_bam]
Required Description
input_bam The name of the input BAM file or Data Set from which reads will be read.
output_bam The name of the output BAM file or Data Set where filtered reads will be written to.
(Default = None)
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file LOG_FILE Writes the log to file. (Default = None, writes to stdout.)
--log-level Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR, CRITICAL].
(Default = WARNING)
--debug Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to CRITICAL to suppress output. (Default = False)
-v, --verbose Sets the verbosity level. (Default = NONE)
--show-zmws Prints a list of ZMWs and exits. (Default = False)
--include INCLUDE LIST Specifies the ZMWs to include in the output. This can be a comma-separated list of
ZMWs, or a file containing a list of ZMWs (one hole number per line), or a BAM/
Data Set file. (Default = NONE)

Page 5
Examples
Pulling out two ZMWs from a BAM file:
bamsieve --include 111111,222222 full.subreads.bam sample.subreads.bam
Pulling out two ZMWs from a Data Set file:
bamsieve --include 111111,222222 full.subreadset.xml sample.subreadset.xml
Using a text list:
bamsieve --include zmws.txt full.subreads.bam sample.subreads.bam
Using another BAM or Data Set as a list:
bamsieve --include mapped.alignmentset.xml full.subreads.bam mappable.subreads.bam
Generating a list of ZMWs from a Data Set:
bamsieve --show-zmws mapped.alignmentset.xml > mapped_zmws.txt
Anonymizing a Data Set:
bamsieve --include zmws.txt --ignore-metadata --anonymize full.subreadset.xml
anonymous_sample.subreadset.xml
Removing a read:
bamsieve --exclude 111111 full.subreadset.xml filtered.subreadset.xml
Selecting 0.1% of reads:
bamsieve --percentage 0.1 full.subreads.bam random_sample.subreads.bam
Selecting a different 0.1% of reads:
bamsieve --percentage 0.1 --seed 98765 full.subreads.bam random_sample.subreads.bam
--exclude EXCLUDE LIST Specifies the ZMWs to exclude from the output. This can be a comma-separated
list of ZMWs, or a file containing a list of ZMWs (one hole number per line), or a
BAM/Data Set file that specifies ZMWs. (Default = NONE)
--percentage PERCENTAGE Specifies a percentage of a SMRT
®
Cell to recover (Range = 1-100) rather than a
specific list of reads. (Default = NONE)
-n COUNT, --count COUNT Specifies a specific number of ZMWs picked at random to recover.
(Default = NONE)
-s SEED, --seed SEED Specifies a random seed for selecting a percentage of reads. (Default = NONE)
--ignore-metadata Discard the input Data Set metadata. (Default = False)
--barcodes Specifies that the include/exclude list contains barcode indices instead of ZMW
numbers. (Default = False)
Options Description

Page 6
Selecting just two ZMWs/reads at random:
bamsieve --count 2 full.subreads.bam two_reads.subreads.bam
Selecting by barcode:
bamsieve --barcodes --include 4,7 full.subreads.bam two_barcodes.subreads.bam
Generating a tiny BAM file that contains only mappable reads:
bamsieve --include mapped.subreads.bam full.subreads.bam mappable.subreads.bam
bamsieve --count 4 mappable.subreads.bam tiny.subreads.bam
Splitting a Data Set into two halves:
bamsieve --percentage 50 full.subreadset.xml split.1of2.subreadset.xml
bamsieve --exclude split.1of2.subreadset.xml full.subreadset.xml
split.2of2.subreadset.xml
Extracting Unmapped Reads:
bamsieve --exclude mapped.alignmentset.xml movie.subreadset.xml unmapped.subreadset.xml
Page 7
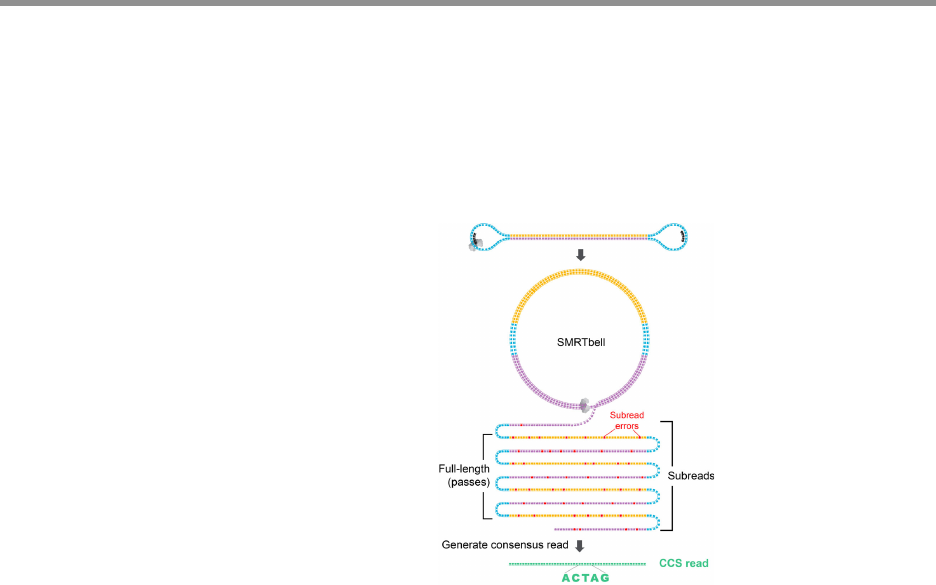
ccs
Circular Consensus Sequencing (CCS) Analysis computes consensus
sequences from multiple “passes” around a circularized single DNA
molecule (SMRTbell
®
template). CCS analysis uses the Arrow framework
to achieve optimal consensus results given the number of passes
available.
CCS analysis workflow
1. Initial filtering
– Filter ZMWs: Remove ZMWs with signal-to-noise ratio (SNR) below
--min-snr.
– Filter subreads: Remove subreads with lengths <50% or >200% of
the median subread length. Stop if the number of full-length
subreads is fewer than
--min-passes.
2. Generate draft
– The polish stage iteratively improves upon a candidate template
sequence. Because polishing is very compute-intensive, it is
desirable to start with a template that is as close as possible to the
true sequence of the molecule to reduce the number of iterations
until convergence. The
ccs software does not pick a full-length
subread as the initial template to be polished, but instead generates
an approximate draft consensus sequence using our improved
implementation of the
Sparc graph consensus algorithm. This
algorithm depends on a subread-to-backbone alignment that is
generated by the
pancake mapper developed by PacBio, using
edlib as the core aligner. Typically, subreads have accuracy of
around 90% and the draft consensus has a higher accuracy, but is
still below 99%.
– Stop if the draft length is shorter than
--min-length and longer
than
--max-length.
3. Alignment
– Align subreads to the draft consensus using
pancake with KSW2 for
downstream windowing and filtering.

Page 8
4. Windowing
– Divide the subread-to-draft alignment into overlapping windows with
a target size of 22 bp with ±2 bp overlap. Avoid breaking windows at
simple repeats (homopolymers to 4-mer repeats) to reduce edge
cases at window borders. Windowing reduces the algorithm run
time from quadratic to linear in the insert size.
5. Single-strand artifacts
– Identify heteroduplexes, where one strand of the SMRTbell differs
significantly from the reverse complement of the other strand.
Subread orientation is inferred from the alignment. Small
heteroduplexes, such as single base
A paired with a matching G, are
retained and the ambiguity is reflected in base quality. Molecules
with a single difference longer than 20 bp between the strands are
removed and recorded as heteroduplexes in the
<outputPrefix>.ccs_report.txt file.
6. Trim large insertions
– Insertions in the subreads relative to the draft that are longer than
--max-insertion-size, default 30 bp, are trimmed since they
typically represented spurious sequencing activity.
7. Filter candidates
– For each window, a heuristic is used to find those positions that
likely need polishing. In addition, homopolymers are always
polished. Skipping unambiguous positions makes the polishing at
least twice as fast.
8. Polishing
– The core polishing uses the
arrow algorithm, a left-right Hidden
Markov-Model (HMM) that models the enzymatic and photophysical
events in SMRT sequencing. Emission and transition parameters are
estimated by a dinucleotide template context. Transition
parameters form a homogeneous Markov chain. The transition
parameters do not depend on the position within the template, only
the pulse width of a base call, the dinucleotide context of the
template, and the SNR of the ZMW.
Arrow computes the log-
likelihood that the subread originates from the template,
marginalizing over all possible alignments of the subread to the
template. For every position in the template that is a candidate for
polishing,
arrow tests if the log-likelihood is improved by
substituting one of the other three nucleotides, inserting one of the
four nucleotides after the position, or deleting the position itself.
Once
arrow does not find any further beneficial mutations to the
template in an iteration, it stops.
9. QV calculation
– The log-likelihood ratio between the most likely template sequence
and all of its mutated counterparts is used to calculate a quality for
each base in the final consensus. The average of the per-base
qualities is the read accuracy,
rq.
10. Final steps

Page 9
– The per-window consensus template sequences and base qualities
are concatenated and overhangs (overlaps between adjacent
windows) are trimmed. If the predicted read accuracy is at least
--min-rq, then the consensus read is written to the output.
Input files
•One .subreads.bam file containing the subreads for each SMRTbell
®
template sequenced.
Output files
• A BAM file with one entry for each consensus sequence derived from a
productive ZMW. BAM is a general file format for storing sequence
data, which is described fully by the SAM/BAM working group. The
CCS analysis output format is a version of this general format, where
the consensus sequence is represented by the "Query Sequence".
Several tags were added to provide additional meta information. An
example BAM entry for a consensus as seen by
samtools is shown
below.
m64009_201008_223950/1/ccs 4 * 0 255 * * 0 0 ATCGCCTACC
~|~t~R~~r~ RG:Z:a773c1f2 fi:B:C,26,60,21,41,33,26,63,45,73,33 fn:i:6
fp:B:C,11,18,21,35,8,18,31,8,23,11 np:i:12
ri:B:C,17,37,24,4,70,21,12,44,21,32 rn:i:6 rp:B:C,16,56,17,9,23,19,10,10,23,12
rq:f:0.999651 sn:B:f,10.999,16.2603,3.964,7.17746 we:i:9816064 ws:i:20
zm:i:1
Following are some of the common fields contained in the output BAM
file:
Field Description
Query Name Movie Name / ZMW # /ccs
FLAG Required by the format but meaningless in this context. Always set to
4 to indicate the
read is unmapped.
Reference Name Required by the format but meaningless in this context. Always set to *.
Mapping Start Required by the format but meaningless in this context. Always set to 0.
Mapping Quality Required by the format but meaningless in this context. Always set to 255.
CIGAR Required by the format but meaningless in this context. Always set to *.
RNEXT Required by the format but meaningless in this context. Always set to *.
PNEXT Required by the format but meaningless in this context. Always set to 0.
TLEN Required by the format but meaningless in this context. Always set to 0.
Consensus Sequence The consensus sequence generated.
Quality Values The per-base parametric quality metric. For details see “Interpreting QUAL values” on
page 12.
RG Tag The read group identifier.
bc Tag A 2-entry array of upstream-provided barcode calls for this ZMW.
bq Tag The quality of the barcode call. (Optional: Depends on barcoded inputs.)
Page 10
Usage
ccs [OPTIONS] INPUT OUTPUT
Example
ccs --all myData.subreads.bam myResult.bam
np Tag The number of full passes that went into the subread. (Optional: Depends on barcoded
inputs.)
rq Tag The predicted read quality.
zm Tag The ZMW hole number.
ma Tag Bitmask storing whether a SMRTbell adapter is missing on either side of the molecule
that produced a CCS read. 0 indicates that neither end has a confirmed missing adapter.
ac Tag An array containing the counts of detected and missing SMRTbell adapters on either
side of the molecule that produced a CCS read:
• Detected adapters on left/start
• Missing adapters on left/start
• Detected adapters on right/end
• Missing adapter on right/end
Field Description
Required Description
Input File Name The name of a single subreads.bam or a subreadset.xml file to be processed.
(Example = myData.subreads.bam)
Output File Name The name of the output BAM file; comes after all other options listed. Valid output
files are the BAM and the Dataset .xml formats. (Example = myResult.bam)
Options Description
--version Prints the version number.
--report-file Contains a result tally of the outcomes for all ZMWs that were processed. If no file
name is given, the report is output to the file ccs_report.txt. In addition to the
count of successfully-produced consensus sequences, this file lists how many
ZMWs failed various data quality filters (SNR too low, not enough full passes, and
so on) and is useful for diagnosing unexpected drops in yield.
--min-snr Removes data that is likely to contain deletions. SNR is a measure of the strength
of signal for all 4 channels (A, C, G, T) used to detect base pair incorporation. This
value sets the threshold for minimum required SNR for any of the four channels.
Data with SNR < 2.5 is typically considered lower quality. (Default = 2.5)
--min-length Specifies the minimum length requirement for the minimum length of the draft
consensus to be used for further polishing. If the targeted template is known to be a
particular size range, this can filter out alternative DNA templates. (Default = 10)
--max-length Specifies the maximum length requirement for the maximum length of the draft
consensus to be used for further polishing. For robust results while avoiding
unnecessary computation on unusual data, set to ~20% above the largest expected
insert size. (Default = 50000)
--min-passes Specifies the minimum number of passes for a ZMW to be emitted. This is the
number of full passes. Full passes must have an adapter hit before and after the
insert sequence and so do not include any partial passes at the start and end of the
sequencing reaction. It is computed as the number of passes mode across all
windows. (Default = 3)
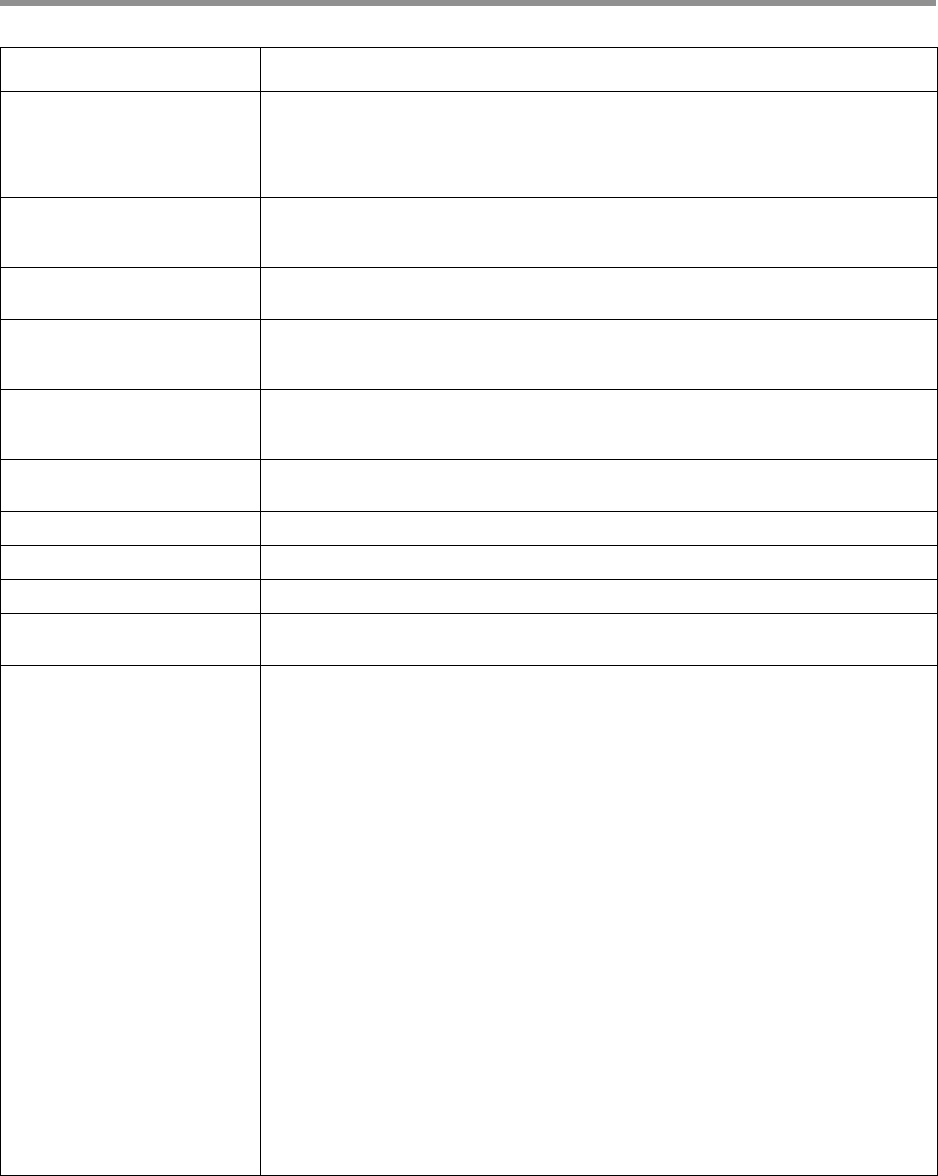
Page 11
--min-rq Specifies the minimum predicted accuracy of a read. CCS analysis generates an
accuracy prediction for each read, defined as the expected percentage of matches
in an alignment of the consensus sequence to the true read. A value of 0.99
indicates that only reads expected to be 99% accurate are emitted.
(Default = 0.99)
--num-threads Specifies how many threads to use while processing. By default, CCS analysis uses
as many threads as there are available cores to minimize processing time, but
fewer threads can be specified here.
--log-file The name of a log file to use. If none is given, the logging information is printed to
STDERR. (Example: mylog.txt)
--log-level Specifies verbosity of log data to produce. By setting --logLevel=DEBUG, you
can obtain detailed information on what ZMWs were dropped during processing, as
well as any errors which may have appeared. (Default = INFO)
--skip-polish After constructing the draft consensus, do not proceed with the polishing steps.
This is significantly faster, but generates less accurate data with no RQ or QUAL
values associated with each base.
--by-strand Separately generates a consensus sequence from the forward and reverse strands.
Useful for identifying heteroduplexes formed during sample preparation.
--chunk Operates on a single chunk. Format i/N, where i in [1,N]. Examples: 3/24 or 9/9.
--max-chunks Determines the maximum number of chunks, given an input file.
--model-path Specifies the path to a model file or directory containing model files.
--model-spec Specifies the name of the chemistry or model to use, overriding the default
selection.
--all Generates one representative sequence per ZMW, irrespective of quality and
passes. --min-passes 0 --min-rq 0 --max-length 0 are set implicitly and
cannot be changed; --all also deactivates the maximum draft length filter.
Filtering has to be performed downstream.
The ccs --all option changes the workflow as follows:
1. There is special behavior for low-pass ZMWs. If a ZMW has fewer than 2 full-
length subreads, use the subread of median length as representative
consensus, optionally with its kinetic information as forward orientation using
--all-kinetics, and do not polish.
2. Only polish ZMWs with at least two full-length subreads mapping back to the
draft. Otherwise, set predicted accuracy rq tag to -1 to indicate that the
predicted accuracy was not calculated, and populate per-base QVs with + (QV
10) the approximate raw accuracy. Kinetic information is not available for
unpolished drafts.
3. Instead of using an unpolished draft without kinetic information as a
representative consensus sequence, if --subread-fallback is used, fall
back to a representative subread with kinetic information.
How is --all different from explicitly setting --min-passes 0 --min-rq 0?
• Setting --min-passes 0 --min-rq 0 is a brute-force combination that
polishes every ZMW, even those that only have one partial subread, with
polishing making no difference. In contrast, --all is a bit smarter and only
polishes ZMWs with at least one full-length subread and one additional partial
subread.
Options Description

