For Research Use Only. Not for use in diagnostic procedures. © Copyright 2021 by Pacific Biosciences of California, Inc. All rights reserved PN 102-094-100 Version 01 (April 2021)
HiFi Sequencing and Software v10.1 Release:
Technical Overview for Sequel II System & Sequel IIe System Users
Sequel II and IIe Systems ICS v10.1 / SMRT Link v10.1

HiFi Sequencing and Software v10.1 Release:
Technical Overview for Sequel II & IIe Systems Users
A. Sequel II and IIe Systems v10.1 Release Overview
Summary Overview of Key Features & Improvements
Sequel II and IIe System Instrument Control Software v10.1 Updates
Sequel II and IIe System Consumables and Sample Preparation Workflow Updates
SMRT Link Sample Setup, Run Design & Run QC Updates
Example HiFi Library Sequencing Performance Data
B. SMRT Link v10.1 Release Overview
Summary Overview of Key Features & Improvements
CCS Analysis Application Features & Reports
SMRT Link v10.1 Cloud
SMRT Link Applications Updates
SMRT Link General Usability Improvements
SMRT Link Fixed & Known Issues
C. Sequel II and IIe Systems Applications Support Resources

HIFI SEQUENCING AND SOFTWARE V10.1 RELEASE:
KEY FEATURES & IMPROVEMENTS
▪ New consumables enable improved
HiFi data quality
▪ Updated HiFi sample prep protocol for
WGS applications enables reduced
DNA input requirements and
higher sample throughput / yr
▪ Updated Sequel II and IIe System
Instrument Control Software v10.1 enables
on-instrument sequencing
workflow improvements
▪ Updated SMRT Link v10.1 software
features new analysis applications
and improves Sample Setup & Run
Design ease of use
3

New Consumables
- SMRTbell Enzyme Clean Up Kit 2.0 (101-932-600) (NEW)
- Sequencing Primer v5 (102-067-400) (NEW)
- Polymerase Binding Kit 2.2 (101-894-200) (NEW)
Updated HiFi Sample Prep Protocol for De Novo Assembly
and Variant Detection
- Enables reduced minimum input gDNA (≥5 µg) for running multiple
SMRT Cells
- Supports high-throughput sample processing and automation
Sequel II and IIe System Instrument Control Software v10.1
- Updated on-instrument robotic workflow for improved fluidic handling
- Supports identification of new barcoded overhang adapters
- Supports new Adaptive Loading (AL) feature
SMRT Link v10.1
- Support for new consumables
- Application-specific Sample Setup and Run Design*
- New Adaptive Loading feature in Run Design for WGS applications
- New SARS-CoV-2 analysis application for COVID-19 surveillance
- Usability and user experience improvements
HIFI SEQUENCING AND SOFTWARE V10.1 RELEASE:
KEY FEATURES & IMPROVEMENTS
▪ New consumables enable improved
HiFi data quality
▪ Updated HiFi sample prep protocol for
WGS applications enables reduced
DNA input requirements and
higher sample throughput / yr
▪ Updated Sequel II and IIe System
Instrument Control Software v10.1 enables
on-instrument sequencing
workflow improvements
▪ Updated SMRT Link v10.1 software
features new analysis applications
and improves Sample Setup & Run
Design ease of use
4
* Feature first introduced with SMRT Link v10.0 limited release

For Research Use Only. Not for use in diagnostic procedures. © Copyright 2021 by Pacific Biosciences of California, Inc. All rights reserved.
Sequel II and IIe Systems v10.1 Release Overview
Subhead should be no longer than 1 line

SEQUEL II AND IIe SYSTEMS V10.1 RELEASE:
SUMMARY OVERVIEW OF KEY FEATURES & IMPROVEMENTS
▪ New consumables enable improved
HiFi data quality
▪ Updated HiFi sample prep protocol for
WGS applications enables reduced
DNA input requirements and
higher sample throughput / yr
▪ Updated Sequel II and IIe System
Instrument Control Software v10.1 enables
on-instrument sequencing
workflow improvements
Updated HiFi Sample Prep Protocol for De Novo Assembly
and Variant Detection
- Enables reduced minimum input gDNA (≥5 µg) for running multiple
SMRT Cells
- Supports high-throughput sample processing and automation
Sequel II and IIe System Instrument Control Software v10.1
- Updated on-instrument robotic workflow for improved fluidic handling
- Identification of new barcoded overhang adapters
- Supports new Adaptive Loading (AL) feature
New Consumables
- SMRTbell Enzyme Clean Up Kit 2.0 (101-932-600) (NEW)
- Sequencing Primer v5 (102-067-400) (NEW)
- Polymerase Binding Kit 2.2 (101-894-200) (NEW)
6

Sequel II and IIe System Instrument Control
Software v10.1 Updates

SEQUEL II AND IIE SYSTEM INSTRUMENT CONTROL SOFTWARE
V10.1 UPDATES
Updated Sequel II and IIe System ICS v10.1 enables several sequencing
workflow improvements
- Updated on-instrument robotic workflow enables
improved fluidic handling during reagent/sample
aspiration & dispense steps to minimize the impact of
drying in low-humidity environments and results in
more uniform loading across the SMRT Cell 8M
surface area
- Enables new Adaptive Loading (AL) feature to
monitor kinetics of immobilization of polymerase
complexes to ZWWs leading to reduced loading
variability and reduced risk of sample overloading
conditions
- Enables support for new barcoded overhang adapters
leading to more streamlined analysis of multiplexed
samples. (Note: New redesigned barcoded overhang
adapter sequences will become available in a future
protocol release.)
8

SEQUEL II AND IIE SYSTEM INSTRUMENT CONTROL SOFTWARE
V10.1 UPDATES (CONT.)
Updated Sequel II and IIe System ICS v10.1 enables several sequencing
workflow improvements
- Enables improved environmental systems control
leading to increased instrument reliability
▪ Note: Reagent chiller no longer cools the work deck
when the instrument is idle and no sequencing kit is
detected by the NFC reader
▪ NFC reader will scan for the presence of unused reagents
in the sequencing plate upon instrument power up, upon
door closure, and upon end of a sequencing run
▪ A sample plate should not be loaded on the work deck
without a sequencing plate present since there is no NFC
tag on sample plates.
Improved environmental
systems control helps
extend the service life
of the reagent chiller.
9

Sequel II and IIe System Consumables and
Sample Preparation Workflow Updates

NEW SEQUEL II AND IIE SYSTEM REAGENT KIT PRODUCT
DESCRIPTIONS
SMRTbell Enzyme Clean Up Kit 2.0 (101-932-600)
- Improved formulation enables more efficient removal of damaged/incomplete
SMRTbell template constructs from final library samples
- For use with HiFi WGS de novo assembly and variant detection applications
Sequencing Primer v5 (102-067-400)
- Sequencing Primer v5 is recommended for use with Sequel II Binding Kit 2.2
and enables more processive sequencing of SMRTbell DNA templates leading
to improved HiFi data quality
Sequel II Binding Kit 2.2 and DNA Internal Control 1.0 (102-089-000)
- Faster Sequel II Polymerase 2.2 resulting in more subread passes and
improved HiFi data quality
- No change to spike-in DNA Internal Control
11