Page 12
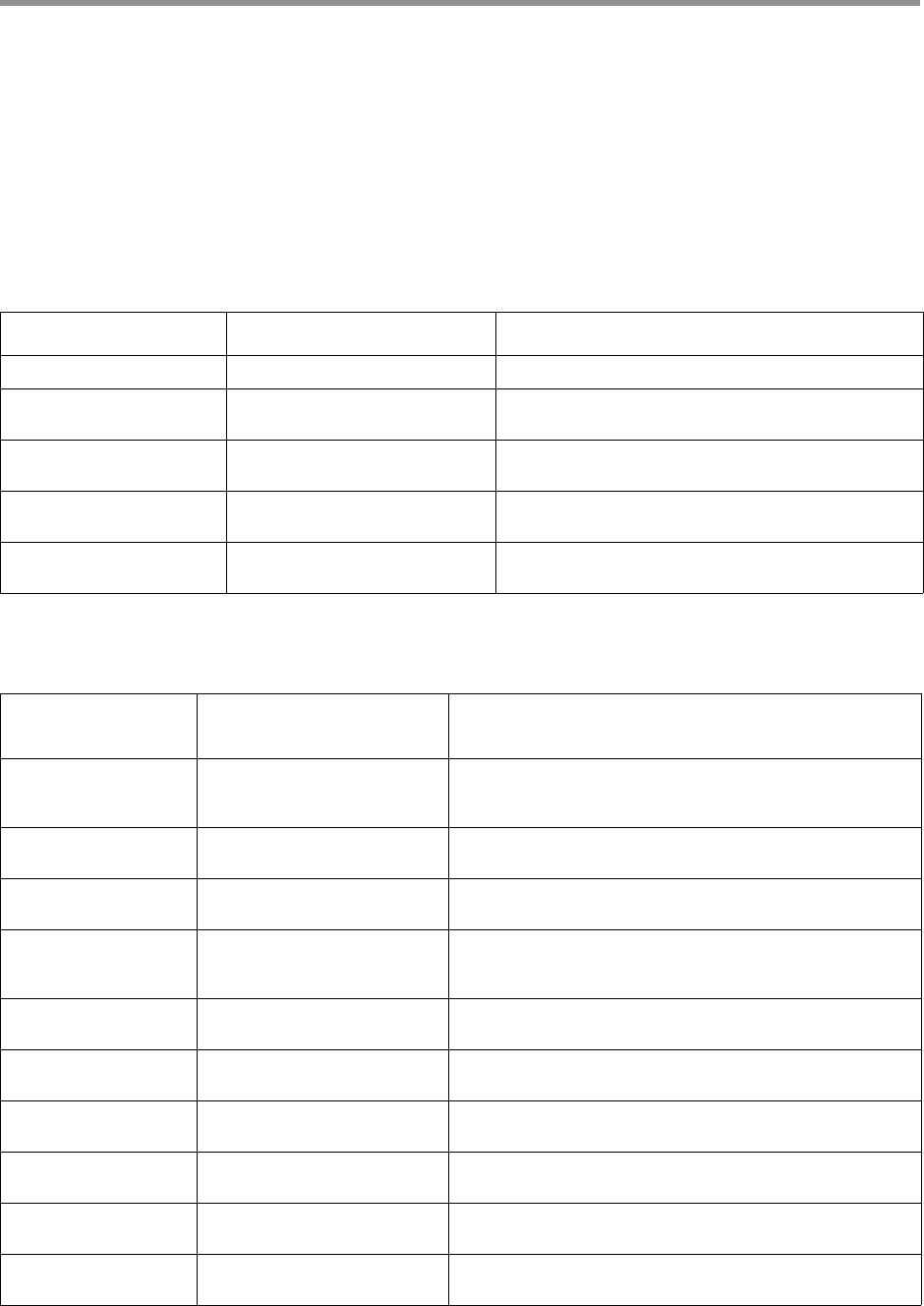
Interpreting QUAL values
The QUAL value of a read is a measure of the posterior likelihood of an
error at a particular position. Increasing QUAL values are associated with
a decreasing probability of error. For indels and homopolymers, there is
ambiguity as to which QUAL value is associated with the error probability.
Shown below are different types of alignment errors, with an
* indicating
which sequence BP should be associated with the alignment error.
Mismatch
*
ccs: ACGTATA
ref: ACATATA
Deletion
*
ccs: AC-TATA
ref: ACATATA
Insertion
*
ccs: ACGTATA
ref: AC-TATA
Homopolymer insertion or deletion
Indels should always be left-aligned, and the error probability is only given
for the first base in a homopolymer.
* *
ccs: ACGGGGTATA ccs: AC-GGGTATA
ref: AC-GGGTATA ref: ACGGGGTATA
--hifi-kinetics Generates averaged kinetic information for polished reads, independently for both
strands of the insert. Forward is defined with respect to the orientation represented
in SEQ and is considered to be the native orientation. As with other PacBio-specific
tags, aligners will not re-orient these fields.
Base modifications can be inferred from per-base pulse width (PW) and inter-pulse
duration (IPD) kinetics.
Minor cases exist where a certain orientation may get filtered out entirely from a
ZMW, preventing valid values from being passed for that record. In these cases,
empty lists are passed for the respective record/orientation, and number of passes
are set to zero.
To facilitate the use of HiFi reads with base modifications workflows, we added an
executable in pbbam called ccs-kinetics-bystrandify which creates a
pseudo --by-strand BAM with corresponding pw and ip tags that imitates a
normal, unaligned subreads BAM.
--all-kinetics Adds kinetic information for all ZMWs, except for unpolished draft consensus.
--subread-fallback When combined with --all, uses a subread instead of a draft as representative
consensus.
--suppress-reports Suppresses the generation of default reports and metric files.
Options Description
Page 13
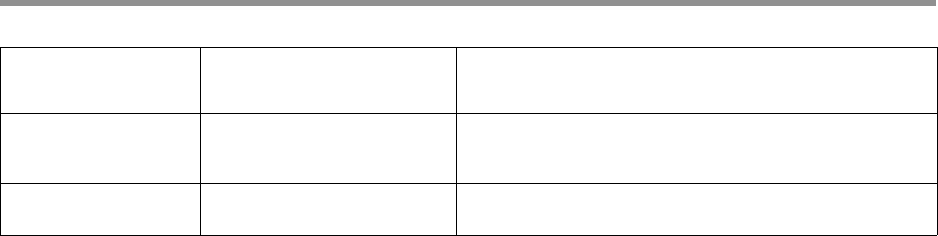
CCS Analysis Yield report
The CCS Analysis Yield report specifies the number of ZMWs that
successfully produced consensus sequences, as well as a count of how
many ZMWs did not produce a consensus sequence for various reasons.
The entries in this report, as well as parameters used to increase or
decrease the number of ZMWs that pass various filters, are shown in the
table below.
The first part is a summary of inputs and outputs:
The second part explains in detail the exclusive ZMW count for those
ZMWs that were filtered:
ZMW results Parameters affecting results Description
ZMWs input None The number of input ZMWs.
ZMWs pass filters All custom processing settings The number of CCS reads successfully produced on
the first attempt, using the fast windowed approach.
ZMWs fail filters All custom processing settings The number of ZMWs reads that failed to produce a
CCS read.
ZMWs shortcut filters -all The number of ZMWs having fewer than 2 full-length
subreads.
ZMWs with tandem
repeats
--min-tandem-repeat-
length
The number of ZMWs with repeats larger than the
specified threshold.
ZMW results
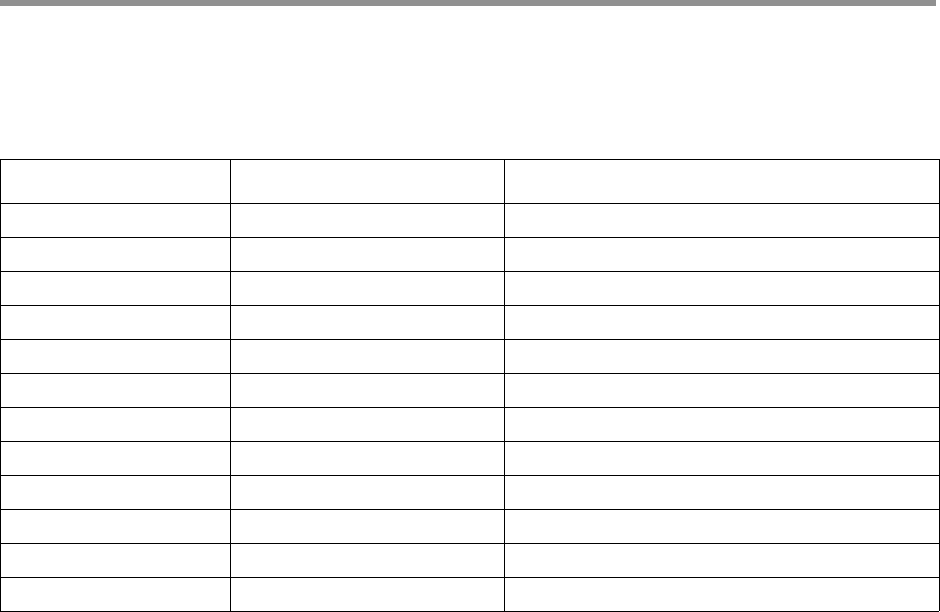
Parameters affecting
results
Description
No usable subreads None The ZMW had no usable subreads. Either there were no
subreads, or all subreads had lengths outside the range
<50% or >200% of the median subread length.
Below SNR threshold --min-snr The ZMW had at least one channel's SNR below the
minimum threshold.
Lacking full passes --min-passes There were not enough subreads that had an adapter at
the start and end of the subread (a "full pass").
Heteroduplexes None The SMRTbell contains a heteroduplex. In this case, it is
not clear what the consensus should be and so the ZMW is
dropped.
Min coverage violation None The ZMW is damaged on one strand and cannot be
polished reliably.
Draft generation error None Subreads do not match the generated draft sequence,
even after multiple tries.
Draft above
--max-length
--max-length The draft sequence was above the maximum length
threshold.
Draft below
--min-length
--min-length The draft sequence was below the minimum length
threshold.
Lacking usable
subreads
None Too many subreads were dropped while polishing.
CCS analysis did not
converge
None The consensus sequence did not converge after the
maximum number of allowed rounds of polishing.
Page 14
How do I read the ccs_report.txt file?
By default, each CCS analysis generates a
ccs_report.txt file. This file
summarizes how many ZMWs generated HiFi reads and how many ZMWs
failed CCS reads generation because of the listed causes. For those
failing, each ZMW contributes to exactly one reason of failure;
percentages are with respect to number of failed ZMWs.
Does CCS analysis dislike low-complexity regions?
Low-complexity comes in many shapes and forms. A particular challenge
for CCS analysis are highly-enriched tandem repeats, such as hundreds of
copies of AGGGGT. Prior to
ccs v5.0, inserts with many copies of a small
repeat likely did not generate a consensus sequence. Since
ccs v5.0, every
ZMW is tested if it contains a tandem repeat of length
--min-tandem-repeat-length 1000. For this, we use symmetric DUST,
specifically this
sdust implementation, but slightly modified. If a ZMW is
flagged as a tandem repeat, internally
--disable-heuristics is
activated for only this ZMW, and various filters that are known to exclude
low-complexity sequences are disabled. This recovers most of the low-
complexity consensus sequences, without impacting run time
performance.
Can I produce one consensus sequence for each strand of a molecule?
Yes, use
--by-strand. Make sure that you have sufficient coverage, as
--min-passes are per-strand in this case. For each strand, CCS analysis
generates one consensus read that has to pass all filters. Read name
suffix indicates strand. Example:
m64011_190714_120746/14/ccs/rev
m64011_190714_120746/35/ccs/fwd
How does --by-strand work? For each ZMW:
• Determine orientation of reads with respect to the one closest to the
median length.
• Sort reads into two buckets, forward and reverse strands.
• Treat each strand as an individual entity as we do with ZMWs:
– Apply all filters per strand individually.
– Create a draft for each strand.
CCS read below
minimum predicted
accuracy
--min-rq Each CCS read has a predicted level of accuracy
associated with it. Reads that are below the minimum
specified threshold are removed.
Unknown error during
processing
None These should not occur.
ZMW results
Parameters affecting
results
Description
Page 15
– Polish each strand.
– Write out each polished strand consensus.
BAM tags generated
Tag Type Description
ec f Effective coverage
fi B,C Forward IPD (Codec V1)
fn i Forward number of complete passes (zero or more)
fp B,C Forward PulseWidth (Codec V1)
np i Number of full-length subreads
ri B,C Reverse IPD (Codec V1)
rn i Reverse number of complete passes (zero or more)
rp B,C Reverse PulseWidth (Codec V1)
rq f Predicted average read accuracy
sn B,F Signal-to-noise ratios for each nucleotide
zm i ZMW hole number
RG z Read group

Page 16
dataset
The dataset tool creates, opens, manipulates and writes Data Set XML
files. The commands allow you to perform operations on the various types
of data held by a Data Set XML: Merge, split, write, and so on.
Usage
dataset [-h] [--version] [--log-file LOG_FILE]
[--log-level {DEBUG,INFO,WARNING,ERROR,CRITICAL} | --debug | --quiet | -v]
[--strict] [--skipCounts]
{create,filter,merge,split,validate,summarize,consolidate,loadstats,newuuid,loadmetada
ta,copyto,absolutize,relativize}
create Command: Create an XML file from a fofn (file-of-file names) or
BAM file. Possible types:
SubreadSet, AlignmentSet, ReferenceSet,
HdfSubreadSet, BarcodeSet, ConsensusAlignmentSet,
ConsensusReadSet, ContigSet
.
dataset create [-h] [--type DSTYPE] [--name DSNAME] [--generateIndices]
[--metadata METADATA] [--novalidate] [--relative]
outfile infile [infile ...]
Example
The following example shows how to use the dataset create command
to create a barcode file:
dataset create --generateIndices --name my_barcodes --type BarcodeSet
my_barcodes.barcodeset.xml my_barcodes.fasta
Options Description
-h, --help Displays help information and exits.
<Command> -h Displays help for a specific command.
-v, --version Displays program version number and exits.
--log-file LOG_FILE Writes the log to file. (Default = None, writes to stdout.)
--log-level Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR,
CRITICAL]. (Default = INFO)
--debug Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to CRITICAL to suppress output.
(Default = False)
-v Sets the verbosity level. (Default = NONE)
--strict Turns on strict tests and display all errors. (Default = False)
--skipCounts Skips updating NumRecords and TotalLength counts.
(Default = False)
Required Description
outfile The name of the XML file to create.
infile The fofn (file-of-file-names) or BAM file(s) to convert into an XML file.
Page 17
filter
Command: Filter an XML file using filters and threshold values.
• Suggested filters:
alignedlength, as, astart, bc, bcf, bcq, bcr,
bq,
cx, length, mapqv, movie, n_subreads, pos, qend, qid,
qname, qstart, readstart, rname, rq, tend, tstart, zm
• More resource-intensive filter: [qs]
Note: Multiple filters with different names are ANDed together. Multiple
filters with the same name are ORed together, duplicating existing
requirements. The filter string should be enclosed in single quotes.
dataset filter [-h] infile outfile filters [filters ...]
Examples
Filter on read quality > 0.99 (Q20):
% dataset filter in.consensusreadset.xml hifi.consensusreadset.xml 'rq >= 0.99'
Filter on read quality and length:
% dataset filter in.consensusreadset.xml filtered.consensusreadset.xml 'rq >= 0.99 AND
length >= 10000'
Filter for very long and very short reads:
% dataset filter in.consensusreadset.xml filtered.consensusreadset.xml 'length >=
40000; length <= 400'
Filter for specific high-quality barcodes:
% dataset filter mixed.consensusreadset.xml samples1-3.consensusreadset.xml 'bc =
[0,1,2] AND bq >= 26'
Options Description
--type DSTYPE Specifies the type of XML file to create. (Default = NONE)
--name DSNAME The name of the new Data Set XML file.
--generateIndices Generates index files (.pbi and .bai for BAM, .fai for FASTA). Requires
samtools/pysam and pbindex. (Default = FALSE)
--metadata METADATA A metadata.xml file (or Data Set XML) to supply metadata.
(Default = NONE)
--novalidate Specifies not to validate the resulting XML. Leaves the paths as they are.
--relative Makes the included paths relative instead of absolute. This is not
compatible with --novalidate.
Required Description
infile The name of the XML file to filter.
outfile The name of the output filtered XML file.
filters The values to filter on. (Example: rq>0.85)
Page 18
merge
Command: Combine XML files.
dataset merge [-h] outfile infiles [infiles ...]
split Command: Split a Data Set XML file.
dataset split [-h] [--contigs] [--barcodes] [--zmws] [--byRefLength]
[--noCounts] [--chunks CHUNKS] [--maxChunks MAXCHUNKS]
[--targetSize TARGETSIZE] [--breakContigs]
[--subdatasets] [--outdir
infile [outfiles...]
validate Command: Validate XML and ResourceId files. (This is an
internal testing functionality that may be useful.)
Note: This command requires that
pyxb (not distributed with SMRT Link)
be installed. If not installed,
validate simply checks that the files pointed
to in
ResourceIds exist.
Required Description
infiles The names of the XML files to merge.
outfile The name of the output XML file.
Required Description
infile The name of the XML file to split.
Options Description
outfiles The names of the resulting XML files.
--contigs Splits the XML file based on contigs. (Default = FALSE)
--barcodes Splits the XML file based on barcodes. (Default = FALSE)
--zmws Splits the XML file based on ZMWs. (Default = FALSE)
--byRefLength Splits contigs by contig length. (Default = TRUE)
--noCounts Updates the Data Set counts after the split. (Default = FALSE)
--chunks x Splits contigs into x total windows. (Default = 0)
--maxChunks x Splits the contig list into at most x groups. (Default = 0)
--targetSize x Specifies the minimum number of records per chunk. (Default = 5000)
--breakContigs Breaks contigs to get closer to maxCounts. (Default = False)
--subdatasets Splits the XML file based on sub-datasets. (Default = False)
--outdir OUTDIR Specifies an output directory for the resulting XML files.
(Default = <in-place>, not the current working directory.)
Page 19
dataset validate [-h] [--skipFiles] infile
summarize Command: Summarize a Data Set XML file.
dataset summarize [-h] infile
consolidate Command: Consolidate XML files.
dataset consolidate [-h] [--numFiles NUMFILES] [--noTmp]
infile datafile xmlfile
loadstats Command: Load an sts.xml file containing pipeline statistics
into a Data Set XML file.
dataset loadstats [-h] [--outfile OUTFILE] infile statsfile
Required Description
infile The name of the XML file to validate.
Options Description
--skipFiles Skips validating external resources. (Default = False)
Required Description
infile The name of the XML file to summarize.
Required Description
infile The name of the XML file to consolidate.
datafile The name of the resulting data file.
xmlfile The name of the resulting XML file.
Options Description
--numFiles x Specifies the number of data files to produce. (Default = 1)
--noTmp Do not copy to a temporary location to ensure local disk use.
(Default = False)
Required Description
infile The name of the Data Set XML file to modify.
statsfile The name of the .sts.xml file to load.
Options Description
--outfile OUTFILE The name of the XML file to output. (Default = None)
Page 20
newuuid
Command: Refresh a Data Set's Unique ID.
dataset newuuid [-h] [--random] infile
loadmetadata Command: Load a .metadata.xml file into a Data Set XML
file.
dataset loadmetadata [-h] [--outfile OUTFILE] infile metadata
copyto Command: Copy a Data Set and resources to a new location.
dataset copyto [-h] [--relative] infile outdir
absolutize Command: Make the paths in an XML file absolute.
dataset absolutize [-h] [--outdir OUTDIR] infile
Required Description
infile The name of the XML file to refresh.
Options Description
--random Generates a random UUID, instead of a hash. (Default = False)
Required Description
infile The name of the Data Set XML file to modify.
metadata The .metadata.xml file to load, or Data Set to borrow from.
Options Description
--outfile OUTFILE Specifies the XML file to output. (Default = None)
Required Description
infile The name of the XML file to copy.
outdir The directory to copy to.
Options Description
--relative Makes the included paths relative instead of absolute. (Default = False)
Required Description
infile The name of the XML file whose paths should be absolute.
Options Description
--outdir OUTDIR Specifies an optional output directory. (Default = None)

Page 21
relativize
Command: Make the paths in an XML file relative.
dataset relativize [-h] infile
Examples - Filter reads
To filter one or more BAM file’s worth of subreads, aligned or otherwise,
and then place them into a single BAM file:
# usage: dataset filter <in_fn.xml> <out_fn.xml> <filters>
dataset filter in_fn.subreadset.xml filtered_fn.subreadset.xml 'rq>0.85'
# usage: dataset consolidate <in_fn.xml> <out_data_fn.bam> <out_fn.xml>
dataset consolidate filtered_fn.subreadset.xml consolidate.subreads.bam
out_fn.subreadset.xml
The filtered Data Set and the consolidated Data Set should be read-for-
read equivalent when used with SMRT
®
Analysis software.
Example - Resequencing pipeline
• Align two movie’s worth of subreads in two SubreadSets to a
reference.
• Merge the subreads together.
• Split the subreads into Data Set chunks by contig.
•Process using
gcpp on a chunkwise basis (in parallel).
1. Align each movie to the reference, producing a Data Set with one BAM
file for each execution:
pbalign movie1.subreadset.xml referenceset.xml movie1.alignmentset.xml
pbalign movie2.subreadset.xml referenceset.xml movie2.alignmentset.xml
2. Merge the files into a FOFN-like Data Set; BAMs are not touched:
# dataset merge <out_fn> <in_fn> [<in_fn> <in_fn> ...]
dataset merge merged.alignmentset.xml movie1.alignmentset.xml movie2.alignmentset.xml
3. Split the Data Set into chunks by contig name; BAMs are not touched:
– Note that supplying output files splits the Data Set into that many
output files (up to the number of contigs), with multiple contigs per
file.
– Not supplying output files splits the Data Set into one output file per
contig, named automatically.
– Specifying a number of chunks instead will produce that many files,
with contig or even subcontig (reference window) splitting.
dataset split --contigs --chunks 8 merged.alignmentset.xml
Required Description
infile The name of the XML file whose paths should be relative.

Page 22
4. Process the chunks:
gcpp --reference referenceset.xml --output
chunk1consensus.fasta,chunk1consensus.fastq,chunk1consensus.vcf,chunk1consensus.gff
chunk1contigs.alignmentset.xml
The chunking works by duplicating the original merged Data Set (no BAM
duplication) and adding filters to each duplicate such that only reads
belonging to the appropriate contigs are emitted. The contigs are
distributed among the output files in such a way that the total number of
records per chunk is about even.
Page 23
Demultiplex
Barcodes
The Demultiplex Barcodes application identifies barcode sequences in
PacBio single-molecule sequencing data.
Demultiplex Barcodes can demultiplex samples that have a unique per-
sample barcode pair and were pooled and sequenced on the same SMRT
®
Cell. There are four different methods for barcoding samples with PacBio
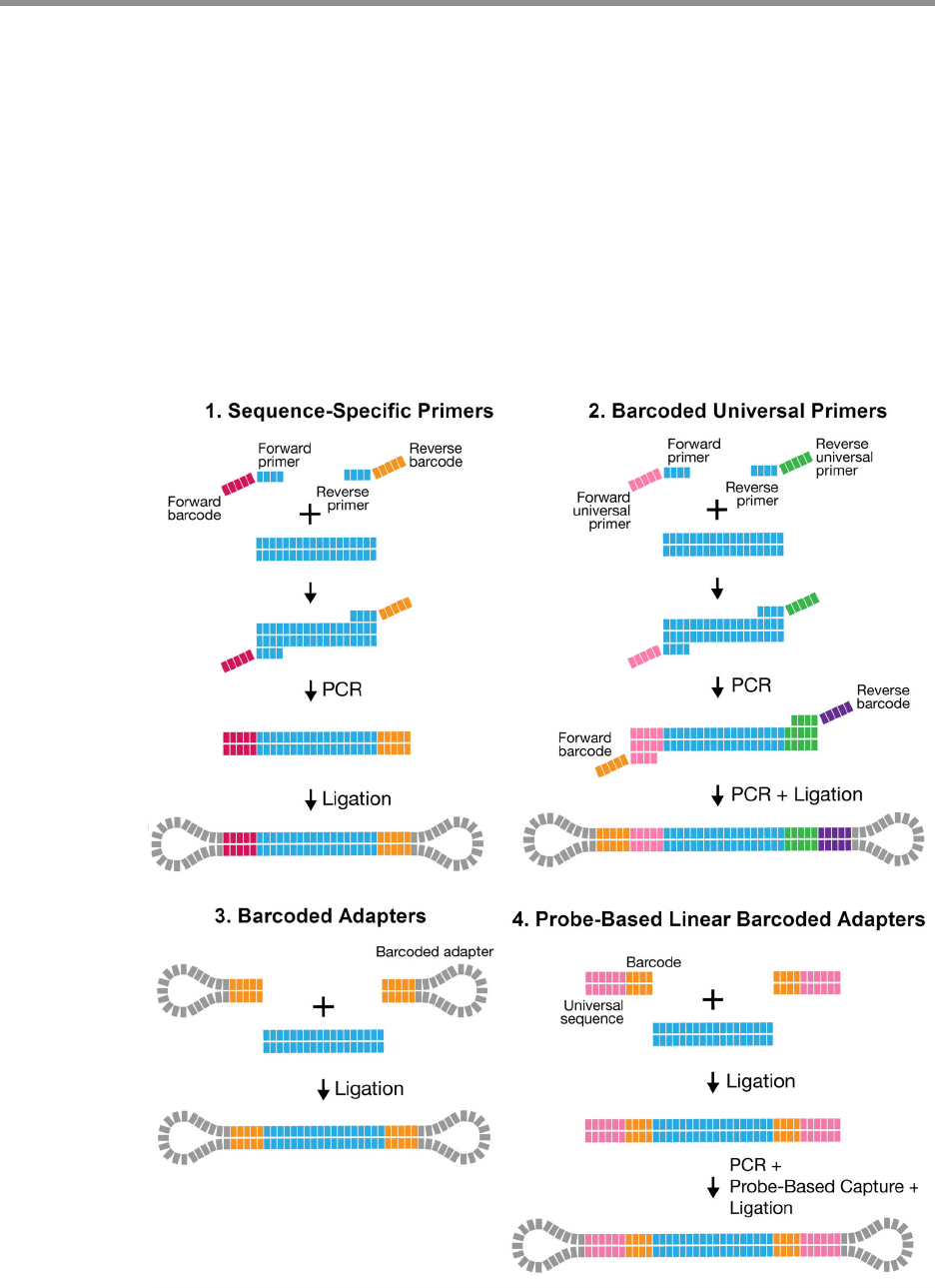
technology:
1. Sequence-specific primers
2. Barcoded universal primers
3. Barcoded adapters
4. Linear Barcoded Adapters for Probe-based Captures
Page 24
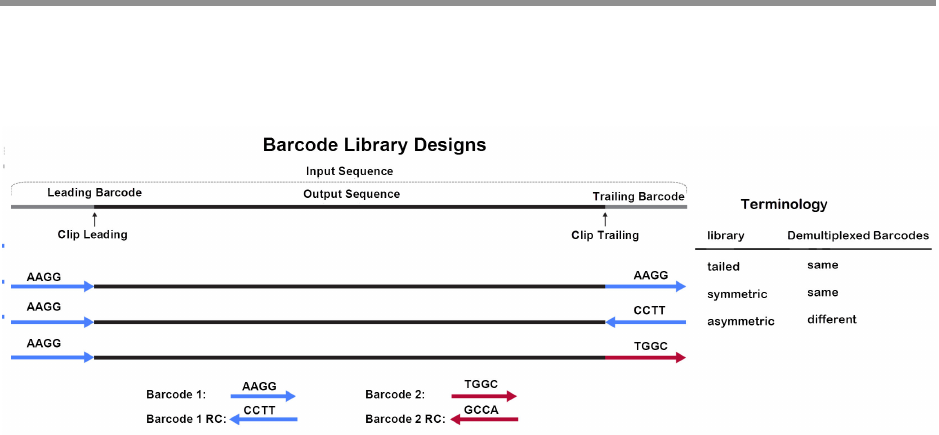
In addition, there are three different barcode library designs. Demultiplex
Barcodes supports raw subread and CCS reads demultiplexing.
In the overview above, the input sequence is flanked by adapters on both
sides. The bases adjacent to an adapter are barcode regions. A read can
have up to two barcode regions, leading and trailing. Either or both adapt-
ers can be missing and consequently the leading and/or trailing region is
not being identified.
For symmetric and tailed library designs, the same barcode is attached to
both sides of the insert sequence of interest. The only difference is the
orientation of the trailing barcode. For barcode identification, one read
with a single barcode region is sufficient.
For the asymmetric design, different barcodes are attached to the sides of
the insert sequence of interest. To identify the different barcodes, a read
with leading and trailing barcode regions is required.
Output barcode pairs are generated from the identified barcodes. The
barcode names are combined using “
--“, for example bc1002--bc1054.
The sort order is defined by the barcode indices, starting with the lowest.
Workflow
By default, Demultiplex Barcodes processes input reads grouped by ZMW,
except if the
--per-read option is used. All barcode regions along the
read are processed individually. The final per-ZMW result is a summary
over all barcode regions. Each ZMW is assigned to a pair of selected
barcodes from the provided set of candidate barcodes. Subreads from the
same ZMW will have the same barcode and barcode quality. For a
particular target barcode region, every barcode sequence gets aligned as
given and as reverse-complement, and higher scoring orientation is
chosen. This results in a list of scores over all candidate barcodes.
•If only same barcode pairs are of interest (symmetric/tailed), use the
--same option to filter out different barcode pairs.

Page 25
•If only different barcode pairs are of interest (asymmetric), use the
--different option to require at least two barcodes to be read, and
remove pairs with the same barcode.
Parameter presets
Recommended parameter combinations are available using
--preset for
HiFi input:
• HIFI-SYMMETRIC
--ccs --min-score 80 --min-end-score 50 --min-ref-span 0.75
--same
• HIFI-ASYMMETRIC
--ccs --min-score 80 --min-end-score 50 --min-ref-span 0.75
--different --min-scoring-regions 2
•NONE (Default)
Half adapters
For an adapter call with only one barcode region, the high-quality region
finder cuts right through the adapter. The preceding or succeeding
subread was too short and was removed, or the sequencing reaction
started/stopped there. This is called a half adapter. Thus, there are also
1.5, 2.5, N+0.5 adapter calls.
ZMWs with half or only one adapter can be used to identify the same
barcode pairs; positive-predictive value might be reduced compared to
high adapter calls. For asymmetric designs with different barcodes in a
pair, at least a single full-pass read is required; this can be two adapters,
two half adapters, or a combination.
Usage:
• Any existing output files are overwritten after execution.
•Always use
--peek-guess to remove spurious barcode hits.
Analysis of subread data:
lima movie.subreads.bam barcodes.fasta prefix.bam
lima movie.subreadset.xml barcodes.barcodeset.xml prefix.subreadset.xml
Analysis of CCS reads:
lima --css movie.ccs.bam barcodes.fasta prefix.bam
lima --ccs movie.consensusreadset.xml barcodes.barcodeset.xml
prefix.consensusreadset.xml
If you do not need to import the demultiplexed data into SMRT Link, use
the
--no-pbi option to minimize memory consumption and run time.
Symmetric or tailed options:
Raw: --same
CCS read: --same --ccs
Page 26
Asymmetric options:
Raw: --different
CCS reads: --different --ccs
Example execution:
lima m54317_180718_075644.subreadset.xml \
Sequel_RSII_384_barcodes_v1.barcodeset.xml \
m54317_180718_075644.demux.subreadset.xml \
--different --peek-guess
Options Description
--same Retains only reads with the same barcodes on both ends of the insert
sequence, such as symmetric and tailed designs.
--different Retains only reads with different barcodes on both ends of the insert
sequence, asymmetric designs. Enforces --min-passes ≥ 1.
--min-length n Omits reads with lengths below n base pairs after demultiplexing. ZMWs
with no reads passing are omitted. (Default = 50)
--max-input-length n Omits reads with lengths above n base pairs for scoring in the
demultiplexing step. (Default = 0, deactivated)
--min-score n Omits ZMWs with average barcode scores below n. A barcode score
measures the alignment between a barcode attached to a read and an
ideal barcode sequence, and is an indicator how well the chosen barcode
pair matches. It is normalized to a range between 0 (no hit) and 100 (a
perfect match).
(Default = 0, PacBio recommends setting it to 26.)
--min-end-score n Specifies the minimum end barcode score threshold applied to the
individual leading and trailing ends. (Default = 0)
--min-passes n Omits ZMWs with less than n full passes, a read with a leading and trailing
adapter. (Default = 0, no full-pass needed) Example:
0 pass : insert - adapter - insert
1 pass : insert - adapter - INSERT - adapter - insert
2 passes: insert - adapter - INSERT - adapter - INSERT -
adapter - insert
--score-full-pass Uses only reads flanked by adapters on both sides (full-pass reads) for
barcode identification.
--min-ref-span Specifies the minimum reference span relative to the barcode length.
(Default = 0.5)
--per-read Scores and tags per subread, instead of per ZMW.
--ccs Sets defaults to -A 1 -B 4 -D 3 -I 3 -X 1.
--peek n Looks at the first n ZMWs of the input and return the mean. This lets you
test multiple test barcode.fasta files and see which set of barcodes
was used.
--guess n This performs demultiplexing twice. In the first iteration, all barcodes are
tested per ZMW. Afterwards, the barcode occurrences are counted and
their mean is tested against the threshold n; only those barcode pairs that
pass this threshold are used in the second iteration to produce the final
demultiplexed output. A prefix.lima.guess file shows the decision
process; --same is being respected.
--guess-min-count Specifies the minimum ZMW count to whitelist a barcode. This filter is
ANDed with the minimum barcode score specified by --guess.
(Default = 0)
Page 27
Input files
Input data in PacBio-enhanced BAM format is either:
• Sequence data - Unaligned subreads, directly from Sequel II systems
and Sequel IIe systems.
• Unaligned CCS reads, generated by CCS analysis.
Barcodes are provided as a FASTA file or BarcodeSet file:
• One entry per barcode sequence.
• No duplicate sequences.
--peek-guess Sets the following options:
--peek 50000 --guess 45 --guess-min-count 10.
Demultiplex Barcodes will run twice on the input data. For the first 50,000
ZMWs, it will guess the barcodes and store the mask of identified
barcodes. In the second run, the barcode mask is used to demultiplex all
ZMWs.
If combined with --ccs then the barcode score threshold is increased by
--guess 75.
--single-side Identifies barcodes in molecules that only have barcodes adjacent to one
adapter.
--window-size-mult
--window-size-bp
The candidate region size multiplier: barcode_length * multiplier.
(Default = 3)
Optionally, you can specify the region size in base pairs using
--window-size-bp. If set, --window-size-mult is ignored.
--num-threads n Spawns n threads; 0 means use all available cores. This option also
controls the number of threads used for BAM and PBI compression.
(Default = 0)
--chunk-size n Specifies that each thread consumes n ZMWs per chunk for processing.
(Default = 10).
--no-bam Does not produce BAM output. Useful if only reports are of interest, as run
time is shorter.
--no-pbi Does not produce a .bam.pbi index file. The on-the-fly .bam.pbi file
generation buffers the output data. If you do not need a .bam.pbi index
file for SMRT Link import, use this option to decrease memory usage to a
minimum and shorten the run time.
--no-reports Does not produce any reports. Useful if only demultiplexed BAM files are
needed.
--dump-clips Outputs all clipped barcode regions generated to the
<prefix>.lima.clips file.
--dump-removed Outputs all records that did not pass the specified thresholds, or are
without barcodes, to the <prefix>.lima.removed.bam file.
--split-bam
--split-bam-named
Specifies that each barcode has its own BAM file called
prefix.idxBest-idxCombined.bam, such as prefix.0-0.bam.
Optionally ,--split-bam-named names the files by their barcode names
instead of their barcode indices.
--isoseq Removes primers as part of the Iso-Seq
®
pipeline.
See “Demultiplexing Iso-Seq
®
data” on page 32 for details.
--bad-adapter-ratio n Specifies the maximum ratio of bad adapters. (Default = 0).
Options Description

Page 28
• All bases must be in upper-case.
• Orientation-agnostic (forward or reverse-complement, but not
reversed.)
Example:
>bc1000
CTCTACTTACTTACTG
>bc1001
GTCGTATCATCATGTA
>bc1002
AATATACCTATCATTA
Note: Name barcodes using an alphabetic character prefix to avoid later
barcode name/index confusion.
Output files
Demultiplex Barcodes generates multiple output files by default, all
starting with the same prefix as the output file, using the suffixes
.bam,
.subreadset.xml, and .consensusreadset.xml. The report prefix is
lima. Example:
lima m54007_170702_064558.subreads.bam barcode.fasta /my/path/
m54007_170702_064558_demux.subreadset.xml
For all output files, the prefix is
/my/path/m54007_170702_064558_demux.
• <prefix>.bam: Contains clipped records, annotated with barcode
tags, that passed filters and respect the
--same option.
•
<prefix>.lima.report: A tab-separated file describing each ZMW,
unfiltered. This is useful information for investigating the
demultiplexing process and the underlying data. A single row contains
all reads from a single ZMW. For
--per-read, each row contains one
subread, and ZMWs might span multiple rows.
•
<prefix>.lima.summary: Lists how many ZMWs were filtered, how
many ZMWs are the same or different, and how many reads were
filtered.
(1)
ZMWs input (A): 213120
ZMWs above all thresholds (B): 176356 (83%)
ZMWs below any threshold (C): 36764 (17%)
(2)
ZMW Marginals for (C):
Below min length : 26 (0%)
Below min score : 0 (0%)
Below min end score : 5138 (13%)
Below min passes : 0 (0%)
Below min score lead : 11656 (32%)
Below min ref span : 3124 (8%)
Without adapter : 25094 (68%)
With bad adapter : 10349 (28%) <- Only with --bad-adapter-ratio

Page 29
Undesired hybrids : xxx (xx%) <- Only with --peek-guess
Undesired same barcode pairs : xxx (xx%) <- Only with --different
Undesired diff barcode pairs : xxx (xx%) <- Only with --same
Undesired 5p--5p pairs : xxx (xx%) <- Only with --isoseq
Undesired 3p--3p pairs : xxx (xx%) <- Only with --isoseq
Undesired single side : xxx (xx%) <- Only with --isoseq
Undesired no hit : xxx (xx%) <- Only with --isoseq
(3)
ZMWs for (B):
With same barcode : 162244 (92%)
With different barcodes : 14112 (8%)
Coefficient of correlation : 32.79%
(4)
ZMWs for (A):
Allow diff barcode pair : 157264 (74%)
Allow same barcode pair : 188026 (88%)
Bad adapter yield loss : 10112 (5%) <- Only with --bad-adapter-ratio
Bad adapter impurity : 10348 (5%) <- Only without --bad-adapter-ratio
(5)
Reads for (B):
Above length : 1278461 (100%)
Below length : 2787 (0%)
Explanation of each block:
1. Number of ZMWs that went into lima, how many ZMWs were passed
to the output file, and how many did not qualify.
2. For those ZMWs that did not qualify: The marginal counts of each
filter. (Filter are described in the Options table.)
When running with
--peek-guess or similar manual option combina-
tion and different barcode pairs are found during peek, the full SMRT
Cell may contain low-abundant different barcode pairs that were
identified during peek individually, but not as a pair. Those unwanted
barcode pairs are called hybrids.
3. For those ZMWs that passed: How many were flagged as having the
same or different barcode pair, as well as the coefficient of variation
for the barcode ZMW yield distribution in percent.
4. For all input ZMWs: How many allow calling the same or different
barcode pair. This is a simplified version of how many ZMW have at
least one full pass to allow a different barcode pair call and how many
ZMWs have at least half an adapter, allowing the same barcode pair
call.
5. For those ZMWs that qualified: The number of reads that are above
and below the specified
--min-length threshold.
•
<prefix>.lima.counts: A .tsv file listing the counts of each
observed barcode pair. Only passing ZMWs are counted.
Page 30
Example: column -t prefix.lima.count
• <prefix>.lima.clips: Contains clipped barcode regions generated
using the
--dump-clips option. Example:
head -n 6 prefix.lima.clips
>m54007_170702_064558/4850602/6488_6512 bq:34 bc:11
CATGTCCCCTCAGTTAAGTTACAA
>m54007_170702_064558/4850602/6582_6605 bq:37 bc:11
TTTTGACTAACTGATACCAATAG
>m54007_170702_064558/4916040/4801_4816 bq:93 bc:10
• <prefix>.lima.removed.bam: Contains records that did not pass the
specified thresholds, or are without barcodes, using the option
--dump-removed. lima does not generate a .pbi, nor Data Set for this
file. This option cannot be used with any splitting option.
•
<prefix>.lima.guess: A .tsv file that describes the barcode
subsetting process activated using the
--peek and --guess options.
• One DataSet,
.subreadset.xml, or .consensusreadset.xml file is
generated per output BAM file.
•
.pbi: One PBI file is generated per output BAM file.
What is a universal spacer sequence and how does it affect
demultiplexing?
For library designs that include an identical sequence between adapter
and barcode, such as probe-based linear barcoded adapters samples,
Demultiplex Barcodes offers a special mode that is activated if it finds a
shared prefix sequence among all provided barcode sequences.
Example:
>custombc1
ACATGACTGTGACTATCTCACACATATCAGAGTGCG
>custombc2
ACATGACTGTGACTATCTCAACACACAGACTGTGAG
In this case, Demultiplex Barcodes detects the shared prefix
IdxFirst IdxCombined IdxFirstNamed IdxCombinedNamed Counts MeanScore
0 0 bc1001 bc1001 1145 68
1 1 bc1002 bc1002 974 69
2 2 bc1003 bc1003 1087 68
IdxFirst IdxCombined IdxFirstNamed IdxCombinedNamed NumZMWs MeanScore Picked
0 0 bc1001t bc1001t 1008 50 1
1 1 bc1002t bc1002t 1005 60 1
2 2 bc1003t bc1003t 5 24 0
3 3 bc1004t bc1004t 555 61 1

Page 31
ACATGACTGTGACTATCTCA
and removes it internally from all barcodes.
Subsequently, it increases the window size by the length
L of the prefix
sequence.
•If
--window-size-bp N is used, the actual window size is L + N.
•If
--window-size-mult M is used, the actual window size is
(L + |bc|) * M.
Because the alignment is semi-global, a leading reference gap can be
added without any penalty to the barcode score.
What are bad adapters?
In the
subreads.bam file, each subread has a context flag cx. The flag
specifies, among other things, whether a subread has flanking adapters,
before and/or after. Adapter-finding was improved and can also find
molecularly-missing adapters, or those obscured by a local decrease in
accuracy. This may lead to missing or obscured bases in the flanking
barcode. Such adapters are labelled "bad", as they don't align with the
adapter reference sequence(s). Regions flanking those bad adapters are
problematic, because they can fully or partially miss the barcode bases,
leading to wrong classification of the molecule.
lima can handle those
adapters by ignoring regions flanking bad adapters. For this,
lima
computes the ratio of number of bad adapters divided by number of all
adapters.
By default,
--bad-adapter-ratio is set to 0 and does not perform any
filtering. In this mode, bad adapters are handled just like good adapters.
But the
*.lima.summary file contains one row with the number of ZMWs
that have at least 25% bad adapters, but otherwise pass all other filters.
This metric can be used as a diagnostic to assess library preparation.
If
--bad-adapter-ratio is set to non-zero positive (0,1), bad adapter
flanking barcode regions are treated as missing. If a ZMW has a higher
ratio of bad adapters than provided, the ZMW is filtered and consequently
removed from the output. The
*.lima.summary file contains two
additional rows.
With bad adapter : 10349 (28%)
Bad adapter yield loss : 10112 (5%)
The first row counts the number of ZMWs that have bad adapter ratios
that are too high; the percentage is with respect to the number of all ZMW
not passing. The second row counts the number of ZMWs that are
removed solely due to bad adapter ratios that are too high; the percentage
is with respect the number of all input ZMWs and consequently is the
effective yield loss caused by bad adapters.

Page 32
If a ZMW has ~50% bad adapters, one side of the molecule is molecularly-
missing an adapter. For 100% bad adapter, both sides are missing
adapters. A lower than ~40% percentage indicates decreased local
accuracy during sequencing leading to adapter sequences not being
found. If a high percentage of ZMWs is molecularly-missing adapters, you
should improve library preparation.
Demultiplexing Iso-Seq
®
data
Demultiplex Barcodes is used to identify and remove Iso-Seq cDNA
primers. If the Iso-Seq sample is barcoded, the barcodes should be
included as part of the primer. Only by using the command-line can users
use
lima with the --isoseq option for demultiplexing Iso-Seq data.
The input Iso-Seq data format for demultiplexing is
.ccs.bam. Users must
first generate a CCS reads BAM file for an Iso-Seq Data Set before running
lima. The recommended parameters for running CCS analysis for Iso-Seq
are
min-pass=1, min accuracy=0.9, and turning Polish to OFF.
1. Primer IDs must be specified using the suffix
_5p to indicate 5’ cDNA
primers and the suffix
_3p to indicate 3’ cDNA primers. The 3’ cDNA
primer should not include the Ts and is written in reverse complement.
2. Below are four example primer sets. The first is unbarcoded, the
second has barcodes (shown in lower case) adjacent to the 3’ primer.
Example 1: The Iso-Seq cDNA Primer primer set, included with the SMRT
Link installation.
Users following the standard Iso-Seq Express protocol without
multiplexing, or running a Data Set that has already been demultiplexed
(either using Run Design or the SMRT
®
Analysis application) should use
this default option.
>IsoSeq_5p
GCAATGAAGTCGCAGGGTTGGG
>IsoSeq_3p
GTACTCTGCGTTGATACCACTGCTT
Example 2: The Iso-Seq 12 Barcoded cDNA Primers set, included with the
SMRT Link installation.
Users using barcoded cDNA primers listed in the Appendix 3 -
Recommended barcoded NEBNext single cell cDNA PCR primer and Iso-
Seq Express cDNA PCR primer sequences section of the document

Page 33
Procedure & checklist - Preparing Iso-Seq
®
libraries using SMRTbell
Prep Kit 3.0, should select this option.
>bc1001_5p
CACATATCAGAGTGCGGCAATGAAGTCGCAGGGTTGGGG
>bc1002_5p
ACACACAGACTGTGAGGCAATGAAGTCGCAGGGTTGGGG
…
(There are a total of 24 sequence records, representing 12 pairs of F/R
barcoded cDNA primers.)
Example 3: An example of a custom cDNA primer set. 4 tissues were
multiplexed using barcodes on the 3’ end only.
>IsoSeq_5p
GCAATGAAGTCGCAGGGTTGGG
>dT_BC1001_3p
AAGCAGTGGTATCAACGCAGAGTACCACATATCAGAGTGCG
>dT_BC1002_3p
AAGCAGTGGTATCAACGCAGAGTACACACACAGACTGTGAG
>dT_BC1003_3p
AAGCAGTGGTATCAACGCAGAGTACACACATCTCGTGAGAG
>dT_BC1004_3p
AAGCAGTGGTATCAACGCAGAGTACCACGCACACACGCGCG
Example 4: Special Handling for the TeloPrime cDNA Kit
The Lexogen TeloPrime cDNA kit contains As in the 3’ primer that cannot
be differentiated from the polyA tail. For best results, remove the As from
the 3’ end as shown below:
>TeloPrimeModified_5p
TGGATTGATATGTAATACGACTCACTATAG
>TeloPrimeModified_3p
CGCCTGAGA
3. Use the --isoseq mode. Note that this cannot be combined with the
--guess option.
4. The output will be only different pairs with a 5p and 3p combination:
demux.5p--tissue1_3p.bam
demux.5p--tissue2_3p.bam
The --isoseq parameter set is very conservative for removing any
spurious and ambiguous calls, and guarantees that only proper
asymmetric (barcoded) primer are used in downstream analyses. Good
libraries reach >75% CCS reads passing the Demultiplex Barcodes filters.
BAM tags
In SMRT Link v11.0, LB and SM tags are set by the user in Run Design. The
SM tag can also be set in Demultiplex Barcodes in SMRT Analysis.

Page 34
Non-demultiplex case:
•
LB: Well Sample Name.
•
SM: Bio Sample Name.
Multiplexed case, BAM pre-demultiplexing:
•
LB: Well Sample Name.
•
SM: Tag removed.
Multiplexed case, BAMs post-demultiplexing:
•
LB: Well Sample Name for all child barcode BAMs.
•
SM: Each individual Bio Sample Name for the specific barcode.
•
BC: Barcode sequence or hyphenated barcode sequences of the pair.
•
DS: Appends barcode information used in demultiplexing: BarcodeFile,
BarcodeHash, BarcodeCount, BarcodeMode, BarcodeQuality.
• Example read group header after demultiplexing:
@RG
ID:66d5a6af/3--3
PL:PACBIO
DS:READTYPE=SUBREAD;
Ipd:CodecV1=ip;
PulseWidth:CodecV1=pw;
BINDINGKIT=101-500-400;
SEQUENCINGKIT=101-427-800;
BASECALLERVERSION=5.0.0;
FRAMERATEHZ=100.000000;
BarcodeFile=Sequel_16_barcodes_v3.barcodeset.xml;
BarcodeHash=f2b1fa0b43eb6ccbb30749883bb550e3;
BarcodeCount=16;
BarcodeMode=Symmetric;
BarcodeQuality=Score
PU:m54010_200212_162236
SM:MySampleName
PM:SEQUEL
BC:ACAGTCGAGCGCTGCGT
Page 35
export-datasets
The export-datasets tool takes one or more PacBio Data Set XML files
and packages all contents (including index files and supplemental Data
Sets) into a single ZIP archive. Data Set resources, such as BAM files, are
reorganized and renamed to flatten the directory structure, avoid
redundant file writes, and convert all resource paths from absolute paths
to relative paths. Where multiple Data Sets are provided, the contents of
each is nested in a directory named after the
UniqueId attribute in the
XML.
The resulting archive is primarily intended to be directly imported into
SMRT Link using the Data Management interface, but it may also be
unpacked manually and used on the command line.
Usage
export-datasets [options] <dataset>...
Input files
• One or more PacBio Dataset XML files.
Output file
•One output ZIP file.
Examples
export-datasets m64001_200704_012345.subreadset.xml
export-datasets sample1.consensusreadset.xml sample2.consensusreadset.xml
\sample3.consensusreadset.xml -o barcoded_ccs.zip
export-datasets /opt/smrtlink/jobs/0000/0000001/0000001234/outputs/
mapped.alignmentset.xml
Options Description
-o, --output Name of output ZIP file. (Default = datasets_<timestamp>.zip)
--keep-parent-ref Keeps the reference to the parent Data Set when archiving a
demultiplexed child Data Set.
--no-scraps Excludes the scraps.bam file if present in the XML file.
-h, --help Displays help information and exits.
--log-file Writes the log to a file. (Default = stderr)
--log-level Specifies the log level; values are [ERROR,DEBUG, INFO, WARN].
(Default = WARN)
--logback Override all logger configuration using a specified logback.xml file.
--log2stdout If True, log output is displayed to the console. (Default = False)
--debug Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to ERROR. (Default = False)
--verbose Alias for setting the log level to INFO. (Default = False)

Page 36
export-job
The export-job tool packages a SMRT Link Analysis job for export to
another system, usually for reimportation into another SMRT Link
instance. All internal paths in job output files are converted from absolute
to relative paths, and many of the internal details of the Cromwell
workflows are omitted. The export is not a complete record of the job, but
rather a collection of job output files and metadata.
Note that
export-job will include any external Data Sets referenced in
output Data Sets inside the job, for example ReferenceSets associated
with mapped Data Sets, or BarcodeSets associated with demultiplexed
Data Sets. However, these Data Sets will not be imported along with the
job. The exported job does not include the input reads used to run the job;
these may be exported separately using the
export-datasets tool.
Important: Only SMRT Link v10.0 or later generates the necessary
metadata files for
export-job to save a full record of job execution. Jobs
created with older versions of SMRT Link will still be archived, but the
metadata will be empty and/or incorrect.
Usage
export-job [options] <job_dir>
Input
• A path to a job directory.
Output file
•One output ZIP file.
Examples
export-job /path/to/smrtlink/jobs-root/0000/0000000/0000000860 -o job860.zip
Options Description
<job_dir> Path to a SMRT Link job directory.
-o, --output Name of output ZIP file. (Default = job_<timestamp>.zip)
-h, --help Displays help information and exits.
--log-file Writes the log to a file. (Default = stderr)
--log-level Specifies the log level; values are [ERROR,DEBUG, INFO, WARN].
(Default = WARN)
--logback Override all logger configuration using a specified logback.xml file.
--log2stdout If True, log output is displayed to the console. (Default = False)
--debug Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to ERROR. (Default = False)
--verbose Alias for setting the log level to INFO. (Default = False)

Page 37
To reimport on another system:
pbservice import-job job860.zip

Page 38
gcpp
gcpp is a variant-calling tool provided by the GCpp package which provides
several variant-calling algorithms for PacBio sequencing data.
Usage
gcpp -j8 --algorithm=arrow \
-r lambdaNEB.fa \
-o variants.gff \
aligned_subreads.bam
This example requests variant-calling, using 8 worker processes and the
Arrow algorithm, taking input from the file
aligned_subreads.bam, using
the FASTA file
lambdaNEB.fa as the reference, and writing output to
variants.gff.
A particularly useful option is
--referenceWindow/-w; which allows the
variant-calling to be performed exclusively on a window of the reference
genome.
Input files
• A sorted file of reference-aligned reads in PacBio’s standard BAM
format.
• A FASTA file that follows the PacBio FASTA file convention. If
specifying an input FASTA file, a FASTA index file (
.fai) with the same
name and path is required. If the
.fai file is not supplied, gcpp exits
and displays an error message.
Note: The
--algorithm=arrow option requires that certain metrics be in
place in the input BAM file. It requires per-read SNR metrics, and the per-
base
PulseWidth metric for Sequel data.
The selected algorithm will stop with an error message if any features that
it requires are unavailable.
Output files
Output files are specified as comma-separated arguments to the -o flag.
The file name extension provided to the
-o flag is meaningful, as it
determines the output file format. For example:
gcpp aligned_subreads.bam -r lambda.fa -o myVariants.gff,myConsensus.fasta
will read input from aligned_subreads.bam, using the reference
lambda.fa, and send variant call output to the file myVariants.gff, and
consensus output to
myConsensus.fasta.
The file formats currently supported (using extensions) are:
•
.gff: PacBio GFFv3 variants format; convertible to BED.
•
.vcf: VCF 4.2 variants format (that is compatible with v4.3.)
•
.fasta: FASTA file recording the consensus sequence calculated for
each reference contig.
Page 39
• .fastq: FASTQ file recording the consensus sequence calculated for
each reference contig, as well as per-base confidence scores.
Available algorithms
At this time there are three algorithms available for variant calling:
plurality, poa and arrow.
•
plurality is a simple and very fast procedure that merely tallies the
most frequent read base or bases found in alignment with each
reference base, and reports deviations from the reference as potential
variants. This approach is prone to insertion and deletion errors.
•
poa uses the partial order alignment algorithm to determine the
consensus sequence. It is a heuristic algorithm that approximates a
multiple sequence alignment by progressively aligning sequences to
an existing set of alignments.
•
arrow uses the per-read SNR metric and the per-pulse pulsewidth
metric as part of its likelihood model.
Confidence values
The arrow and plurality algorithms make a confidence metric available
for every position of the consensus sequence. The confidence should be
interpreted as a phred-transformed posterior probability that the
consensus call is incorrect; such as:
gcpp clips reported QV values at 93; larger values cannot be encoded in a
standard FASTQ file.
Options Description
-j Specifies the number of worker processes to use.
--algorithm= Specifies the variant-calling algorithm to use; values are plurality,
arrow and poa. (Default = arrow)
-r Specifies the FASTA reference file to use.
-o Specifies the output file format; values are .gff, .vcf, .fasta, and
.fastq.
--maskRadius When using the arrow algorithm, setting this option to a value N greater
than 0 causes gcpp to pass over the data a second time after masking out
regions of reads that have >70% errors in 2*N+1 bases. This setting has
little to no effect at low coverage, but for high-coverage datasets (>50X),
setting this parameter to 3 may improve final consensus accuracy. In rare
circumstances, such as misassembly or mapping to the wrong reference,
enabling this parameter may cause worse performance.
--minConfidence MINCONFIDENCE
-q MINCONFIDENCE
Specifies the minimum confidence for a variant call to be output to
variants.{gff,vcf} (Default = 40)
--minCoverage MINCOVERAGE
-x MINCOVERAGE
Specifies the minimum site coverage for variant calls and consensus to be
calculated for a site. (Default = 5)

Page 40
Chemistry specificity
The --algorithm=arrow parameter is trained per-chemistry. arrow
identifies the sequencing chemistry used for each run by looking at
metadata contained in the input BAM data file. This behavior can be
overridden by a command-line option.
When multiple chemistries are represented in the reads in the input file,
the Arrow will model reads appropriately using the parameter set for its
chemistry, thus yielding optimal results.

Page 41
Genome
Assembly
The Genome Assembly application generates de novo assemblies using
HiFi reads. The application is fast, produces contiguous assemblies, and
is suitable for genomes of any size.
The Genome Assembly application is powered by the IPA HiFi genome
assembler and includes the following features:
• Separates haplotypes during assembly using a novel phasing stage
(Nighthawk).
• Polishes the contigs with phased reads using
Racon.
• Improves haplotype separation using the
purge_dups tool.
Workflow of the Genome Assembly application
Analysis steps are highly optimized to produce assemblies of large
genomes efficiently.
The workflow consists of seven stages:
1. Sequence database construction.
2. Fast overlap computation using the
Pancake tool.
3. A dedicated phasing stage using the
Nighthawk tool.
4. Filtering chimeras and residual repeats.
5. Layout based on the string graph.
6. Polishing using the
Racon tool.
7. Purging haplotype duplicates from the primary assembly using the
third-party tool
purge_dups.
The workflow accepts HiFi XML Data Sets as input.
IPA HiFi genome assembler
• Scales well on a cluster.
• The workflow has an embedded downsampling feature:
– If the genome size and the desired coverage are specified, the initial
stage (sequence database construction) downsamples the input
Data Set to the desired coverage.
– Otherwise, the full coverage is used.
Usage
The Genome Assembly application is run using the pbcromwell run
command, with the
pb_assembly_hifi parameter to specify the
application. See “pbcromwell” on page 75 for details.

Page 42
To view information on the available Genome Assembly options, enter:
pbcromwell show-workflow-details pb_assembly_hifi
The minimum command needed to run the workflow requires the input
and the number of threads. The following example uses 16 threads:
pbcromwell run pb_assembly_hifi -e <input.xml> --nproc 16
The following example performs an assembly using an input XML Data
Set, and uses all default settings, including 1 CPU:
pbcromwell run pb_assembly_hifi -e <input.consensusreadset.xml>
Note: The default options for this workflow are equivalent to the following
command:
pbcromwell run pb_assembly_hifi \
-e <input.consensusreadset.xml> \
--task-option reads=None \
--task-option ipa2_genome_size=0 \
--task-option ipa2_downsampled_coverage=0 \
--task-option ipa2_advanced_options="" \
--task-option ipa2_run_polishing=True \
--task-option ipa2_run_phasing=True \
--task-option ipa2_run_purge_dups=True \
--task-option ipa2_ctg_prefix="ctg." \
--task-option ipa2_reads_db_prefix="reads" \
--task-option ipa2_cleanup_intermediate_files=True \
--task-option dataset_filters="" \
--task-option filter_min_qv=20 \
--nproc 8
The default options for this workflow should work well for any genome
types.
If the assembly is run on a single local node with high CPU count, such as
64 cores, we recommend that the job submission for
pbcromwell is
configured so that it uses 4 concurrent jobs and 16 threads per job.
We found this to be more efficient than using 64 threads and 1 concurrent
job, as many steps are very data I/O-dependent.
You can apply a similar principle for compute environments with more or
fewer cores. For example, for a machine with 80 cores, one can use 20
threads and 4 concurrent jobs.
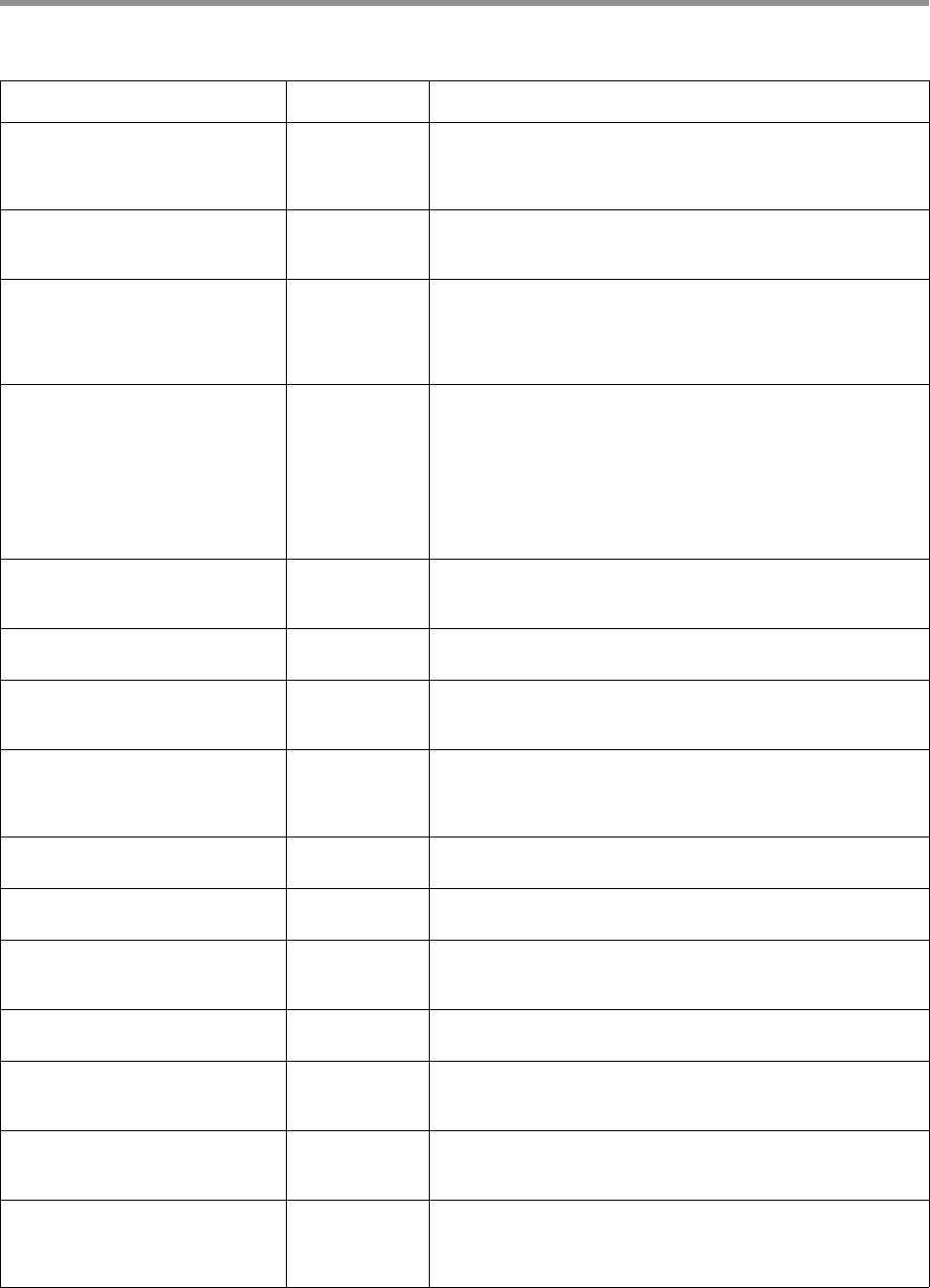
Page 43
Genome Assembly parameters input files
Option Default value Description
-e, --eid_ccs NONE Optional parameter, required if --task-option reads
<input> is not specified. This is a SMRT Link-specific input
parameter and supports only PacBio Consensusreadset
XML files as input.
--task-option reads NONE Optional parameter, required if -e <input> is not specified.
Supports multiple input formats: FASTA, FASTQ, BAM, XML,
FOFN and gzipped versions of FASTA/FASTQ.
--task-option
ipa2_genome_size
0 The approximate number of base pairs expected in the
genome. This is used only for downsampling; if the value is ≤
0, downsampling is disabled. Note: It is better to slightly
overestimate rather than underestimate the genome length to
ensure good coverage across the genome.
--task-option
ipa2_downsampled_coverage
0 The input Data Set can be downsampled to a desired
coverage, provided that both the
ipa2_downsampled_coverage and ipa2_genome_size
options are specified and >0.
Downsampling applies to the entire assembly process,
including polishing.
This parameter selects reads randomly, using a fixed random
seed for reproducibility.
--task-option
ipa2_advanced_options
NONE A semicolon-separated list of KEY=VALUE pairs. New line
characters are not accepted. (These are described later in
this document.)
--task-option
ipa2_run_polishing
TRUE Enables or disables the polishing stage of the workflow.
Polishing can be disabled to perform fast draft assemblies.
--task-option
ipa2_run_phasing
TRUE Enables or disables the phasing stage of the workflow.
Phasing can be disabled to assemble haploid genomes, or to
perform fast draft assemblies.
--task-option
ipa2_run_purge_dups
TRUE Enables or disables identification of “duplicate” alternate
haplotype contigs which may be assembled in the primary
contig file, and moves them to the associate contig (haplotig)
file.
--task-option
ipa2_ctg_prefix
.ctg The prefix used to label the output generated contigs.
--task-option
ipa2_reads_db_prefix
reads The prefix of the sequence and seed databases which will be
used internally for assembly.
--task-option
ipa2_cleanup_intermediate
_files
TRUE Removes intermediate files from the run directory to save
space.
--task-option
dataset_filters
NONE (General pbcromwell option)
A semicolon-separated (not
comma-separated) list of other filters to add to the Data Set.
--task-option
filter_min_qv
20 (General pbcromwell option) Phred-scale integer QV cutoff
for filtering HiFi reads. The default for all applications is 20
(QV 20), or 99% predicted accuracy.
--task-option
downsample_factor
0 Downsampling factor applied directly to the input Data Set.
This parameter is not related to
ipa2_downsampled_coverage.
--task-option
mem_scale_factor
8 Controls the amount of requested memory for individual
Cromwell tasks in the workflow. The default value of 8 is
good for larger genomes, but it may be too much memory for
smaller genomes.
Page 44
• *.bam file containing PacBio data.
•
*.fasta or *.fastq file containing PacBio data.
•
*.xml file containing PacBio data.
•
*.fofn files with file names of files containing PacBio data.
Output files
• final_purged_primary.fasta file containing assembled primary
contigs.
•
final_purged_haplotigs.fasta file containing assembled
haplotigs.
Advanced parameters
Advanced parameters should be rarely modified. For the special cases
when that is required, advanced parameters are documented below.
Advanced parameters specified on the command line:
•Are in the form of
key = value pairs.
• Each pair is separated by a semicolon (
;) character.
• The full set of advanced parameters is surrounded by one set of
double quotes.
• The specified value of a parameter overwrites the default options for
that key. All desired options of that parameter must be explicitly listed,
not just the ones which should change from the default.
• Setting an empty value clears the parameter; it does not reset the
value back to default.
Example:
--task-option ipa2_advanced_options="config_seeddb_opt=-k 28;config_block_size=2048"
--config NONE (General pbcromwell option) Java configuration file for
running Cromwell.
--nproc 1 (General pbcromwell option) Number of processors, (except
per task).
Option Default value Description
Page 45
Complete list of available advanced parameters and default values:
Advanced parameters Default value Description
config_genome_size 0 The approximate number of base pairs expected in the
genome, used to determine the coverage cutoff. This is
only used for downsampling; 0 turns downsampling off.
Note: It is better to slightly overestimate rather than
underestimate the genome length to ensure good
coverage across the genome.
config_coverage 0 The input Data Set can be downsampled to a desired
coverage, provided that both the Downsampled
coverage and Genome Length parameters are
specified and above 0.
Downsampling applies to the entire assembly process,
including polishing. This feature selects reads randomly,
using a fixed random seed for reproducibility.
config_polish_run 1 Enables or disables the polishing stage of the workflow.
Polishing can be disabled to perform fast draft
assemblies. 0 disables this feature; 1 enables it.
config_phase_run 1 Enables or disables the phasing stage of the workflow.
Phasing can be disabled to assemble haploid genomes,
or to perform fast draft assemblies. 0 disables this
feature; 1 enables it.
config_purge_dups_run 1 Enables or disables the purge_dups stage of the
workflow. 0 disables this feature; 1 enables it.
config_autocomp_max_cov 1 If enabled, the maximum allowed overlap coverage at
either the 5’ or the 3’ end of every read is automatically
determined based on the statistics computed from the
overlap piles. This value is appended to the
config_ovl_filter_opt value internally, and
supersedes the manually specified --max-cov and
--max-diff values of that parameter. These options
are used to determine potential repeats and filter out
those reads before the string graph is constructed.
0 disables this feature; 1 enables it.
config_block_size 4096 The overlapping process is performed on pairs of blocks
of input sequences, where each block contains the
number of sequences which crop up to this size (in
Mbp). Note: The number of pairwise comparisons grows
quadratically with the number of blocks (meaning more
cluster jobs), but also the larger the block size the more
resources are required to execute each pairwise
comparison.
config_existing_db_prefix NONE Allows injection of an existing SeqDB, so that one
doesn’t have to be built from scratch. The provided
existing DB is symbolically linked and used for assembly.
(This option is intended for debugging purposes.)
Page 46
config_ovl_filter_opt --max-diff 80
--max-cov 100
--min-cov 2
--bestn 10
--min-len 4000
--gapFilt
--minDepth 4
--idt-stage2 98
Overlap filter options.
--gapFilt - Enables the chimera filter, which analyzes
each overlap pile, and determines whether a pread is
chimeric based on the local coverage across the pread.
--minDepth - Option for the chimera filter. The chimera
filter is ignored when a local region of a read has
coverage lower than this value.
The other parameters are:
--min-cov - Minimum allowed coverage at either the 5'
or the 3' end of a read. If the coverage is below this
value, the read is blacklisted and all of the overlaps it is
incident with are ignored. This helps remove potentially
chimeric reads.
--max-cov - Maximum allowed coverage at either the 5'
or the 3' end of a read. If the coverage is above this
value, the read is blacklisted and all of the overlaps it is
incident with are ignored. This helps remove repetitive
reads which can make tangles in the string graph. Note
that this value is a heuristic which works well for ~30x
seed length cutoff. If the cutoff is set higher, we advise
that this value be also increased. Alternatively, using the
autocompute_max_cov option can automatically
estimate the value of this parameter, which can improve
contiguity (for example, in cases when the input genome
size or the seed coverage were overestimated).
--max-diff - Maximum allowed difference between the
coverages at the 5' and 3' ends of any particular read. If
the coverage is above this value, the read is blacklisted
and all of the overlaps it is incident with are ignored. If the
autocompute_max_cov option is used, then the same
computed value is supplied to this parameter as well.
--bestn - Keep at most this many overlaps on the 5'
and the 3' side of any particular read.
--min-len - Filter overlaps where either A-read or the
B-read are shorter than this value.
--idt-stage2 - Filter overlaps with identity below 98%.
--high-copy-sample-rate - Controls the
downsampling of reads from high copy elements to the
expected coverage determined by maxCov*rate, where
rate is the value of this parameter. If rate is 0, then
these high coverage reads are discarded.
config_ovl_min_idt 98 The final overlap identity threshold. Applied during the
final filtering stage, right before the overlaps are passed
to the layout stage.
config_ovl_min_len 1000 The minimum length of either A-read or a B-read to keep
the overlap. Applied during the final filtering stage, right
before the overlaps are passed to the layout stage.
config_ovl_opt --one-hit-per-
target
--min-idt 96
Overlapping options for the pancake overlapping tool.
The options set by this parameter here are passed
directly to pancake. For details on pancake options,
use pancake -h.
The defaults used here are: --one-hit-per-target
which keeps only the best hit in case there are multiple
possible overlaps between a pair of reads (tandem
repeats); and --min-idt 96 which will filter out any
overlap with identity lower than 96%.
config_phasing_opt
NONE Options for the phasing tool nighthawk. The options
set by this parameter are passed directly to nighthawk.
For details on nighthawk options, use nighthawk -h.
Advanced parameters Default value Description
Page 47
config_phasing_split_opt --split-type
noverlaps
--limit 3000000
Options that control the chunking of the phasing jobs,
and through that regulate the time and memory
consumption of each individual chunk.
The defaults are: --split-type noverlaps which
splits the chunks by the number of overlaps; and
--limit 3000000 which allow at most approximately
3 million overlaps per chunk.
Empirically, the current defaults keep the maximum
memory consumption (RSS) of the phasing jobs under 4
GB per chunk.
config_seeddb_opt -k 28
-w 120
--space 1
Options to control the seed computation. These options
are passed directly to the pancake seeddb command.
Defaults: -k 28 is the k-mer size of 28 bp; -w 120 is the
minimizer window size of 120 bp; and --space 1
specifies the spacing for spaced seed construction, with
1 gap in between every two bases of the seed.
For more details on these and other options, use
pancake seeddb -h.
config_seqdb_opt --compression 1 Options to control the construction of the sequence
database. These options are passed directly to the
pancake seqdb command.
Current default is --compression 1 which turns on the
2-bit encoding compression of the sequences.
For more details on these and other options, use
pancake seqdb -h.
config_use_hpc 0 This parameter enables (1) or disables (0) an
experimental Homopolymer Compression feature.
If this feature is enabled, the overlaps are computed
from homopolymer-compressed sequences. The layout
stage is somewhat slower because the sequences have
to be aligned to determine the correct homopolymer-
expanded coordinates.
config_use_seq_ids 1 This feature is mostly useful for debugging purposes. If
0 is specified, then the overlaps contain original
sequence names instead of their numerical IDs.
The default of 1 uses the numerical IDs to represent
reads, which uses memory much more efficiently.
config_purge_map_opt --min-map-len
1000
--min-idt 98.0
--bestn 5
This option is used to control the mapping of the reads to
contigs for the purge_dups tool. The mapper used is
pancake, and the options set by this parameter are used
directly by pancake. For details on pancake options,
use pancake -h.
Option --min-map-len 1000 removes any alignment
which did not span more than 1000 bp during the
mapping process; --min-idt 98.0 removes any
alignment with identity below 98.0%, and --bestn 5
keeps at most 5 top scoring alignments for each query
read (one primary alignment and at most 4 secondary
alignments).
Advanced parameters Default value Description
Page 48
config_purge_dups_calcuts NONE The third-party tool purge_dups can accept user-
defined cutoffs for purging. On some genomes, the
automated computation of the cutoffs in purge_dups
can result in suboptimal values, and in this case a user
can specify them manually.
This option is passed directly to the
purge_dups_calcuts tool. For details on the possible
values that can be passed to this tool, use
ipa_purge_dups_calcuts without parameters.
Relevant parameters include:
-l INT Lower bound for read depth.
-m INT Transition between haploid and diploid.
-u INT Upper bound for read depth.
config_m4filt_high_copy_s
ample_rate
1.0 This option is passed to the --high-copy-sample-
rate parameter of the overlap filter, which controls the
downsampling of reads from high copy elements to the
expected coverage determined by maxCov*rate, where
rate is the value of this parameter. If 0, then these high
coverage reads are discarded.
Note: This parameter supersedes the
config_ovl_filter_opt options.
config_max_polish_block_m
b
100 During the polishing stage, contigs are grouped into
chunks of approximate size specified by this parameter
(in megabases). Each chunk is processed separately and
in parallel (depending on the system configuration).
config_layout_opt NONE This value is passed directly to the assembly layout
stage. To get a list of valid options for this parameter,
enter ipa2_ovlp_to_graph -h on the command line.
Advanced parameters Default value Description

Page 49
HiFiViral SARS-
CoV-2 Analysis
Use this application to analyze multiplexed samples sequenced with the
HiFiViral SARS-CoV-2 Kit. For each sample, this analysis provides:
• Consensus sequence (FASTA).
• Variant calls (VCF).
• HiFi reads aligned to the reference (BAM).
• Plot of HiFi read coverage depth across the SARS-CoV-2 genome.
Across all samples, this analysis provides:
• Job summary table including passing sample count at 90 and 95%
genome coverage.
• Sample summary table including, for each sample: Count of variable
sites, genome coverage, read coverage, and probability of multiple
strains, and other metrics.
• Plate QC graphical summary of performance across samples in assay
plate layout.
• Plot of HiFi read depth of coverage for all samples.
Notes:
• The application accepts HiFi reads (BAM format) as input. HiFi reads
are reads generated with CCS analysis that have a quality value equal
to or greater than Phred-scaled Q20.
• This application is for SARS-CoV-2 analysis only and is not
recommended for other viral studies. The Wuhan reference genome is
included with SMRT Link and used by default, but advanced users may
specify other reference genomes. We have not tested the application
with reference genomes other than the Wuhan reference genome.
• The application is intended to identify variable sites and call a single
consensus sequence per sample. The output consensus sequence is
produced based on the dominant variant observed. Minor variant
information that passes through a default threshold may be encoded
in the raw VCF, but does not get propagated into the consensus
sequence FASTA.
• The HiFiViral SARS-CoV-2 Analysis application can be run using the
Auto Analysis feature available in Run Design. This feature allows
users to complete all necessary analysis steps immediately after
sequencing without manual intervention. The Auto Analysis workflow
includes CCS, Demultiplex Barcodes, and HiFiViral SARS-CoV-2
Analysis.
HiFiViral SARS-CoV-2 application workflow
1. Process the reads using the
mimux tool to trim the probe arm
sequences.
2. Align the reads to the reference genome using
pbmm2.
3. Call and filter variants using
bcftools, generating the raw variant calls
in VCF file format. Filtering in this step removes low-quality calls (less
than Q20), and normalizes indels.

Page 50
4. Filter low-frequency variants using vcfcons and generate a consensus
sequence by injecting variants into the reference genome. At each
position, a variant is called only if both the base coverage exceeds the
minimum base coverage threshold (Default = 4) and the fraction of
reads that support this variant is above the minimum variant frequency
threshold (Default = 0.5). See here for details.
Preparing input data for the HiFiViral SARS-CoV-2 Analysis
application
1. Run the Demultiplex Barcodes cromwell workflow, where the input to
that application are HiFi reads, and the primers are multiplexed
barcode primers. See “Demultiplex Barcodes” on page 23 for details. If
HiFi reads have not been generated on the instrument, run CCS analy-
sis first. See “ccs” on page 7 for details.
– Provide the proper barcode sequences:
Barcoded M13 Primer Plate.
– Use the task option
lima_symmetric_barcodes=false. (The
barcode pairs are asymmetric.)
• Provide the correctly-formatted barcode pair-to-Bio Sample CSV file.
For details, see the
--task-option sample_wells_csv option in the
table further down this page.
Input files
• movie.consensusreadset.xml: Previously-demultiplexed HiFi reads,
packaged as separate BAM files wrapped in an XML Data Set. (See
“Preparing input data for the HiFiViral SARS-CoV-2 Analysis
application” on page 50 for details.)
•
sars_cov2.referenceset.xml: The Wuhan reference genome.
•
HiFiViral_SARS-CoV-2_Enrichment_Probes.barcodeset.xml:
Dataset XML specifying the probe sequence file in FASTA format.
•[Optional]
Plate QC csv: Four-column CSV file that includes barcode,
biosample, plateID and wellID.
To specify this optional file, add the following option to
pbservice:
--task-option sample_wells_csv=<path to CSV file>
Output files
• pb_sars_cov2_kit.probe_counts_zip: Zipped TSV files with probe
counts, per sample.
•
pb_sars_cov2_kit.variants_csv: CSV variant calls for all samples.
•
pb_sars_cov2_kit.vcf_zip: Zipped VCF files containing the final
variant calls, per sample.
•
pb_sars_cov2_kit.raw_vcf_zip: Zipped VCF files containing the
raw variant calls, per sample.
•
pb_sars_cov2_kit.fasta_zip: Zipped final consensus sequences,
by sample, in FASTA format. This is a single consensus sequence with
Ns for each sample.
•
pb_sars_cov2_kit.frag_fasta_zip: Zipped file of consensus
sequences, split on
Ns, in FASTA format, by sample.
Page 51
• pb_sars_cov2_kit.mapped_zip: Zipped BAM files containing the
output from mapping HiFi reads to the reference genome, by sample.
•
samples.consensus_mapped.bam.zip: Zipped BAM files containing
the output from mapping consensus FASTA files to the reference
genome, by sample.
•
samples.coverage.png.zip: Zipped per-sample coverage graphs in
png format.
•
hifi_reads.fastq.zip: Zipped file of per-sample trimmed HiFi reads
in FASTQ format.
•
sample_summary.csv: Sample Summary file in CSV format, with one
row per sample.
Running the SARS-CoV-2 Analysis application
pbcromwell run pb_sars_cov2_kit \
-e <movie.consensusreadset.xml> \
-e $SMRT_ROOT/current/bundles/smrtinub/current/private/pacbio/barcodes/ HiFiViral_SARS-
CoV-2_Enrichment_Probes.barcodeset.xml \
-e eid_ref_dataset_2:$SMRT_ROOT/current/bundles/smrtinub/current/private/pacbio/
canneddata/referenceset/SARS-CoV-2/sars_cov2.referenceset.xml
--task-option min_alt_freq=0.5 \
--task-option min_bq=80 \
--task-option mimux_overrides="--max-len=800 --same" \
--task-option sample_wells_csv=None \
--config cromwell.conf \
--nproc 8
Options Description
--task-option
sample_wells_csv
Specifies a 4-column CSV file used to generate the Plate QC Report, which
displays analysis results for each sample in the assay plate. The CSV file
must contain barcode pairs, Bio Sample name, Plate IDs, and Well IDs. The
report is useful for diagnosing sample issues based on plate location.
(Default = None)
--task-option min_coverage Specifies the minimum read coverage. Below this value, the consensus
sequence will be set to Ns and no variants are called. (Default = 4)
--task-option min_alt_freq Specifies that only variants whose frequency is greater than this value are
reported. This frequency is determined based on the read depth (DP) and
allele read count (AD) information in the VCF output file. We recommend
using the default value to properly call the dominant alternative variant
while also filtering out potential artifacts. (Default = 0.5)
--task-option min_bq Specifies that reads with barcode scores below this minimum value are
not included in analysis. (Default = 80)
--task-option mimux_overrides Specifies additional options to pass to the mimux preprocessing tool for
trimming and filtering reads by probe sequences. Options should be
entered in space-separated format.
Available options include:
--max-len: Specifies the maximum sequence length. (Default = 800)
--same: Specifies that only reads with arms sequences from the same
probe are used.
--task-option probes_fasta Specifies a FASTA file containing probes sequences if using probes other
than those supplied with the HiFiViral SARS-CoV-2 kit.

Page 52
ipdSummary
The ipdSummary tool detects DNA base-modifications from kinetic
signatures. It is part of the
kineticsTool package.
kineticsTool loads IPDs observed at each position in the genome,
compares those IPDs to value expected for unmodified DNA, and outputs
the result of this statistical test. The expected IPD value for unmodified
DNA can come from either an in-silico control or an amplified control. The
in-silico control is trained by PacBio and shipped with the package. It
predicts the IPD using the local sequence context around the current
position. An amplified control Data Set is generated by sequencing
unmodified DNA with the same sequence as the test sample. An amplified
control sample is usually generated by whole-genome amplification of the
original sample.
Modification detection
The basic mode of kineticsTool does an independent comparison of IPDs
at each position on the genome, for each strand, and outputs various
statistics to CSV and GFF files (after applying a significance filter).
Modifications identification
kineticsTool also has a Modification Identification mode that can decode
multi-site IPD “fingerprints” into a reduced set of calls of specific
modifications. This feature has the following benefits:
• Different modifications occurring on the same base can be
distinguished; for example, 6mA and 4mC.
• The signal from one modification is combined into one statistic,
improving sensitivity, removing extra peaks, and correctly centering
the call.
Algorithm: Synthetic control
Studies of the relationship between IPD and sequence context reveal that
most of the variation in mean IPD across a genome can be predicted from
a 12-base sequence context surrounding the active site of the DNA
polymerase. The bounds of the relevant context window correspond to the
window of DNA in contact with the polymerase, as seen in DNA/
polymerase crystal structures. To simplify the process of finding DNA
modifications with PacBio data, the tool includes a pre-trained lookup
table mapping 12-mer DNA sequences to mean IPDs observed in C2
chemistry.
Algorithm: Filtering and trimming
kineticsTool uses the Mapping QV generated by pbmm2 and stored in the
cmp.h5 or BAM file (or AlignmentSet) to ignore reads that are not
confidently mapped. The default minimum Mapping QV required is 10,
implying that
pbmm2 has 90% confidence that the read is correctly mapped.
Because of the range of read lengths inherent in PacBio data, this can be
changed using the
--mapQvThreshold option.

Page 53
There are a few features of PacBio data that require special attention to
achieve good modification detection performance.
kineticsTool
inspects the alignment between the observed bases and the reference
sequence for an IPD measurement to be included in the analysis. The
PacBio read sequence must match the reference sequence for
k around
the cognate base. In the current module,
k=1. The IPD distribution at some
locus can be thought of as a mixture between the “normal” incorporation
process IPD, which is sensitive to the local sequence context and DNA
modifications, and a contaminating “pause” process IPD, which has a
much longer duration (mean > 10 times longer than normal), but happen
rarely (~1% of IPDs).
Note: Our current understanding is that pauses do not carry useful
information about the methylation state of the DNA; however a more
careful analysis may be warranted. Also note that modifications that
drastically increase the roughly 1% of observed IPDs are generated by
pause events. Capping observed IPDs at the global 99
th
percentile is
motivated by theory from robust hypothesis testing. Some sequence
contexts may have naturally longer IPDs; to avoid capping too much data
at those contexts, the cap threshold is adjusted per context as follows:
capThreshold = max(global99, 5*modelPrediction,
percentile(ipdObservations, 75))
Algorithm: Statistical testing
We test the hypothesis that IPDs observed at a particular locus in the
sample have longer means than IPDs observed at the same locus in
unmodified DNA. If we have generated a Whole Genome Amplified Data
Set, which removes DNA modifications, we use a case-control, two-
sample t-test. This tool also provides a pre-calibrated “synthetic control”
model which predicts the unmodified IPD, given a 12-base sequence
context. In the synthetic control case we use a one-sample t-test, with an
adjustment to account for error in the synthetic control model.
Usage
To run using a BAM input, and output GFF and HDF5 files:
ipdSummary aligned.bam --reference ref.fasta m6A,m4C --gff basemods.gff \
--csv_h5 kinetics.h5
To run using cmp.h5 input, perform methyl fraction calculation, and output
GFF and CSV files:
ipdSummary aligned.cmp.h5 --reference ref.fasta m6A,m4C --methylFraction \
--gff basemods.gff --csv kinetics.csv

Page 54
Input files
• A standard PacBio alignment file - either AlignmentSet XML, BAM, or
cmp.h5 - containing alignments and IPD information.
• Reference sequence used to perform alignments. This can be either a
FASTA file or a ReferenceSet XML.
Output files
The tool provides results in a variety of formats suitable for in-depth
statistical analysis, quick reference, and consumption by visualization
tools. Results are generally indexed by reference position and reference
strand. In all cases the strand value refers to the strand carrying the
modification in the DNA sample. Remember that the kinetic effect of the
modification is observed in read sequences aligning to the opposite
strand. So reads aligning to the positive strand carry information about
modification on the negative strand and vice versa, but the strand
containing the putative modification is always reported.
•
modifications.gff: Compliant with the GFF Version 3 specification.
Each template position/strand pair whose probability value exceeds
the probability value threshold appears as a row. The template
position is 1-based, per the GFF specifications. The strand column
refers to the strand carrying the detected modification, which is the
opposite strand from those used to detect the modification. The GFF
confidence column is a Phred-transformed probability value of
detection.
The auxiliary data column of the GFF file contains other statistics
which may be useful for downstream analysis or filtering. These
include the coverage level of the reads used to make the call, and +/-
20 bp sequence context surrounding the site.
•
modifications.csv: Contains one row for each (reference position,
strand) pair that appeared in the Data Set with coverage at least
x.
x defaults to 3, but is configurable with the --minCoverage option. The
reference position index is 1-based for compatibility with the GFF file
in the R environment. Note that this output type scales poorly and is
not recommended for large genomes; the HDF5 output should perform
much better in these cases.
Output options Description
--gff FILENAME GFF format.
--csv FILENAME Comma-separated value format.
--bigwig FILENAME BigWig file format.
Page 55
Output columns: In-silico control mode
Output columns: Case control mode
Column Description
refId Reference sequence ID of this observation.
tpl 1-based template position.
strand Native sample strand where kinetics were generated. 0 is the strand of the original
FASTA, 1 is opposite strand from FASTA.
base The cognate base at this position in the reference.
score Phred-transformed probability value that a kinetic deviation exists at this position.
tMean Capped mean of normalized IPDs observed at this position.
tErr Capped standard error of normalized IPDs observed at this position
(standard deviation/sqrt(coverage)).
modelPrediction Normalized mean IPD predicted by the synthetic control model for this sequence
context.
ipdRatio tMean/modelPrediction.
coverage Count of valid IPDs at this position.
frac Estimate of the fraction of molecules that carry the modification.
fracLow 2.5% confidence bound of the frac estimate.
fracUpp 97.5% confidence bound of the frac estimate.
Column Description
refId Reference sequence ID of this observation.
tpl 1-based template position.
strand Native sample strand where kinetics were generated. 0 is the strand of the
original FASTA, 1 is opposite strand from FASTA.
base The cognate base at this position in the reference.
score Phred-transformed probability value that a kinetic deviation exists at this
position.
caseMean Mean of normalized case IPDs observed at this position.
controlMean Mean of normalized control IPDs observed at this position.
caseStd Standard deviation of case IPDs observed at this position.
controlStd Standard deviation of control IPDs observed at this position.
ipdRatio tMean/modelPrediction.
testStatistic T-test statistic.
coverage Mean of case and control coverage.
controlCoverage Count of valid control IPDs at this position.
caseCoverage Count of valid case IPDs at this position.

Page 56
isoseq3
The isoseq3 tool enables analysis and functional characterization of
transcript isoforms for sequencing data generated on PacBio instruments.
The analysis can be performed de novo, without a reference genome. If a
reference genome is available, an optional collapse step to produce
unique isoform files based on genomic coordinates is available. The input
to
isoseq3 tools should be HiFi (CCS) reads in BAM format.
Usage
isoseq3 <tool>
Typical workflow
1. Visualize primers, then remove primers and demultiplex:
cat primers.fasta
>5p
GCAATGAAGTCGCAGGGTTGGGG
>3p
GTACTCTGCGTTGATACCACTGCTT
lima movie.ccs.bam primers.fasta demux.bam --isoseq
See “Demultiplex Barcodes” on page 23 for details on the lima tool.
2. Identify and remove polyA tails; also remove artificial concatemers.
The output are full-length, non-concatemer (FLNC) reads:
isoseq3 refine demux.5p--3p.bam primers.fasta flnc.bam --require-polya
3. Cluster FLNC reads at the isoform levels to generate consensus tran-
script isoform sequences. This generates
unpolished.hq.bam and
unpolished.hq.fasta.gz files, which are the high-quality (HQ)
transcripts that should be analyzed further.
isoseq3 cluster flnc.bam unpolished.bam --use-qvs
4. (Optional) Map transcripts to the genome and collapse HQ transcripts
based on genomic mapping:
pbmm2 align unpolished.bam reference.fasta aligned.sorted.bam --preset ISOSEQ --sort
isoseq3 collapse aligned.sorted.bam out.gff or
isoseq3 collapse aligned.sorted.bam movie.ccs.bam out.gff
See “pbmm2” on page 84 for details.
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits
Page 57
refine
Tool: Remove polyA and concatemers from full-length (FL) reads
and generate full-length non-concatemer (FLNC) transcripts (FL to FLNC).
Usage
isoseq refine [options] <ccs.demux.bam|xml> <primer.fasta|xml> <flnc.bam|xml>
cluster Tool: Cluster FLNC reads and generate transcripts.
Usage
isoseq3 cluster [options] input output
Example
isoseq3 cluster flnc.bam unpolished.bam --use-gvs
Custom BAM tags
isoseq3 cluster adds the following custom PacBio tags to the output
BAM file:
•
ib: Barcode summary: triplets delimited by semicolons, each triplet
contains two barcode indices and the ZMW counts, delimited by
commas. Example:
0,1,20;0,3,5
• im: ZMW names associated with this isoform.
Inputs/outputs Description
ccs.demux.bam|xml Input demultiplexed CCS reads BAM or ConsensusReadSet XML file.
This is usually the output from running lima with the --isoseq option,
such as demux.5p--3p.bam.
primer.fasta|xml Input primer FASTA or BarcodeSet XML file.
flnc.bam|xml Output FLNC BAM or ConsensusReadSet XML file.
Preprocessing Description
--min-polya-length Specifies the minimum poly(A) tail length. (Default = 20)
--require-polya Requires reads to have a poly(A) tail and remove it.
Options Description
--help, -h Displays help information and exits.
--version Displays program version number and exits.
--verbose, -v Sets the verbosity level.
-j,--num-threads Specifies the number of threads to use when processing; 0 means
autodetection. (Default = 0)
--log-file Writes the log to a file. (Default = stderr)
--log-level Specifies the log level; values are [DEBUG, INFO, WARN, TRACE, FATAL].
(Default = WARN)
Page 58
• is: Number of ZMWs associated with this isoform.
summarize Tool: Create a .csv-format barcode overview from transcripts.
Usage
isoseq3 summarize [options] input output
Example
isoseq3 summarize unpolished.bam summary.csv
Inputs/outputs Description
input flnc.bam file or movie.consensusreadset.xml file.
output unpolished.bam file prefix (the actual output files will be
unpolished.hq.bam and unpolished.lq-bam) or
unpolished.transcriptset.xml file.
Options Description
--s1 Specifies the number of seeds for minimer-only clustering.
(Default = 1000)
--s2 Specifies the number of seeds for DP clustering. (Default = 1000)
--poa-cov Specifies the maximum number of CCS reads used for POA consensus.
(Default = 10)
--use-qvs Use CCS analysis Quality Values; sets --poa-cov to 100.
--split-bam Splits BAM output files into a maximum of N files; 0 means no splitting.
(Default = 0)
--min-subreads-split Subread threshold for High-Quality/Low-Quality split; only works with
--use-qvs. (Default = 7)
--log-level Specifies the log level; values are [DEBUG, INFO, WARN, ERROR,
CRITICAL]. (Default = WARN)
-v,--verbose Uses verbose output.
-j,--num-threads Specifies the number of threads to use; 0 means autodetection.
(Default = 0)
--log-file Writes the log to a file. (Default = stdout)
Inputs/outputs Description
input unpolished.bam file. (The output from isoseq3 cluster.)
output summary.csv file.
Options Description
--log-level Specifies the log level; values are [DEBUG, INFO, WARN, ERROR,
CRITICAL]. (Default = WARN)
-v,--verbose Uses verbose output.
--log-file Writes the log to file. (Default = stdout)
Page 59
collapse
Tool: Collapse transcripts based on genomic mapping.
Usage
isoseq3 collapse [options] <alignments.bam|xml> <ccs.bam|xml> <out.fastq>
Examples:
isoseq3 collapse aligned.sorted.bam out.gff
or
isoseq3 collapse aligned.sorted.bam ccs.bam out.gff
Inputs/outputs Description
alignments Alignments mapping transcripts to the reference genome. (BAM or XML
file).
ccs.bam Optional input BAM file containing CCS reads.
out.fastq Collapsed transcripts in FASTQ format.
Options Description
--min-aln-coverage Ignores alignments with less than the Minimum Query Coverage.
(Default = 0.95)
--min-aln-identity Ignores alignments with less than the Minimum Alignment Identity.
(Default = 0.50)
--max-fuzzy-junction Ignores mismatches or indels shorter than or equal to N. (Default = 5)
--version Displays program version number and exits.
--log-file Writes the log to file. (Default = stderr)
--log-level Specifies the log level; values are [DEBUG, INFO, WARN, ERROR,
CRITICAL]. (Default = WARN)
-j,--num-threads Specifies the number of threads to use; 0 means autodetection.
(Default = 0)

Page 60
juliet
juliet is a general-purpose minor variant caller that identifies and phases
minor single nucleotide substitution variants in complex populations. It
identifies codon-wise variants in coding regions, performs a reference-
guided de novo variant discovery, and annotates known drug-resistance
mutations. Insertion and deletion variants are currently ignored; support
will be added in a future version. There is no technical limitation with
respect to the target organism or gene.
The underlying model is a statistical test, the Bonferroni-corrected Fisher's
Exact test. It compares the number of observed mutated codons to the
number of expected mutations at a given position.
juliet uses JSON target configuration files to define different genes in
longer reference sequences, such as overlapping open reading frames in
HIV. These predefined configurations ease batch applications and allow
immediate reproducibility. A target configuration may contain multiple
coding regions within one reference sequence and optional drug
resistance mutation positions.
Notes:
• The preinstalled target configurations are meant for a quick start. It is
the user's responsibility to ensure that the target configurations used
are correct and up-to-date.
• If the target configuration
none was specified, the provided reference
is assumed to be in-frame.
Performance
At a coverage of 6,000 CCS reads with a predicted accuracy (RQ) of
≥
0.99, the false positive and false negative rates are below 1% and
0.001% (10
-5
), respectively.
Usage
juliet --config "HIV" data.align.bam patientZero.html
Required Description
input_file.bam Input aligned BAM file containing CCS reads, which must be PacBio-
compliant, that is, cigar M is forbidden.
output_file.html Output report HTML file.
Configuration Description
--config,-c Path to the target configuration JSON file, predefined target configuration
tag, or the JSON string.
--mode-phasing,-p Phase variants and cluster haplotypes.
Page 61
Input files
• BAM-format files containing CCS reads. These must be PacBio-
compliant, that is,
cigar M is forbidden.
• Input CCS reads should have a minimal predicted accuracy of
0.99.
• Reads should be created with CCS analysis using the
--richQVs
option. Without the
--richQVs information, the number of false
positive calls might be higher, as
juliet is missing information to
filter actual heteroduplexes in the sample provided.
•
juliet currently does not demultiplex barcoded data; you must
provide one BAM file per barcode.
Output files
A JSON and/or HTML file:
juliet data.align.bam patientZero.html
juliet data.align.bam patientZero.json
juliet data.align.bam patientZero.html patientZero.json
The HTML file includes the same content as the JSON file, but in more
human-readable format. The HTML file contains four sections:
Restrictions Description
--region,-r Specifies the genomic region of interest; reads are clipped to that region.
Empty means all reads.
--drm-only,-k Only reports DRM positions specified in the target configuration. Can be
used to filter for drug-resistance mutations - only known variants from the
target configuration are called.
--min-perc,-m Specifies the minimum variant percentage to report.
Example: --min-perc 1 will only show variant calls with an observed
abundance of more than 1%. (Default = 0)
--max-perc,-n Specifies the maximum variant percentage to report.
Example: --max-perc 95 will only show variant calls with an observed
abundance of less than 95%. (Default = 100)
Chemistry override (specify both) Description
--sub,-s Specifies the substitution rate. Use to override the learned rate.
(Default = 0)
--del,-d Specifies the deletion rate. Use to override the learned rate. (Default = 0)
Options Description
--help, -h Displays help information and exits.
--verbose, -v Sets the verbosity level.
--version Displays program version number and exits.
--debug Returns all amino acids, irrespective of their relevance.
--mode-phasing,-p Phases variants and cluster haplotypes.
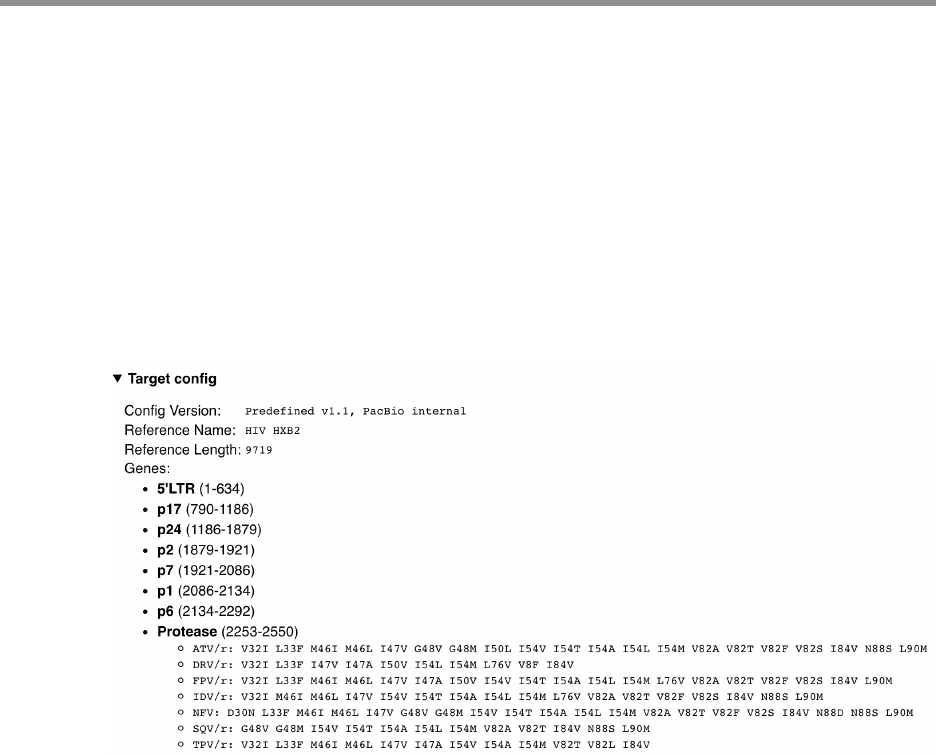
Page 62
1. Input data
Summarizes the data provided, the exact call for
juliet, and juliet
version for traceability purposes.
2. Target config
Summarizes details of the provided target configuration for traceability.
This includes the configuration version, reference name and length, and
annotated genes. Each gene name (in bold) is followed by the reference
start, end positions, and possibly known drug resistance mutations.
3. Variant discovery
For each gene/open reading frame, there is one overview table.
Each row represents a variant position.
• Each variant position consists of the reference codon, reference amino
acid, relative amino acid position in the gene, mutated codon,
percentage, mutated amino acid, coverage, and possible affected
drugs.
• Clicking the row displays counts of the multiple-sequence alignment
counts of the -3 to +3 context positions.

Page 63
4. Drug summaries
Summarizes the variants grouped by annotated drug mutations:
Predefined target configuration
juliet ships with one predefined target configuration, for HIV. Following
is the command syntax for running that predefined target configuration:
juliet --config "HIV" data.align.bam patientZero.html

Page 64
• Note: For the predefined configuration HIV, use the HIV HXB2
complete genome for alignment.
Customized target configuration
To define your own target configuration, create a JSON file. The root child
genes contains a list of coding regions, with begin and end, the name of
the gene, and a list of drug resistant mutations. Each DRM consists of its
name and the positions it targets. The
drms field is optional. If provided,
the
referenceSequence is used to call mutations, otherwise it will be
tested against the major codon. All indices are with respect to the
provided alignment space, 1-based, begin-inclusive and end-exclusive
[).
Target configuration: Example 1- A customized
json target configuration
file named
my_customized_hiv.json:
{
"genes": [
{
"begin": 2550,
"drms": [
{
"name": "fancy drug",
"positions": [ "M41L" ]
}
],
"end": 2700,
"name": "Reverse Transcriptase"
}
],
"referenceName": "my seq",
"referenceSequence": "TGGAAGGGCT...",

Page 65
"version": "Free text to version your config files"
"databaseVersion": "DrugDB version x.y.z (last updated YYYY-MM-DD)"
}
Run with a customized target configuration using the --config option:
juliet --config my_customized_hiv.json data.align.bam patientZero.html
Valid formats for DRMs/positions
103 Only the reference position.
M130 Reference amino acid and reference position.
M103L Reference aa, reference position, mutated aa.
M103LKA Reference aa, reference position, list of possible mutated aas.
103L Reference position and mutated aa.
103LG Reference position and list mutated aas.
Missing amino acids are processed as wildcard (*).
Example:
{ "name": "ATV/r", "positions": [ "V32I", "L33", "46IL",
"I54VTALM", "V82ATFS", "84" ] }
Target configuration: Example 2 - BCR-ABL:
For BCR-ABL, using the ABL1 gene with the following reference
NM_005157.5, a typical target configuration looks like this:
{
"genes": [
{
"name": "ABL1",
"begin": 193,
"end": 3585,
"drms": [
{
"name": "imatinib",
"positions": [
"T315AI","Y253H","E255KV","V299L","F317AICLV","F359CIV" ]
},
{
"name": "dasatinib",
"positions": [ "T315AI","V299L","F317AICLV" ]
},
{
"name": "nilotinib",
"positions": [ "T315AI","Y253H","E255KV","F359CIV" ]
},
{
"name": "bosutinib",
"positions": [ "T315AI" ]
}
]
}
],
"referenceName": "NM_005157.5",
"referenceSequence": "TTAACAGGCGCGTCCC..."

Page 66
No target configuration
If no target configuration is specified, either make sure that the sequence
is in-frame, or specify the region of interest to mark the correct reading
frame, so that amino acids are correctly translated. The output is labeled
with
unknown as the gene name:
juliet data.align.bam patientZero.html
Phasing
The default mode is to call amino-acid/codon variants independently.
Using the
--mode-phasing option, variant calls from distinct haplotypes
are clustered and visualized in the HTML output.
• The row-wise variant calls are "transposed" onto per-column
haplotypes. Each haplotype has an ID:
[A-Z]{1}[a-z]?.
• For each variant, colored boxes in this row mark haplotypes that
contain this variant.
• Colored boxes per haplotype/column indicate variants that co-occur.
Wild type (no variant) is represented by plain dark gray. A color palette
helps to distinguish between columns.
• The JSON variant positions has an additional
haplotype_hit boolean
array with the length equal to the number of haplotypes. Each entry
indicates if that variant is present in the haplotype. A haplotype block
under the root of the JSON file contains counts and read names. The
order of those haplotypes matches the order of all
haplotype_hit
arrays.

Page 67
There are two types of tooltips in the haplotype section of the table.
The first tooltip is for the Haplotypes % and shows the number of reads
that count towards (A) Actually reported haplotypes, (B) Haplotypes that
have less than 10 reads and are not being reported, and (C) Haplotypes
that are not suitable for phasing. Those first three categories are mutually
exclusive and their sum is the total number of reads going into
juliet.
For (C), the three different marginals provide insights into the sample
quality; as they are marginals, they are not exclusive and can overlap. The
following image shows a sample with bad PCR conditions:
The second type of tooltip is for each haplotype percentage and shows the
number of reads contributing to this haplotype:

Page 68
Microbial
Genome
Analysis
The Microbial Genome Analysis application is powered by the IPA HiFi
genome assembler and includes a base modification detection feature
performed after the assembly.
Workflow of the Microbial Genome Analysis application
The workflow consists of several steps around 2 main stages:
1. Chromosomal stage: Assemble large contigs using IPA, the HiFi
genome assembly tool.
2. Separate reads that were used for accurate large contigs from all other
reads.
3. Plasmid stage: Assemble plasmids (using IPA) from the reads sepa-
rated in the previous step.
4. De-duplicate plasmids.
5. Collect all contigs into a single FASTA file.
6. Rotate circular contigs.
7. Align the input Data Set to the assembled contigs.
8. Polish assembled contigs using
Racon.
9. Perform base modification detection.
The application accepts HiFi XML Data Sets as input, and has an
embedded downsampling feature:
• If the genome size and the desired coverage are specified, both stages
of assembly are downsampled, as with the Genome Assembly
application.
• Otherwise, the full coverage is used.
The embedded downsampling feature is not applied to the alignment
stage; all input reads will be aligned against the assembled contigs.
Usage
The Microbial Genome Analysis application is run using the pbcromwell
run command, with the
pb_microbial_analysis parameter to specify the
application. See “pbcromwell” on page 75 for details.
To view information on the available Microbial Genome Analysis options,
enter:
pbcromwell show-workflow-details pb_microbial_analysis
The minimum command needed to run the workflow requires the input
Data Set.
The following example performs assembly and base modification
detection using an input XML Data Set, and uses all default settings,
including 1 CPU:
pbcromwell run pb_microbial_analysis -e <input.consensusreadset.xml>
Page 69
Note: To specify different task options on the command line, consider the
following example:
pbcromwell run pb_microbial_analysis \
-e <input.consensusreadset.xml> \
--task-option ipa2_genome_size=0 \
--task-option ipa2_downsampled_coverage=0 \
--task-option microasm_plasmid_contig_len_max=300000
--task-option ipa2_cleanup_intermediate_files=True \
--task-option dataset_filters="" \
--task-option filter_min_qv=20 \
--nproc 8
The default options for this application should work well for any genome
type.
As microbes are relatively small, we rarely find much advantage to using
more than 4 threads, or more than 2 concurrent jobs.
Microbial Genome Analysis parameters
Option Default value Description
-e, --eid_ccs NONE This is a SMRT Link-specific input parameter and supports
only PacBio Consensusreadset XML files as input.
--task-option reads NONE Optional parameter, required if -e <input> is not specified.
Supports multiple input formats: FASTA, FASTQ, BAM, XML,
FOFN and gzipped versions of FASTA/FASTQ.
--task-option
ipa2_genome_size
10M The approximate number of base-pairs expected in the
chromosomal genome. Used only for downsampling in the
assembly stages (that is, not in polishing). If value <=0, then
downsampling is off.
Default: 10M (10 Mega basepairs).
Note: ipa2_genome_size is currently the only option that
accepts a metric suffix. Commas and decimals are not
accepted anywhere. ipa2_genome_size actually accepts a
String, which can be an integer followed by one of these
metric suffixes: k/M/G. For example: 4500k means “4,500
kilobases” or “4,500,000”. M stands for Mega and G stands for
Giga.
--task-option
ipa2_downsampled_coverage
100 The maximum coverage after downsampling with respect to
the estimated genome_size. (The default genome_size
was chosen to be larger than most realistic microbes.)
If value <=0, then downsampling is off.
Default: 100
Example: If your genome is 1G in length, and you specify
ipa2_genome_size=2G, you have over-estimated by 2x. If
you also specify ipa2_downsampled_coverage=100, your
data will be downsampled to 200x coverage, simply because
of the over-estimate. The cost of extra coverage is greater
runtime. The defaults are usually fine.
Page 70
--task-option
ipa2_advanced_options_chro
m
See Description
column
A semicolon-separated list of KEY=VALUE pairs. New line
characters are not accepted. (These are described later in
this document.)
config_block_size = 100;
config_seeddb_opt = -k 28 -w 20 --space 0 --use-
hpc-seeds-only;
config_ovl_opt = --one-hit-per-target --min-idt 98
--traceback --mask-hp --mask-repeats --trim --trim-
window-size 30 --trim-match-frac 0.75
--task-option
ipa2_advanced_options_plas
mid
See Description
column
A semicolon-separated list of KEY=VALUE pairs. New line
characters are not accepted. (These are described later in
this document.)
con-fig_block_size = 100;
con-fig_ovl_filter_opt = --max-diff 80 --max-cov
100 --min-cov 2 --bestn 10 --min-len 500 --gapFilt
--minDepth 4 --idt-stage2 98;
con-fig_ovl_min_len = 500;
con-fig_seeddb_opt = -k 28 -w 20 --space 0 --use-
hpc-seeds-only;
config_ovl_opt = --one-hit-per-target --min-idt 98
--min-map-len 500 --min-anchor-span 500 --traceback
--mask-hp --mask-repeats --trim --trim-window-size
30 --trim-match-frac 0.75 --smart-hit-per-target --
secondary-min-ovl-frac 0.05; con-fig_layout_opt = -
-allow-circular;
--task-option
ipa2_cleanup_intermediate_
files
TRUE Removes intermediate files from the run directory to save
space.
--task-option
microasm_plasmid_contig_le
n_max
300000 After the chromosomal stage, in task
filter_draft_contigs, separates long contigs
(presumed to be chromosomal) from shorter contigs (to be
re-assembled in the plasmid stage). Then, after the plasmid
stage, in dedup_plasmids, contigs less than this value are
ignored. The default value is usually fine.
--task-option
microasm_run_secondary_pol
ish
TRUE Specifies that an additional round of polishing will be applied
after the two stages of assembly have completed.
--task-option run_basemods TRUE Specifies that base modification analysis be performed.
--task-option
kineticstools_identify_mod
s
m4C,m6A Specify the base modifications to identify, in a comma-
separated list.
--task-option
kineticstools_p_value
0.001 Specifies the probability value cutoff for detecting base
modifications.
--task-option
motif_min_score
35 Specifies the minimum QMod score used to identify a motif.
--task-option
motif_min_fraction
0.30 Specifies the minimum methylated fraction to identify a motif.
--task-
optionrun_find_motifs
TRUE Specifies that motif-finding be performed.
--task-option
dataset_filters
NONE (General pbcromwell option) A semicolon-separated (not
comma-separated) list of other filters to add to the Data Set.
--task-option
filter_min_qv
20 (General pbcromwell option) Phred-scale integer QV cutoff
for filtering HiFi reads. The default for all applications is 20
(QV 20), or 99% predicted accuracy.
Option Default value Description
Page 71
--task-option
ipa2_advanced_options_chro
m
See Description
column
A semicolon-separated list of KEY=VALUE pairs. New line
characters are not accepted. (These are described later in
this document.)
config_block_size = 100;
config_seeddb_opt = -k 28 -w 20 --space 0 --use-
hpc-seeds-only;
config_ovl_opt = --one-hit-per-target --min-idt 98
--traceback --mask-hp --mask-repeats --trim --trim-
window-size 30 --trim-match-frac 0.75
--task-option
ipa2_advanced_options_plas
mid
See Description
column
A semicolon-separated list of KEY=VALUE pairs. New line
characters are not accepted. (These are described later in
this document.)
con-fig_block_size = 100;
con-fig_ovl_filter_opt = --max-diff 80 --max-cov
100 --min-cov 2 --bestn 10 --min-len 500 --gapFilt
--minDepth 4 --idt-stage2 98;
con-fig_ovl_min_len = 500;
con-fig_seeddb_opt = -k 28 -w 20 --space 0 --use-
hpc-seeds-only;
config_ovl_opt = --one-hit-per-target --min-idt 98
--min-map-len 500 --min-anchor-span 500 --traceback
--mask-hp --mask-repeats --trim --trim-window-size
30 --trim-match-frac 0.75 --smart-hit-per-target --
secondary-min-ovl-frac 0.05; con-fig_layout_opt = -
-allow-circular;
--task-option
ipa2_cleanup_intermediate_
files
TRUE Removes intermediate files from the run directory to save
space.
--task-option
microasm_plasmid_contig_le
n_max
300000 After the chromosomal stage, in task
filter_draft_contigs, separates long contigs
(presumed to be chromosomal) from shorter contigs (to be
re-assembled in the plasmid stage). Then, after the plasmid
stage, in dedup_plasmids, contigs less than this value are
ignored. The default value is usually fine.
--task-option
microasm_run_secondary_pol
ish
TRUE Specifies that an additional round of polishing will be applied
after the two stages of assembly have completed.
--task-option run_basemods TRUE Specifies that base modification analysis be performed.
--task-option
kineticstools_identify_mod
s
m4C,m6A Specify the base modifications to identify, in a comma-
separated list.
--task-option
kineticstools_p_value
0.001 Specifies the probability value cutoff for detecting base
modifications.
--task-option
motif_min_score
35 Specifies the minimum QMod score used to identify a motif.
--task-option
motif_min_fraction
0.30 Specifies the minimum methylated fraction to identify a motif.
--task-
optionrun_find_motifs
TRUE Specifies that motif-finding be performed.
--task-option
dataset_filters
NONE (General pbcromwell option) A semicolon-separated (not
comma-separated) list of other filters to add to the Data Set.
--task-option
filter_min_qv
20 (General pbcromwell option) Phred-scale integer QV cutoff
for filtering HiFi reads. The default for all applications is 20
(QV 20), or 99% predicted accuracy.
Option Default value Description
Page 72
Input files
• *.bam file containing PacBio data.
•
*.xml file containing PacBio data.
Output files
• final_assembly.fasta: File containing all assembled contigs,
rotated.
•
assembly.rotated.polished.renamed.fsa: File for NCBI, but
otherwise identical to
final_assembly.fasta; only the headers are
changed.
•
pb_microbial_analysis.motifs_csv: File containing motifs and
modifications.
Advanced assembly options
Note: Advanced parameters should be rarely modified. Advanced
parameters for the Microbial Genome Assembly workflow are the same as
those for the Genome Assembly workflow. See “Advanced parameters” on
page 44 for details.
Advanced parameters specified on the command line:
•Are in the form of
key = value pairs.
• Each pair is separated by a semicolon (
;) character.
• The full set of advanced parameters is surrounded by one set of
double quotes.
• The specified value of a parameter overwrites the default options for
that key. All desired options of that parameter must be explicitly listed,
not just the ones which should change from the default.
• Setting an empty value clears the parameter; it does not reset the
value back to default.
Example:
--task-option ipa2_advanced_options_chrom="config_seeddb_opt=-k
28;config_block_size=2048"
--task-option log_level INFO Specifies the logging (verbosity) level for tools which support
this feature. Values are: [TRACE, DEBUG, INFO, WARN,
FATAL]
--task-option nproc 1 Specified the number of CPU threads to use.
--task-option max_nchunks 40 Specifies the maximum number of chunks per task.
--task-option tmp_dir /tmp Specifies an optional temporary directory for used by several
subtools, such as sort.
Option Default value Description

Page 73
motifMaker
The motifMaker tool identifies motifs associated with DNA modifications
in prokaryotic genomes. Modified DNA in prokaryotes commonly arises
from restriction-modification systems that methylate a specific base in a
specific sequence motif. The canonical example is the m6A methylation
of adenine in GATC contexts in E. coli. Prokaryotes may have a very large
number of active restriction-modification systems present, leading to a
complicated mixture of sequence motifs.
PacBio SMRT sequencing is sensitive to the presence of methylated DNA
at single base resolution, via shifts in the polymerase kinetics observed in
the real-time sequencing traces. For more background on modification
detection, see here.
Algorithm
Existing motif-finding algorithms such as MEME-chip and YMF are sub-
optimal for this case for the following reasons:
• They search for a single motif, rather than attempting to identify a
complicated mixture of motifs.
• They generally don't accept the notion of aligned motifs - the input to
the tools is a window into the reference sequence which can contain
the motif at any offset, rather than a single center position that is
available with kinetic modification detection.
• Implementations generally either use a Markov model of the reference
(MEME-chip), or do exact counting on the reference, but place
restrictions on the size and complexity of the motifs that can be
discovered.
Following is a rough overview of the algorithm used by
motifMaker:
Define a motif as a set of tuples: (position relative to methylation, required
base). Positions not listed in the motif are implicitly degenerate. Given a
list of modification detections and a genome sequence, define the
following objective function on motifs:
Motif score(motif) = (# of detections matching motif) / (# of genome sites matching
motif) * (Sum of log-pvalue of detections matching motif) = (fraction methylated) * (sum
of log-pvalues of matches)
Then, search (close to exhaustively) through the space of all possible
motifs, progressively testing longer motifs using a branch-and-bound
search. The “fraction methylated” term must be less than 1, so the
maximum achievable score of a child node is the sum of scores of
modification hits in the current node, allowing pruning of all search paths
whose maximum achievable score is less than the best score discovered
so far.
Usage
Run the find command, and pass the reference FASTA and the
modifications.gff (.gz) file output by the PacBio modification detection
workflow.
Page 74
The reprocess subcommand annotates the GFF file with motif
information for better genome browsing.
MotifMaker [options] [command] [command options]
find Command: Run motif-finding.
find [options]
reprocess Command: Update a modifications.gff file with motif
information based on new Modification QV thresholds.
reprocess [options]
Output files
Using the find command:
• Output CSV file: This file has the same format as the standard Fields
included in motif_summary.csv described in the Methylome Analysis
White Paper.
Using the
reprocess command:
• Output GFF file: The format of the output file is the same as the input
file, and is described in the Methylome Analysis White Paper under
Fields included in the modifications.gff file.
Options Description
-h, --help Displays help information and exits.
* -f, --fasta Reference FASTA file.
* -g, --gff Modifications.gff or .gff.gz file.
-m, --minScore Specifies the minimum Qmod score to use in motif finding.
(Default = 40.0)
* -o, --output Outputs motifs.csv file.
-x, --xml Outputs motifs XML file.
Options Description
-c, --csv Raw modifications.csv file.
* -f, --fasta Reference FASTA file.
* -g, --gff Modifications.gff or .gff.gz file.
-m, --minFraction Specifies that only motifs above this methylated fraction are used.
(Default = 0.75)
-m, --motifs Motifs.csv file.
* -o, --output Reprocessed modifications.gff file.
Page 75
pbcromwell
The pbcromwell tool is PacBio’s wrapper for the cromwell scientific
workflow engine used to power SMRT Link. pbcromwell includes
advanced utilities for interacting directly with a Cromwell server.
pbcromwell is designed primarily for running workflows distributed and
supported by PacBio, but it is written to handle any valid WDL source
(version 1.0), and is very flexible in how it takes input. PacBio workflows
are expected to be found in the directory defined by the
SMRT_PIPELINE_BUNDLE_DIR environment variable, which is
automatically defined by the SMRT Link distribution.
Note that pbcromwell does not interact with SMRT Link services; to run a
Cromwell workflow as a SMRT Link job, use pbservice. (For details, see
“pbservice” on page 91.)
Note: Interaction with the
Cromwell server is primarily intended for
developers and power users.
Usage:
pbcromwell run [-h] [--output-dir OUTPUT_DIR] [--overwrite] [-i INPUTS]
[-e ENTRY_POINTS] [-n NPROC] [-c MAX_NCHUNKS]
[--target-size TARGET_SIZE] [--queue QUEUE] [-o OPTIONS]
[-t TASK_OPTIONS] [-b BACKEND] [-r MAX_RETRIES]
[--tmp-dir TMP_DIR] [--config CONFIG] [--dry-run]
[run,show-workflows,show-workflow-details,configure,submit,get-
job,abort,metadata,show-running,wait]
Options Description
--output-dir OUTPUT_DIR Specifies the output directory for cromwell output.
(Default = cromwell_out)
--overwrite Overwrites the output directory, if it exists. (Default = False)
-i INPUTS, --inputs INPUTS Specifies cromwell inputs and settings as JSON files. (Default = None)
-e ENTRY_POINTS, --entry
ENTRY_POINTS
Specifies the entry point Data Set; may be repeated for workflows that
take more than one input Data Set. Note that all PacBio workflows require
at least one such entry point.
-n NPROC, --nproc NPROC Specifies the number of processors per task. (Default = 1)
-c MAX_NCHUNKS, --max-nchunks
MAX_NCHUNKS
Specifies the maximum number of chunks per task. (Default = None)
--target-size TARGET_SIZE Specifies the target chunk size. (Default = None)
--queue QUEUE Specifies the cluster queue to use. (Default = None)
-o OPTIONS, --options OPTIONS Specifies additional cromwell engine options, as a JSON file.
(Default = None)
-t TASK_OPTIONS, --task-
option TASK_OPTIONS
Specifies workflow- or task-level option as key=value strings, specific to
the application. May be specified multiple times for multiple options.
(Default = [])
-b BACKEND, --backend BACKEND Specifies the backend to use for running tasks. (Default = None)
-r MAX_RETRIES, --maxRetries
MAX_RETRIES
Specifies the maximum number of times to retry a failing task.
(Default = 1)
Page 76
Enter pbcromwell {command} -h for a command's options.
Examples:
Show available PacBio-developed workflows:
pbcromwell show-workflows
Show details for a PacBio workflow:
pbcromwell show-workflow-details pb_ccs
Generate cromwell.conf with HPC settings:
pbcromwell configure --default-backend SGE --output-file cromwell.conf
Launch a PacBio workflow:
pbcromwell run pb_ccs -e /path/to/movie.subreadset.xml --nproc 8 --config /full/ path/
to/cromwell.conf
pbcromwell run Command: Run a Cromwell workflow. Multiple input
modes are supported, including a
pbsmrtpipe-like set of arguments (for
PacBio workflows only), and JSON files already in the native Cromwell
format.
All PacBio workflows have similar requirements to the
pbsmrtpipe
pipelines in previous SMRT Link versions:
--tmp-dir TMP_DIR Specifies an optional temporary directory for cromwell tasks, which
must exist on all compute hosts. (Default = None)
--config CONFIG Specifies a Java configuration file for running cromwell. (Default = None)
--dry-run Specifies that cromwell is not executed, but instead writes out final
inputs and then exits. (Default = True)
workflow Specifies the workflow ID (such as pb_ccs or
cromwell.workflows.pb_ccs for PacBio workflows only) or a path to
a Workflow Description Language (WDL) source file.
Options Description
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file LOG_FILE Writes the log to file. (Default = None, writes to stdout.)
--log-level=INFO Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR,
CRITICAL.] (Default = INFO)
--debug Alias for setting the log level to DEBUG. (Default = False)
--verbose, -v Sets the verbosity level. (Default = None)

Page 77
1. One or more PacBio dataset XML entry points, usually a SubreadSet or
ConsensusReadSet (
--entry-point <FILE>.)
2. Any number of workflow-specific task options
(--task-option
<OPTION>
.)
3. Engine options independent of the workflow, such as number of pro-
cessors per task (
--nproc), or compute backend (--backend).
Output is directed to a new directory:
--output-dir, which defaults to
cromwell_out. This includes final inputs for the Cromwell CLI, and
subdirectories for logs (workflow and task outputs), links to output files,
and the Cromwell execution itself, which has a complex nested directory
structure. Detailed information about the workflow execution can be found
in the file
metadata.json, in the native Cromwell format.
Note that output file links do not include the individual resource files of
Data Sets and reports (BAM files, index files, plot PNGs, and so on.) Follow
the symbolic links to their real path (for example using
readlink -f) to
find report plots.
For further information about Cromwell, see the official documentation
here.
Workflow examples:
Run the CCS analysis:
pbcromwell run pb_ccs -e <SUBREADS> --nproc 8 --config /full/path/to/cromwell.conf
Run the Iso-Seq workflow, including mapping to a reference, and execute
on SGE:
pbcromwell run pb_isoseq3 -e <SUBREADS> -e <PRIMERS> -e <REFERENCE> --nproc 8 -- config
/full/path/to/cromwell.conf
Run a user-defined workflow:
pbcromwell run my_workflow.wdl -i inputs.json -o options.json --config /full/path/to/
cromwell.conf
Set up input files but exit before starting Cromwell:
pbcromwell run pb_ccs -e <SUBREADS> --nproc 8 --dry-run
Print details about the named PacBio workflow, including input files and
task options. Note: The prefix
cromwell.workflows. is optional.
pbcromwell show-workflow-details pb_ccs
pbcromwell show-workflow-details cromwell.workflows.pb_ccs
Page 78
pbcromwell show-workflow-details
Command: Display details about a
specified PacBio workflow, including input files and task options.
Usage:
pbcromwell show-workflow-details [-h] [--inputs-json INPUTS_JSON]
workflow_id
pbcromwell configure Command: Generate the Java configuration file
used by
cromwell to define backends and other important engine options
that cannot be set at runtime. You can pass this to
pbcromwell run using the --config argument.
Usage:
pbcromwell configure [-h] [--port PORT]
[--local-job-limit LOCAL_JOB_LIMIT]
[--jms-job-limit JMS_JOB_LIMIT]
[--db-port DB_PORT] [--db-user DB_USER]
[--db-password DB_PASSWORD]
[--default-backend DEFAULT_BACKEND]
[--max-workflows MAX_WORKFLOWS]
[--output-file OUTPUT_FILE] [--timeout TIMEOUT]
[--cache] [--no-cache]
Options Description
workflow_id Specifies the workflow ID, such as pb_ccs or
cromwell.workflows.pb_ccs for PacBio workflows only.
--inputs-json INPUTS_JSON Writes a JSON template containing workflow inputs to the specified file.
(Default = None)
Options Description
--port PORT Specifies the port that cromwell should listen to. (Default = 8000)
--local-job-limit
LOCAL_JOB_LIMIT
Specifies the maximum number of local jobs/tasks that can be run at
once. (Default = 10)
--jms-job-limit JMS_JOB_LIMIT Specifies the maximum number of jobs/tasks that can be submitted to the
queueing system at one time. (Default = 500)
--db-port DB_PORT Specifies the database port for cromwell to use; if undefined, database
configuration is omitted. (Default = None)
--db-user DB_USER Specifies the user name used to connect to Postgres.
(Default = smrtlink_user)
--db-password DB_PASSWORD Specifies the password used to connect to Postgres.
--default-backend
DEFAULT_BACKEND
Specifies the default job execution backend. Choices are: Local, SGE,
Slurm, PBS, or LSF. (Default = Local)
--max-workflows MAX_WORKFLOWS Specifies the maximum number of workflows that cromwell can run at
once. (Default = 100)
--output-file OUTPUT_FILE Specifies the name of the output configuration file.
(Default = cromwell.conf)
--timeout TIMEOUT Specifies the time to wait for task completion before checking the cluster
job status. (Default = 600)

Page 79
--cache Enables call caching. (Default = True)
--no-cache Disables call caching. (Default = True)
Options Description

Page 80
pbindex
The pbindex tool creates an index file that enables random access to
PacBio-specific data in BAM files.
Usage
pbindex <input>
Input file
• *.bam file containing PacBio data.
Output file
• *.pbi index file, with the same prefix as the input file name.
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.

Page 81
pbmarkdup
The pbmarkdup tool marks PCR duplicates in CCS reads Data Sets from
amplified libraries. PCR duplicates are different reads that arose from
amplifying a single-source molecule.
pbmarkdup can also optionally
remove the duplicate reads.
Note:
pbmarkdup only works with CCS reads, not with Subreads.
pbmarkdup uses a reference-free comparison method. Duplicates are
identified as pairs of reads that:
1. Have the same length - within 10 bp, and
2. Have high percent identity alignments at the molecule ends at >98%
identity of the first and last 250 bp.
Clusters are formed from sets of two or more duplicate reads, and a single
read is selected as the representative of each cluster. Other reads in the
cluster are considered duplicates.
How are duplicates marked?
In FASTA and FASTQ formats, reads from duplicate clusters have
annotated names. The following is a FASTA example:
>m64013_191117_050515/67104/ccs DUPLICATE=m64013_191117_050515/3802014/ccs DS=2
This shows a marked duplicate read m64013_191117_050515/67104/ccs
that is a duplicate of
m64013_191117_050515/3802014/ccs in a cluster
with 2 reads (
DS value). Accordingly, the following is the read selected as
the representative of the molecule:
>m64013_191117_050515/3802014/ccs DS=2
In BAM format, duplicate reads are flagged with 0x400. The read-level tag
ds provides the number of reads in a cluster (like the DS attribute
described above for FASTA/FASTQ), and the
du tag provides the name of
the representative read (like the
DUPLICATE attribute described above for
FASTA/FASTQ).
Usage
pbmarkdup [options] <INFILE.bam|xml|fa|fq|fofn> <OUTFILE.bam|xml|fa.gz|fq.gz>
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file Logs to a file, instead of stderr.

Page 82
Input files
CCS reads from one or multiple movies in any of the following formats:
• PacBio BAM (
.ccs.bam)
• PacBio dataset (
.dataset.xml)
•File of File Names (
.fofn)
• FASTA (.fasta,.fasta.gz)
•
FASTQ (.fastq,.fastq.gz)
Output files
CCS reads with duplicates marked in a format inferred from the file
extension:
• PacBio BAM (
.ccs.bam)
• PacBio dataset (
.dataset.xml), which also generates a
corresponding BAM file.
• FASTA (.fasta.gz)
•
FASTQ (.fastq.gz)
--log-level Specifies the log level; values are [TRACE,DEBUG,INFO,WARN, FATAL]
(Default = WARN)
--log-level INFO produces a summary report such as:
LIBRARY READS UNIQUE MOLECULES DUPLICATE READS
----------------------------------------------------------------------------------------------------------
<Unnamed> 25000 24948 (99.8%) 52 (0.2%)
SS-lib 496 493 (99.4%) 3 (0.6%)
----------------------------------------------------------------------------------------------------------
TOTAL 25496 25441 (99.8%) 55 (0.2%)
-j,--num-threads Specifies the number of threads to use, 0 means autodetection.
(Default = 0)
Duplicate marking options Description
--cross-library, -x Identifies duplicate reads across sequencing libraries. Libraries are
specified in the BAM read group LB tag.
Output options Description
-rmdup, -r Excludes duplicates from OUTFILE. (This is redundant when
--dup-file is specified.)
--dup-file FILE Stores duplicate reads in an extra file other than OUTFILE. The format of
this file can be different from the output file.
--clobber, -f Overwrites OUTFILE if it exists.
Options Description
Page 83
Allowed input/output combinations
Examples
Run on a single movie:
pbmarkdup movie.ccs.bam output.bam
Run on multiple movies:
pbmarkdup movie1.fasta movie2.fasta output.fasta
Run on multiple movies and output duplicates in separate file:
pbmarkdup movie1.ccs.bam movie2.fastq uniq.fastq --dup-file dups.fasta
Input File Output BAM Output Dataset Output FASTQ Output FASTA
Input BAM
Allowed Allowed Allowed Allowed
Input Dataset
Allowed Allowed Allowed Allowed
Input FASTQ
Not Allowed Not Allowed Allowed Allowed
Input FASTA
Not Allowed Not Allowed Not Allowed Allowed

Page 84
pbmm2
The pbmm2 tool aligns native PacBio data, outputs PacBio BAM files, and
is a SMRT minimap2 wrapper for PacBio data.
pbmm2 supports native PacBio input and output, provides sets of
recommended parameters, generates sorted output on-the-fly, and post-
processes alignments. Sorted output can be used directly for polishing
using GenomicConsensus, if BAM has been used as input to pbmm2.
pbmm2 adds the following SAM tag to each aligned record:
•
mg, stores gap-compressed alignment identity, defined as
nM/(nM + nMM + nInsEvents + nDelEvents).
Usage
pbmm2 <tool>
index
Command: Indexes references and stores them as .mmi files.
Indexing is optional, but recommended if you use the same reference with
the same
--preset multiple times.
Usage:
pbmm2 index [options] <ref.fa|xml> <out.mmi>
Input file
• *.fasta, *.fa file containing reference contigs or
*.referenceset.xml.
Output file
• out.mmi (minimap2 index file.)
Notes:
• You can use existing minimap2 .mmi files with pbmm2 align.
• If you use an index file, you cannot override parameters
-k, -w, nor -u
in
pbmm2 align.
•The
minimap2 parameter -H (homopolymer-compressed k-mer) is
always on for SUBREAD and UNROLLED presets, and can be disabled
using
-u.
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.

Page 85
align
Command: Aligns PacBio reads to reference sequences. The
output argument is optional; if not provided, the BAM output is streamed
to
stdout.
Usage:
pbmm2 align [options] <ref.fa|xml|mmi> <in.bam|xml|fa|fq> [out.aligned.bam|xml]
Input files
• *.fasta file containing reference contigs, or *.referenceset.xml, or
*.mmi index file.
•
*.bam, *.subreadset.xml, *.consensusreadset.xml,
*.transcriptset.xml, *.fasta, *.fa, *.fastq, or *.fastq file
containing PacBio data.
Output files
• *.bam aligned reads in BAM format.
•
*.alignmentset, *.consensusalignmentset.xml, or
*.transcriptalignmentset.xml if XML output was chosen.
The following Data Set Input/output combinations are allowed:
SubreadSet > AlignmentSet
pbmm2 align hg38.referenceset.xml movie.subreadset.xml hg38.movie.alignmentset.xml
ConsensusReadSet > ConsensusAlignmentSet
pbmm2 align hg38.referenceset.xml movie.consensusreadset.xml
hg38.movie.consensusalignmentset.xml --preset CCS
Options Description
--preset Specifies the alignment mode:
• "SUBREAD" -k 19 -w 10
• "CCS" -k 19 -w 10 -u
• "ISOSEQ" -k 15 -w 5 -u
• "UNROLLED" -k 15 -w 15
The option is not case-sensitive. (Default = SUBREAD)
-k Specifies the k-mer size, which cannot be larger than 28. (Default = -1)
-w Specifies the Minimizer window size. (Default = -1)
-u,--no-kmer-compression Disables homopolymer-compressed k-mer. (Compression is on by default
for the SUBREAD and UNROLLED presets.)
--short-sa-cigar Populates the SA tag with the short cigar representation.
Page 86
TranscriptSet > TranscriptAlignmentSet
pbmm2 align hg38.referenceset.xml movie.transcriptset.xml
hg38.movie.transcriptalignmentset.xml --preset ISOSEQ
FASTA/FASTQ input
In addition to native PacBio BAM input, reads can also be provided in
FASTA and FASTQ formats.
Attention: The resulting output BAM file cannot be used as input into
GenomicConsensus!
With FASTA/FASTQ input, the
--rg option sets the read group. Example:
pbmm2 align hg38.fasta movie.Q20.fastq hg38.movie.bam --preset CCS --rg
'@RG\tID:myid\tSM:mysample'
All three reference file formats .fasta, .referenceset.xml, and .mmi
can be combined with FASTA/FASTQ input.
Options Description
-h, --help Displays help information and exits.
--chunk-size Processes
N records per chunk. (Default = 100)
--sort Generates a sorted BAM file.
-m,--sort-memory Specifies the memory per thread for sorting. (Default = 768M)
-j,--alignment-threads Specifies the number of threads used for alignment. 0 means
autodetection. (Default = 0)
-J,--sort-threads Specifies the number of threads used for sorting. 0 means 25% of -j, with
a maximum of 8. (Default = 0)
--sample Specifies the sample name for all read groups. Defaults, in order of
precedence: A) SM field in the input read group B) Biosample name
C) Well sample name D) "UnnamedSample".
--rg Specifies the read group header line such as '@RG\tID:xyz\tSM:abc'.
Only for FASTA/Q inputs.
-y,--min-gap-comp-id-perc Specifies the minimum gap-compressed sequence identity, in percent.
(Default = 70)
-l,--min-length Specifies the minimum mapped read length, in base pairs. (Default = 50)
-N,--best-n Specifies the output at maximum N alignments for each read. 0 means no
maximum. (Default = 0)
--strip Removes all kinetic and extra QV tags. The output cannot be polished.
--split-by-sample Specifies one output BAM file per sample.
--no-bai Omits BAI index file generation for sorted output.
--unmapped Specifies that unmapped records be included in the output.
--median-filter Picks one read per ZMW of median length.
--zmw Processes ZMW Reads; subreadset.xml input is required. This
activates the UNROLLED preset.
Page 87
Examples:
Generate an index file for reference and reuse it to align reads:
pbmm2 index ref.fasta ref.mmi
pbmm2 align ref.mmi movie.subreads.bam ref.movie.bam
Align reads and sort on-the-fly, with 4 alignment and 2 sort threads:
pbmm2 align ref.fasta movie.subreads.bam ref.movie.bam --sort -j 4 -J 2
Align reads, sort on-the-fly, and create a PBI:
--hqregion Processes the HQ region of each ZMW; subreadset.xml input is
required. This activates the UNROLLED preset.
Parameter set options and
overrides
Description
--preset Specifies the alignment mode:
• "SUBREAD" -k 19 -w 10 -o 5 -O 56 -e 4 -E 1 -A 2 -B 5
-z 400 -Z 50 -r 2000 -L 0.5
• "CCS" -k 19 -w 10 -u -o 5 -O 56 -e 4 -E 1 -A 2 -B 5 -
z 400 -Z 50 -r 2000 -L 0.5
• "ISOSEQ" -k 15 -w 5 -u -o 2 -O 32 -e 1 -E 0 -A 1 -B 2
-z 200 -Z 100 -C 5 -r 200000 -G 200000 -L 0.5
• "UNROLLED" -k 15 -w 15 -o 2 -O 32 -e 1 -E 0 -A 1 -B 2
-z 200 -Z 100 -r 2000 -L 0.5
(Default = SUBREAD)
-k Specifies the k-mer size, which cannot be larger than 28. (Default = -1)
-w Specifies the Minimizer window size. (Default = -1)
-u,--no-kmer-compression Disables homopolymer-compressed k-mer. (Compression is on by default
for the SUBREAD and UNROLLED presets.)
-A Specifies the matching score. (Default = -1)
-B Specifies the mismatch penalty. (Default = -1)
-z Specifies the Z-drop score. (Default = -1)
-Z Specifies the Z-drop inversion score. (Default = -1)
-r Specifies the bandwidth used in chaining and DP-based alignment.
(Default = -1)
-o,--gap-open-1 Specifies the gap open penalty 1. (Default = -1)
-O,--gap-open-2 Specifies the gap open penalty 2. (Default = -1)
-e,--gap-extend-1 Specifies the gap extension penalty 1. (Default = -1)
-E,--gap-extend-2 Specifies the gap extension penalty 2. (Default = -1)
-L,--lj-min-ratio Specifies the long join flank ratio. (Default = -1)
-G Specifies the maximum intron length; this changes -r. (Default = -1)
-C Specifies the cost for a non-canonical GT-AG splicing. (Default = -1)
--no-splice-flank Specifies that you do not prefer splicing flanks GT-AG.
Options Description

Page 88
pbmm2 align ref.fasta movie.subreadset.xml ref.movie.alignmentset.xml --sort
Omit the output file and stream the BAM output to stdout:
pbmm2 align hg38.mmi movie1.subreadset.xml | samtools sort > hg38.movie1.sorted.bam
Align the CCS reads fastq input and sort the output:
pbmm2 align ref.fasta movie.Q20.fastq ref.movie.bam --preset CCS --sort --rg
'@RG\tID:myid\tSM:mysample'
Alignment parallelization
The number of alignment threads can be specified using the -j,
--alignment-threads
option. If not specified, the maximum number of
threads are used, minus one thread for BAM I/O and minus the number of
threads specified for sorting.
Sorting
Sorted output can be generated using the --sort option.
• By default, 25% of threads specified with the
-j option (Maximum = 8)
are used for sorting.
• To override the default percentage, the
-J,--sort-threads option
defines the explicit number of threads used for on-the-fly sorting. The
memory allocated per sort thread is defined using the
-m,--sort-
memory
option, accepting suffixes M,G.
Benchmarks on human data show that 4 sort threads are recommended,
but that no more than 8 threads can be effectively leveraged, even with 70
cores used for alignment. We recommend that you provide more memory
to each of a few sort threads to avoid disk I/O pressure, rather than
providing less memory to each of many sort threads.
What are parameter sets and how can I override them?
Per default, pbmm2 uses recommended parameter sets to simplify the
multitudes of possible combinations. See the available parameter sets in
the option table shown earlier.
What other special parameters are used implicitly?
We implicitly use the following minimap2 parameters:
• Soft clipping with
-Y.
• Long cigars for tag CG with -L.
• X/= cigars instead of M with --eqx.
• No overlapping query intervals with repeated matches trimming.
• No secondary alignments are produced using the
--secondary=no
option.
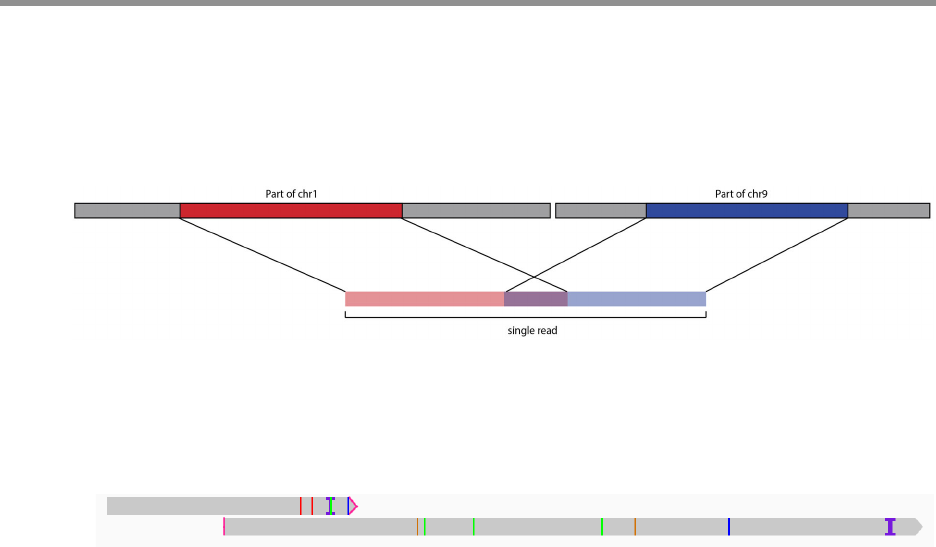
Page 89
What is repeated matches trimming?
A repeated match occurs when the same query interval is shared between
a primary and supplementary alignment. This can happen for
translocations, where breakends share the same flanking sequence:
And sometimes, when a LINE gets inserted, the flanks are duplicated,
leading to complicated alignments, where we see a split read sharing a
duplication. The inserted region itself, mapping to a random other LINE in
the reference genome, may also share sequence similarity to the flanks:
To get the best alignments,
minimap2 decides that two alignments may
use up to 50% (default) of the same query bases. This does not work for
PacBio, as
pbmm2 requires that a single base may never be aligned twice.
Minimap2 offers a feature to enforce a query interval overlap to 0%. If a
query interval gets used in two alignments, one or both get flagged as
secondary and get filtered. This leads to yield loss, and more importantly,
missing SVs in the alignment.
Papers (such as this) present dynamic programming approaches to find
the optimal split to uniquely map query intervals, while maximizing
alignment scores. We don't have per base alignment scores available, thus
our approach is much simpler. We align the read, find overlapping query
intervals, determine one alignment to be maximal reference-spanning,
then trim all others. By trimming,
pbmm2 rewrites the cigar and the
reference coordinates on-the-fly. This allows us to increase the number of
mapped bases, which slightly reduces mapped concordance, but boosts
SV recall rate.
How can I set the sample name?
You can override the sample name (SM field in the RG tag) for all read
groups using the
--sample option. If not provided, sample names derive
from the Data Set input using the following order of precedence: A)
SM
field in the input read group B) Biosample name C) Well sample name
D)
UnnamedSample. If the input is a BAM file and the --sample option was
not used, the
SM field is populated with UnnamedSample.

Page 90
Can I split output by sample name?
Yes, the --split-by-sample option generates one output BAM file per
sample name, with the sample name as the file name prefix, if there is
more than one aligned sample name.
Can I remove all those extra per-base and per-pulse tags?
Yes, the --strip option removes the following extraneous tags if the
input is BAM:
dq, dt, ip, iq, mq, pa, pc, pd, pe, pg, pm, pq,
pt, pv, pw, px, sf, sq, st
. Note that the resulting output BAM file
cannot be used as input into
GenomicConsensus.
Where are the unmapped reads?
Per default, unmapped reads are omitted. You can add them to the output
BAM file using the
--unmapped option.
Can I output at maximum the N best alignments per read?
Use the option -N, --best-n. If set to 0, (the default), maximum filtering
is disabled.
Is there a way to only align one subread per ZMW?
Using the --median-filter option, only the subread closest to the
median subread length per ZMW is aligned. Preferably, full-length
subreads flanked by adapters are chosen.
Page 91
pbservice
The pbservice tool performs a variety of useful tasks within SMRT Link.
•To get help for
pbservice, use pbservice -h.
• To get help for a specific
pbservice command, use
pbservice <command> -h.
Note: pbservice requires authentication when run from a remote host,
using the same credentials used to log in to the SMRT Link GUI. (This also
routes all requests through HTTPS port 8243, so the services port is not
required if authentication is used.) Access to services running on
localhost will continue to work without authentication.
All
pbservice commands include the following optional parameters:
status Command: Use to get system status.
pbservice status [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO}
[--debug] [--quiet]
import-dataset Command: Import Local Data Set XML. The file location
must be accessible from the host where the services are running; often on
a shared file system.
pbservice import-dataset [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
xml_or_dir
Options Description
--host=http://localhost Specifies the server host. Override the default with the environmental
variable PB_SERVICE_HOST.
--port=8070 Specifies the server port. Override the default with the environmental
variable PB_SERVICE_PORT.
--log-file LOG_FILE Writes the log to file. (Default = None, writes to stdout.)
--log-level=INFO Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR,
CRITICAL.] (Default = INFO)
--debug=False Alias for setting the log level to DEBUG. (Default = False)
--quiet=False Alias for setting the log level to CRITICAL to suppress output.
(Default = False)
--user USERNAME Specifies the user to authenticate as; this is required if the target host is
anything other than localhost.
--ask-pass Prompts the user to enter a password.
--password PASSWORD Supplies the password directly. This exposes the password in the shell
history (or log files), so this option is not recommended unless you are
using a limited account without Unix login access.

Page 92
import-run
Command: Create a SMRT Link Run Design and optional Auto
Analysis jobs from a CSV file. This is equivalent to the Run Design CSV
import feature in the SMRT Link UI; the resulting runs can then be edited in
the UI or executed on-instrument.
pbservice import-run [-h][--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
[--dry-run]
csv_file
import-fasta Command: Import a FASTA file and convert to a
ReferenceSet file. The file location must be accessible from the host
where the services are running; often on a shared file system.
pbservice import-fasta [-h] --name NAME --organism ORGANISM --ploidy
PLOIDY [--block] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
fasta_path
run-analysis Command: Run a secondary analysis pipeline using an
analysis.json file.
pbservice run-analysis [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
Required Description
xml_or_dir Specifies a directory or XML file for the Data Set.
Required Description
csv_file Specifies a Run Design CSV-format file.
Option Description
--dry-run Prints the generated run XML without POSTing to the server.
Required Description
fasta_path Path to the FASTA file to import.
Options Description
--name Specifies the name of the ReferenceSet to convert the FASTA file to.
--organism Specifies the name of the organism.
--ploidy Ploidy.
--block=False Blocks during importing process.
Page 93
[--debug] [--quiet] [--block]
json_path
emit-analysis-template Command: Output an analysis.json
template to
stdout that can be run using the run-analysis command.
pbservice emit-analysis-template [-h] [--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
get-job Command: Get a job summary by Job Id.
pbservice get-job [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
job_id
get-jobs Command: Get job summaries by Job Id.
pbservice get-jobs [-h] [-m MAX_ITEMS] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
get-dataset Command: Get a Data Set summary by Data Set Id or UUID.
pbservice get-dataset [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
id_or_uuid
get-datasets Command: Get a Data Set list summary by Data Set type.
pbservice get-datasets [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet] [-m MAX_ITEMS]
Required Description
json_path Path to the analysis.json file.
Options Description
--block=False Blocks during importing process.
Required Description
job_id Job id or UUID.
Options Description
-m=25, --max-items=25 Specifies the maximum number of jobs to get.
Required Description
id_or_uuid Data Set Id or UUID.

Page 94
[-t DATASET_TYPE]
delete-dataset Command: Delete a specified Data Set.
Note: This is a "soft" delete - the database record is tagged as inactive so it
won't display in any lists, but the files will not be removed.
pbservice delete-dataset [-h] [--host HOST] [--port PORT]
[--log-file LOG_FILE]
[--log-level INFO]
[--debug] [--quiet]
[ID]
Examples
To obtain system status, the Data Set summary, and the job summary:
pbservice status --host smrtlink-release --port 9091
To import a Data Set XML:
pbservice import-dataset --host smrtlink-release --port 9091 \
path/to/subreadset.xml
To obtain a job summary using the Job Id:
pbservice get-job --host smrtlink-release --port 9091 \
--log-level CRITICAL 1
To obtain Data Sets by using the Data Set type subreads:
pbservice get-datasets --host smrtlink-alpha --port 8081 \
--quiet --max-items 1 -t subreads
To obtain Data Sets by using the Data Set type alignments:
pbservice get-datasets --host smrtlink-alpha --port 8081 \
--quiet --max-items 1 -t alignments
To obtain Data Sets by using the Data Set type references:
pbservice get-datasets --host smrtlink-alpha --port 8081 \
--quiet --max-items 1 -t references
To obtain Data Sets by using the Data Set type barcodes:
pbservice get-datasets --host smrtlink-alpha --port 8081 \
--quiet --max-items 1 -t barcodes
Required Description
-t=subreads, --dataset-
type=subreads
Specifies the type of Data Set to retrieve: subreads, alignments,
references, barcodes.
Required Description
ID A valid Data Set ID, either UUID or integer ID, for the Data Set to delete.

Page 95
To obtain Data Sets by using the Data Set UUID:
pbservice get-dataset --host smrtlink-alpha --port 8081 \
--quiet 43156b3a-3974-4ddb-2548-bb0ec95270ee
Page 96
pbsv
pbsv is a structural variant caller for PacBio reads. It identifies structural
variants and large indels (Default: ≥20 bp) in a sample or set of samples
relative to a reference.
pbsv identifies the following types of variants:
Insertions, deletions, duplications, copy number variants, inversions, and
translocations.
pbsv takes as input read alignments (BAM) and a reference genome
(FASTA); it outputs structural variant calls (VCF).
Usage:
pbsv [-h] [--version] [--quiet] [--verbose]
{discover,call}...
pbsv discover Command: Finds structural variant (SV) signatures in
read alignments. The input read alignments must be sorted by chromo-
some position. Alignments are typically generated with
pbmm2. The output
SVSIG file contains SV signatures.
Usage:
pbsv discover [options] <ref.in.bam|xml> <ref.out.svsig.gz>
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file Logs to a file, instead of stdout.
--log-level Specifies the log level; values are [TRACE,DEBUG,INFO, WARN, FATAL.]
(Default = WARN)
discover Finds structural variant signatures in read alignments (BAM to SVSIG).
call Calls structural variants from SV signatures and assign genotypes (SVSIG
to VCF).
Required Description
ref.in.bam|xml Coordinate-sorted aligned reads in which to identify SV signatures.
ref.out.svsig.gz Structural variant signatures output.
Options Description
-h, --help Displays help information and exits.
-s,--sample Overrides sample name tag from BAM read group.
-q,--min-mapq Ignores alignments with mapping quality < N. (Default = 20)
-m,--min-ref-span Ignores alignments with reference length < N bp. (Default = 100)
-w,--downsample-window-length Specifies a window in which to limit coverage, in base pairs.
(Default = 10K)
-a,--downsample-max-
alignments
Considers up to N alignments in a window; 0 means disabled.
(Default = 100)
Page 97
pbsv call
Command: Calls structural variants from SV signatures and
assigns genotypes. The input SVSIG file is generated using
pbsv dis-
cover
. The output is structural variants in VCF format.
Usage:
pbsv call [options] <ref.fa|xml> <ref.in.svsig.gz|fofn>
<ref.out.vcf>
-r,--region Limits discovery to this reference region: CHR|CHR:START-END.
-l,--min-svsig-length Ignores SV signatures with length < N bp. (Default = 7)
-b,--tandem-repeats Specifies tandem repeat intervals for indel clustering, as an input BED file.
-k,--max-skip-split Ignores alignment pairs separated by > N bp of a read or reference.
(Default = 100)
-y,--min-gap-comp-id-perc Ignores alignments with gap-compressed sequence identity < N%.
(Default = 70)
Options Description
Required Description
ref.fa|xml Reference FASTA file or ReferenceSet XML file against which to call
variants.
ref.in.svsig.gz|fofn SV signatures from one or more samples. This can be either an SV
signature SVSIG file generated by pbsv discover, or a FOFN of SVSIG
files.
ref.out.vcf Variant call format (VCF) output file.
Options Description
-h, --help Displays help information and exits.
-j,--num-threads Specifies the number of threads to use, 0 means autodetection.
(Default = 0)
-r,--region Limits discovery to this reference region: CHR|CHR:START-END
-t,--types Calls these SV types: "DEL", "INS", "INV", "DUP", "BND", "CNV".
(Default = “DEL,INS,INV,DUP,BND”)
-m,--min-sv-length Ignores variants with length < N bp. (Default = 20)
--max-ins-length Ignores insertions with length > N bp. (Default = 15K)
--max-dup-length Ignores duplications with length > N bp. (Default = 1M)
--cluster-max-length-perc-
diff
Does not cluster signatures with difference in length > P%. (Default = 25)
--cluster-max-ref-pos-diff Does not cluster signatures > N bp apart in the reference. (Default = 200)
--cluster-min-basepair-perc-
id
Does not cluster signatures with base pair identity < P%. (Default = 10)
-x,--max-consensus-coverage Limits to N reads for variant consensus. (Default = 20)
-s,--poa-scores Scores POA alignment with triplet match,mismatch,gap.
(Default = “1,-2,-2“)
--min-realign-length Considers segments with > N length for realignment. (Default = 100)
Page 98
Following is a typical SV analysis workflow starting from subreads:
1. Align PacBio reads to a reference genome, per movie:
Subreads BAM input:
pbmm2 align ref.fa movie1.subreads.bam ref.movie1.bam --sort --median-filter --sample
sample1
CCS reads BAM input:
pbmm2 align ref.fa movie1.ccs.bam ref.movie1.bam --sort --preset CCS --sample sample1
CCS reads FASTQ input:
pbmm2 align ref.fa movie1.Q20.fastq ref.movie1.bam --sort --preset CCS --sample sample1
--rg '@RG\tID:movie1'
2. Discover the signatures of structural variation, per movie or per
sample:
pbsv discover ref.movie1.bam ref.sample1.svsig.gz
pbsv discover ref.movie2.bam ref.sample2.svsig.gz
3. Call structural variants and assign genotypes (all samples); for CCS
analysis input append
--ccs:
pbsv call ref.fa ref.sample1.svsig.gz ref.sample2.svsig.gz
ref.var.vcf
-A, --call-min-reads-all-
samples
Ignores calls supported by < N reads total across samples. (Default = 3)
-O, --call-min-reads-one-
sample
Ignores calls supported by < N reads in every sample. (Default = 3)
-S, --call-min-reads-per-
strand-all-samples
Ignores calls supported by < N reads per strand total across samples.
(Default = 1)
-B,--call-min-bnd-reads-all-
samples
Ignores BND calls supported by < N reads total across samples.
(Default = 2)
-P, --call-min-read-perc-one-
sample
Ignores calls supported by < P% of reads in every sample. (Default = 20)
--preserve-non-acgt Preserves non-ACGT in REF allele instead of replacing with N.
(Default = false)
--hifi, --ccs Uses options optimized for HiFi reads: -S 0 -P 10. (Default = False)
--gt-min-reads Specifies the minimum supporting reads to assign a sample a non-
reference genotype. (Default = 1)
--annotations Annotates variants by comparing with sequences in FASTA.
(Default annotations are ALU, L1, and SVA.)
--annotation-min-perc-sim Annotates variant if sequence similarity > P%. (Default = 60)
--min-N-in-gap Considers
≥ N consecutive "N" bp as a reference gap. (Default = 50)
--filter-near-reference-gap Flags variants < N bp from a gap as "NearReferenceGap".
(Default = 1000)
--filter-near-contig-end Flags variants < N bp from a contig end as “NearContigEnd”.
(Default = 1K)
Options Description

Page 99
Launching a multi-sample pbsv analysis - requirements
1. Merge multiple Bio Sample SMRT Cells to one Data Set with the Bio
Samples specified.
– Each SMRT Cell must have exactly one Bio Sample name - multiple
Bio Sample names cannot be assigned to one SMRT Cell.
– Multiple SMRT Cells can have the same Bio Sample name.
– All of the inputs need to already have the appropriate Bio Sample
records in their
CollectionMetadata. If they don't, they are treated
as a single sample.
2. Create a ReferenceSet from a FASTA file.
– The ReferenceSet is often already generated and registered in SMRT
Link.
– If the ReferenceSet doesn’t exist, use the
dataset create
command to create one:
dataset create --type ReferenceSet --name reference_name reference.fasta
Launching a multi-sample analysis
# Set subreads and ref FASTA
sample1=sample1.subreadset.xml sample2=sample2.subreadset.xml
ref=reference.fasta
pbmm2 align ${ref} ${sample1} sample1.bam --sort --median-filter --sample Sample1
pbmm2 align ${ref} ${sample2} sample2.bam --sort --median-filter --sample Sample2
samtools index sample1.bam
samtools index sample2.bam
pbindex sample1.bam
pbindex sample2.bam
pbsv discover sample1.bam sample1.svsig.gz
pbsv discover sample2.bam sample2.svsig.gz
pbsv call ${ref} sample1.svsig.gz sample2.svsig.gz out.vcf
out.vcf: A pbsv VCF output file, where columns starting from column 10
represent structural variants of Sample 1 and Sample 2:
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT Sample1 Sample2
chr01 222737 pbsv.INS.1 T TTGGTGTTTGTTGTTTTGTTTT . PASS
SVTYPE=INS;END=222737;SVLEN=21;SVANN=TANDEM GT:AD:DP 0/1:6,4:10 0/1:6,5:11
Page 100
pbvalidate
The pbvalidate tool validates that files produced by PacBio software are
compliant with PacBio’s own internal specifications.
Input files
pbvalidate supports the following input formats:
•BAM
•FASTA
• Data Set XML
See here for further information about each format's requirements.
Usage
pbvalidate [-h] [--version] [--log-file LOG_FILE]
[--log-level {DEBUG,INFO,WARNING,ERROR,CRITICAL} | --debug | --quiet | -v]
[-c] [--quick] [--max MAX_ERRORS]
[--max-records MAX_RECORDS]
[--type {BAM,Fasta,AlignmentSet,ConsensusSet,ConsensusAlignmentSet,
SubreadSet,BarcodeSet,ContigSet,ReferenceSet,HdfSubreadSet}]
[--index] [--strict] [-x XUNIT_OUT] [--unaligned]
[--unmapped] [--aligned] [--mapped]
[--contents {SUBREAD,CCS}] [--reference REFERENCE]
file
Required Description
file Input BAM, FASTA, or Data Set XML file to validate.
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file LOG_FILE Writes the log to file. Default (None) will write to stdout.
--log-level Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR,
CRITICAL.] (Default = CRITICAL)
--debug=False Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to CRITICAL to suppress output.
(Default = False)
--verbose, -v Sets the verbosity level. (Default = None)
--quick Limits validation to the first 100 records (plus file header); equivalent to
--max-records=100. (Default = False)
--max MAX_ERRORS Exits after MAX_ERRORS were recorded.
(Default = None; checks the entire file.)
--max-records MAX_RECORDS Exits after MAX_RECORDS were inspected.
(Default = None; checks the entire file.)
--type Uses the specified file type instead of guessing.
[BAM,Fasta,AlignmentSet,ConsensusSet,ConsensusAlignmen
tSet,SubreadSet,BarcodeSet,ContigSet,ReferenceSet,
HdfSubreadSet] (Default = None)
Page 101
Examples
To validate a BAM file:
pbvalidate in.subreads.bam
To validate a FASTA file:
pbvalidate in.fasta
To validate a Data Set XML file:
pbvalidate in.subreadset.xml
To validate a BAM file and its index file (.pbi):
pbvalidate --index in.subreads.bam
To validate a BAM file and exit after 10 errors are detected:
pbvalidate --max 10 in.subreads.bam
To validate up to 100 records in a BAM file:
pbvalidate --max-records 100 in.subreads.bam
To validate up to 100 records in a BAM file
(equivalent to
--max-records=100):
pbvalidate --quick in.subreads.bam
To validate a BAM file, using a specified log level:
pbvalidate --log-level=INFO in.subreads.bam
--index Requires index files:.fai or .pbi. (Default = False)
--strict Turns on additional validation, primarily for Data Set XML.
(Default = False)
BAM options Description
--unaligned Specifies that the file should contain only unmapped alignments.
(Default = None, no requirement.)
--unmapped Alias for --unaligned. (Default = None)
--aligned Specifies that the file should contain only mapped alignments.
(Default = None, no requirement.)
--mapped Alias for --aligned. (Default = None)
--contents Enforces the read type: [SUBREAD, CCS] (Default = None)
--reference REFERENCE Specifies the path to an optional reference FASTA file, used for additional
validation of mapped BAM records. (Default = None)
Options Description

Page 102
To validate a BAM file and write log messages to a file rather than to
stdout:
pbvalidate --log-file validation_results.log in.subreads.bam
Page 103
primrose
The primrose tool analyzes the kinetic signatures of cytosine bases in
CpG motifs to identify the presence of 5mC.
primrose uses a convolution neural network (CNN) to predict the
methylation state (5mC) of each CpG in a HiFi read. Methylation is
assumed to be symmetric between strands with output reported in the
forward direction with respect to the HiFi read sequence. The output uses
the
Mm and Ml tags as defined in the SAM Format Optional Fields
Specification. (See here for details.)
Algorithm
The CNN is trained using invitro modified controls of methylated and
unmethylated human DNA. The unmethylated control comprises a human
shotgun library that has undergone whole genome amplification (WGA), a
process that includes PCR, and therefore removes all modifications. The
methylated control is generated by subjecting a human WGS library to
invitro methylation using a CpG methyltransferase enzyme (
M. SssI).
Kinetic data, pulse width and inter-pulse distance, over a 16 base pair
window for both the forward and reverse strand, is used as input to the
CNN. The output of the CNN is a probability scale measure of whether the
CpG is symmetrically 5mC-modified.
Usage
primrose [options] <HiFi INPUT> <HiFi OUTPUT>
Example
primrose --log-level INFO movie.hifi.bam movie.hifi_5mc.bam
Required Description
HiFi INPUT Input BAM file or ConsensusReadSet XML file.
HiFi OUPUT Output BAM file or ConsensusReadSet XML file.
Options Description
--keep-kinetics Specifies that the kinetics tracks (IPD and PulseWidth records) fi, fp, fn,
ri, rp and rn are included in the output BAM file.
--min-passes Specifies the minimum number of passes. (Default = 3)
--model Specifies a path to a trained TensorFlow model directory, or to an
exported ONNX model file.
-h, --help Displays help information and exits.
--version Displays program version number and exits.
-j,--num-threads Specifies the number of threads to use when processing; 0 means
autodetection. (Default = 0)
--log-file LOG_FILE Writes the log to file. Default (None) will write to stderr.
--log-level Specifies the log level; values are [TRACE, DEBUG, INFO, WARNING, ,
FATAL.] (Default = WARNING)
Page 104
runqc-reports
The runqc-reports tool generates up to five different Run QC reports,
depending on Data Set type: Raw Data, Adapters, Loading, Control, and
CCS reads. Generating a complete set of reports requires the presence of
an
sts.xml resource in the Data Set, but either the CCS Analysis report (or
a fallback Subreads report) will always be generated. All report JSON and
plot PNG files are written to the current working directory, unless
otherwise specified.
Usage
runqc-reports [-h] [--version] [--log-file LOG_FILE]
[--log-level {DEBUG,INFO,WARNING,ERROR,CRITICAL}]
[| --debug | --quiet | -v]
[-o OUTPUT_DIR]
dataset_xml
Examples
runqc-reports moviename.subreadset.xml
runqc-reports moviename.consensusreadset.xml
Required Description
dataset_xml Input SubreadSet or ConsensusReadSet XML, which must contain an
sts.xml resource for the full Run QC report set to be generated.
-o OUTPUT_DIR Output report directory. (Default = Current working directory)
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file LOG_FILE Writes the log to file. Default (None) will write to stdout.
--log-level Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR,
CRITICAL.] (Default = WARNING)
--debug Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to CRITICAL to suppress output.
(Default = False)
--verbose, -v Sets the verbosity level. (Default = None)

Page 105
summarize
Modifications
The summarizeModifications tool generates a GFF summary file
(
alignment_summary.gff) from the output of base modification analysis
(for example,
ipdSummary) combined with the coverage summary GFF
generated by resequencing pipelines. This is useful for power users
running custom workflows.
Usage
summarizeModifications [-h] [--version]
[--log-file LOG_FILE]
[--log-level {DEBUG,INFO,WARNING,ERROR,CRITICAL} | --debug
| --quiet | -v]
modifications alignmentSummary gff_out
Input files
• modifications: Base Modification GFF file.
•
alignmentSummary: Alignment Summary GFF file.
Output files
• gff_out: Coverage summary for regions (bins) spanning the reference
with Base Modification results for each region
.
Options Description
-h, --help Displays help information and exits.
--version Displays program version number and exits.
--log-file LOG_FILE Writes the log to file. Default (None) will write to stdout.
--log-level Specifies the log level; values are [DEBUG, INFO, WARNING, ERROR,
CRITICAL] (Default = INFO)
--debug Alias for setting the log level to DEBUG. (Default = False)
--quiet Alias for setting the log level to CRITICAL to suppress output.
(Default = False)
--verbose, -v Sets the verbosity level. (Default = None)

Page 106
Appendix A - Application entry points and output files
Note: To print information about a specific PacBio workflow, including
input files and task options, use the
pbcromwell show-workflow-
details
command, which is available for all applications. Example:
pbcromwell show-workflow-details pb_hgap4
pbcromwell show-workflow-details cromwell.workflows.pb_hgap4
(The prefix cromwell.workflows is optional.)
CCS Analysis
Analysis application name: cromwell.workflows.pb_ccs
Entry point
:id: eid_subread
:name: Entry eid_subread
:fileTypeId: PacBio.DataSet.SubreadSet
Key output files
Demultiplex
Barcodes
Analysis application name: cromwell.workflows.pb_demux_ccs
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_barcode
:name: Entry eid_barcode
:fileTypeId: PacBio.DataSet.BarcodeSet
Key output files
File name Datastore SourceId
FASTQ file ccs_fastq_out
FASTA file ccs_fasta_out
<moviename>.hifi.reads.bam file ccs_bam_out
Consensus Sequences pb_ccs.ccsxml
CCS Analysis Statistics pb_ccs.report_ccs
All Reads (BAM) reads_bam
<moviename>.hifi.reads.fasta ccs_fasta_out
<moviename>.hifi.reads.fastq ccs_fastq_out
File name Datastore SourceId
Barcode Report Details pb_demux_ccs.summary_csv
Demultiplexed Datasets pb_demux_ccs.demuxed_files_datastore
Unbarcoded Reads pb_demux_ccs.unbarcoded
Page 107
Export Reads
Analysis application name: cromwell.workflows.pb_export_ccs
Entry point
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
Key output files
Note: If users select a lower cutoff Phred QV value, the string
hifi is
replaced by the QV value in the file names.
Example:
<moviename>.q10.fastq.gz.
Genome
Assembly
Analysis application name: cromwell.workflows.pb_assembly_hifi
Entry point
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
Key output files
HiFi Mapping
Analysis application name: cromwell.workflows.pb_ccs_subreads
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_ref_dataset
:name: Entry eid_ref_dataset
:fileTypeId: PacBio.DataSet.ReferenceSet
Key output files
File name Datastore SourceId
<moviename>.hifi_reads.fastq.gz ccs_fastq_out
<moviename>.hifi_reads.fasta.gz ccs_fasta_out
<moviename>.hifi_reads.bam.gz ccs_bam_out
File name Datastore SourceId
Final Polished Assembly,
Primary Contigs
pb_assembly_hifi.final_primary_contigs_fasta
Final Polished Assembly,
Haplotigs
pb_assembly_hifi.final_haplotigs_fasta
List of Circular Contigs pb_assembly_hifi.circular_contigs
Summary Report pb_assembly_hifi.report_polished_assembly
File name Datastore SourceId
Mapped reads pb_align_ccs.mapped
Coverage summary pb_align_ccs.coverage_gff
Page 108
HiFiViral SARS-
CoV-2 Analysis
Analysis application name: cromwell.workflows.pb_sars_cov2_kit
Entry points
:id: eid_ccs (HiFi reads, demultiplexed as separate BAM files)
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_ref_dataset_2 (Reference Genome)
:name: Entry eid_ref_dataset
:fileTypeId: PacBio.DataSet.ReferenceSet
Key output files
Iso-Seq
Analysis
Analysis application name: cromwell.workflows.pb_isoseq3_ccsonly
Entry points
:id: eid_ccs
:name: Reads
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_barcode
:name: Primers
:fileTypeId: PacBio.DataSet.BarcodeSet
:id: eid_ref_dataset
:name: Reference (Optional)
:fileTypeId: PacBio.DataSet.ReferenceSet
File name Datastore SourceId
All Samples, Probe Counts TSV pb_sars_cov2_kit.probe_counts_zip
All Samples, Raw Variant Calls
VCF
pb_sars_cov2_kit.raw_vcf_zip
All Samples, Variant Call VCF pb_sars_cov2_kit.vcf_zip
All Samples, Variant Calls CSV pb_sars_cov2_kit.variants_csv
All Samples, Consensus
Sequence FASTA
pb_sars_cov2_kit.fasta_zip
All Samples, Consensus
Sequence By Fragments FASTA
pb_sars_cov2_kit.frag_fasta_zip
All Samples, Aligned Reads BAM pb_sars_cov2_kit.mapped_zip
All Samples, Consensus
Sequence Aligned BAM
pb_sars_cov2_kit.aligned_frag_zip
All Samples, Trimmed HiFi reads
FASTQ
pb_sars_cov2_kit.trimmed_zip
Failed Sample Info pb_sars_cov2_kit.sample_failures_csv
Failed Sample Analysis Logs pb_sars_cov2_kit.errors_zip
Demultiplex Summaries pb_sars_cov2_kit.lima_summary_zip
Sample Summary Table CSV pb_sars_cov2_kit.summary_csv
All Samples, Genome Coverage
Plots
pb_sars_cov2_kit.coverage_png_zip
Page 109
Key output files
Mark PCR
Duplicates
Analysis application name: cromwell.workflows.pb_mark_duplicates
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
Key output files
File name Datastore SourceId
Collapsed Filtered Isoforms FASTQ pb_isoseq3_ccsonly.collapse_fastq
Collapsed Filtered Isoforms GFF pb_isoseq3_ccsonly.collapse_gff
Group TXT pb_isoseq3_ccsonly.collapse_group
Abundance TXT pb_isoseq3_ccsonly.collapse_abundance
Read Stat TXT pb_isoseq3_ccsonly.collapse_readstat
Collapsed Isoforms Abundance TXT
(Files are numbered consecutively,
1 for each barcoded sample.)
pb_isoseq3_ccsonly.collapse_abundance
Separate clusters, one per barcoded sample.
Collapsed Isoforms Abundance TXT
(Files are numbered consecutively,
1 for each barcoded sample.)
pb_isoseq3_ccsonly.collapse_abundance
Pooled clusters, one per barcoded sample.
High-Quality Transcripts pb_isoseq3_ccsonly.hq_fastq
Low-Quality Transcripts pb_isoseq3_ccsonly.lq_fastq
High-Quality Transcripts Counts
(Files are numbered consecutively,
1 for each barcoded sample.)
pb_isoseq3_ccsonly.barcode_overview_report
Separate clusters, one per barcoded sample.
High-Quality Transcripts Counts
(Files are numbered consecutively,
1 for each barcoded sample.)
pb_isoseq3_ccsonly.barcode_overview_report
Pooled clusters, one per barcoded sample.
CCS reads FASTQ pb_isoseq3_ccsonly.ccs_fastq_zip
Full-length CCS reads pb_isoseq3._ccsonly.flnc_bam
Polished Report pb_isoseq3._ccsonly.polish_report_csv
Cluster Report pb_isoseq3._ccsonly.report_isoseq
File name Datastore SourceId
PCR Duplicates pb_mark_duplicates.duplicates
Deduplicated reads pb_mark_duplicates.deduplicated
In the SMRT Link UI, this displays as <ORIGINAL_DATASET_NAME>
(deduplicated).
Page 110
Microbial
Genome
Analysis
Analysis application name: cromwell.workflows.pb_microbial_analysis
Entry point
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusreadSet
Key output files
Minor Variants
Analysis
Analysis application name: cromwell.workflows.pb_mv_ccs
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_ref_dataset
:name: Entry eid_ref_dataset
:fileTypeId: PacBio.DataSet.ReferenceSet
Key output files
File name Datastore SourceId
Final Polished Assembly pb_microbial_analysis.assembly_fasta
Final Polished Assembly Index pb_microbial_analysis.assembly_fasta.fai
Final Polished Assembly for NCBI pb_microbial_analysis.ncbi_fasta
Mapped BAM pb_microbial_analysis.mapped
Target sequences used to produce
the Mapped BAM
pb_microbial_analysis.mapped_target_fasta
Target sequence Index used to
produce the Mapped BAM
pb_microbial_analysis.mapped_target_fasta_fai
List of Circular Contigs pb_microbial_analysis.circular_list
Coverage Summary pb_microbial_analysis.coverage_gff
Coverage Report pb_microbial_analysis.report_coverage
Mapping Statistics Report pb_microbial_analysis.report_mapping_stats
Mapped BAM Datastore pb_microbial_analysis.mapped_bam_datastore
Polished Assembly Report pb_microbial_analysis.report_polished_assembly
Full Kinetics Summary pb_microbial_analysis.basemods_gff
IPD Ratios pb_microbial_analysis.basemods_csv
Motifs and Modifications pb_microbial_analysis.motifs_csv
Motifs and Modifications pb_microbial_analysis.motifs_gff
File name Datastore SourceId
Minor Variants HTML Reports pb_mv_ccs.juliet_html
Per-Variant Table pb_mv_ccs.report_csv
Alignments pb_mv_ccs.mapped

Page 111
Structural
Variant Calling
Analysis application name: cromwell.workflows.pb_sv_ccs
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_ref_dataset
:name: Entry eid_ref_dataset
:fileTypeId: PacBio.DataSet.ReferenceSet
Key output files
Trim Ultra-Low
Adapters
Analysis application name: cromwell.workflows.pb_trim_adapters
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_barcode
:name: Entry eid_barcode
:fileTypeId: PacBio.DataSet.BarcodeSet
Note: The barcodes need to be a single primer sequence.
Key output files
5mC CpG
Detection
Analysis application name: cromwell.workflows.pb_detect_methyl
Entry points
:id: eid_ccs
:name: Entry eid_ccs
:fileTypeId: PacBio.DataSet.ConsensusReadSet
:id: eid_barcode
:name: Entry eid_barcode
:fileTypeId: PacBio.DataSet.BarcodeSet
File name Datastore SourceId
Structural Variants pb_sv_ccs.variants
Aligned reads (Bio Sample
Name)
pb_sv_ccs.alignments_by_sample_datastore
File name Datastore SourceId
Reads Missing Adapters pb_trim_adapters.unbarcoded
PCR Adapter Data CSV pb_trim_adapters.summary_csv
Trimmed reads pb_trim_adapters.trimmed
In the SMRT Link UI, this displays as <ORIGINAL_DATASET_NAME>
(trimmed).

Page 112
Key output files
File name Datastore SourceId
HiFi reads with 5mC Calls pb_detect_methyl.ccsbam

Page 113
Appendix B - Third party command-line tools
Following is information on the third-party command-line tools included in
the
smrtcmds/bin subdirectory.
bamtools
• A C++ API and toolkit for reading, writing, and manipulating BAM files.
•See https://sourceforge.net/projects/bamtools/ for details.
cromwell
• Scientific workflow engine used to power SMRT Link.
•See https://cromwell.readthedocs.io/en/stable/ for details.
ipython
• An interactive shell for using the PacBio API.
•See https://ipython.org/ for details.
purge_dups
• Removes haplotigs and contig overlaps in a de novo assembly based
on read depth.
•See https://github.com/dfguan/purge_dups for details.
python
• An object-oriented programming language.
•See https://www.python.org/ for details.
samtools,
BCFtools
• A set of programs for interacting with high-throughput sequencing
data in SAM/BAM/CRAM/VCF/BCF2 formats.
•See http://www.htslib.org/ for details.

Page 114
Appendix C - Microbial Genome Analysis advanced options (HiFi reads
only)
Use this application to generate de novo assemblies of small prokaryotic
genomes between 1.9-10 Mb and companion plasmids between 2 – 220
kb from Hifi (CCS) reads.
The workflow also performs base modification analysis of the assembled
genome.
The Microbial Genome Analysis application:
• Includes chromosomal- and plasmid-level de novo genome assembly,
circularization, polishing, and rotation of the origin of replication for
each circular contig.
• Performs base modification detection (m4C and m6A).
• Facilitates assembly of larger genomes (yeast) as well.
• Accepts HiFi read data (BAM format) as input.
The workflow consists of two assembly stages: chromosomal and
plasmid.
Chromosonal stage: Intended for contig assembly of large sequences.
This stage uses more stringent filtering (using advanced options) to
produce contiguous assemblies of complex regions, but it may miss small
sequences in the input sample (such as plasmids.)
Plasmid stage: Intended for a fine-grained assembly. This stage
assembles only the unmapped and poorly mapped reads. It also relaxes
the overlapping parameters, using advanced options.
Both stages use an automated random subsampling process to reduce
the input Data Set for assembly (by default to 100x). Note that the
subsampling is only applied to the contig construction process, while the
alignment and reports stages of the workflow still uses the full input Data
Set.
Available options for the two assembly stages are identical. The only
differences are:
1. Chromosonal stage parameters are prefixed with:
ipa2_advanced_options_chrom. In the SMRT Link interface for the
Microbial Genome Analysis application, this corresponds to the
Advanced Assembly Options for chromosomal stage field.
2. Plasmid stage parameters are prefixed with:
ipa2_advanced_options_plasmid. In the SMRT Link interface for the
Microbial Genome Analysis application, this corresponds to the
Advanced Assembly Options for plasmid stage field.

Page 115
The same sub-options are available to each stage, but the defaults are
very different. The current defaults are:
ipa2_advanced_options_chrom =
"config_block_size = 100; config_seeddb_opt = -k 28 -w 20 --space 0 --use-hpc-seeds-
only; config_ovl_opt = --one-hit-per-target --min-idt 98 --traceback --mask-hp --mask-
repeats --trim --trim-window-size 30 --trim-match-frac 0.75;"
ipa2_advanced_options_plasmid =
"config_block_size = 100; config_ovl_filter_opt = --max-diff 80 --max-cov 100 --min-cov
2 --bestn 10 --min-len 500 --gapFilt --minDepth 4 --idt-stage2 98; config_ovl_min_len =
500; config_seeddb_opt = -k 28 -w 20 --space 0 --use-hpc-seeds-only; config_ovl_opt = -
-one-hit-per-target --min-idt 98 --min-map-len 500 --min-anchor-span 500 --traceback --
mask-hp --mask-repeats --trim --trim-window-size 30 --trim-match-frac 0.75 --smart-hit-
per-target --secondary-min-ovl-frac 0.05; config_layout_opt = --allow-circular;"
Options are separated by semicolons; within each option, parameters are
separated by spaces.
Users should not need to modify any of default options. If the defaults are
modified, workflow behavior could be very different.
Note: The options available in
ipa2_advanced_options_* are exactly the
same as the
config_* options available for the Genome Assembly tool.
See “Genome Assembly parameters input files” on page 43 for details.
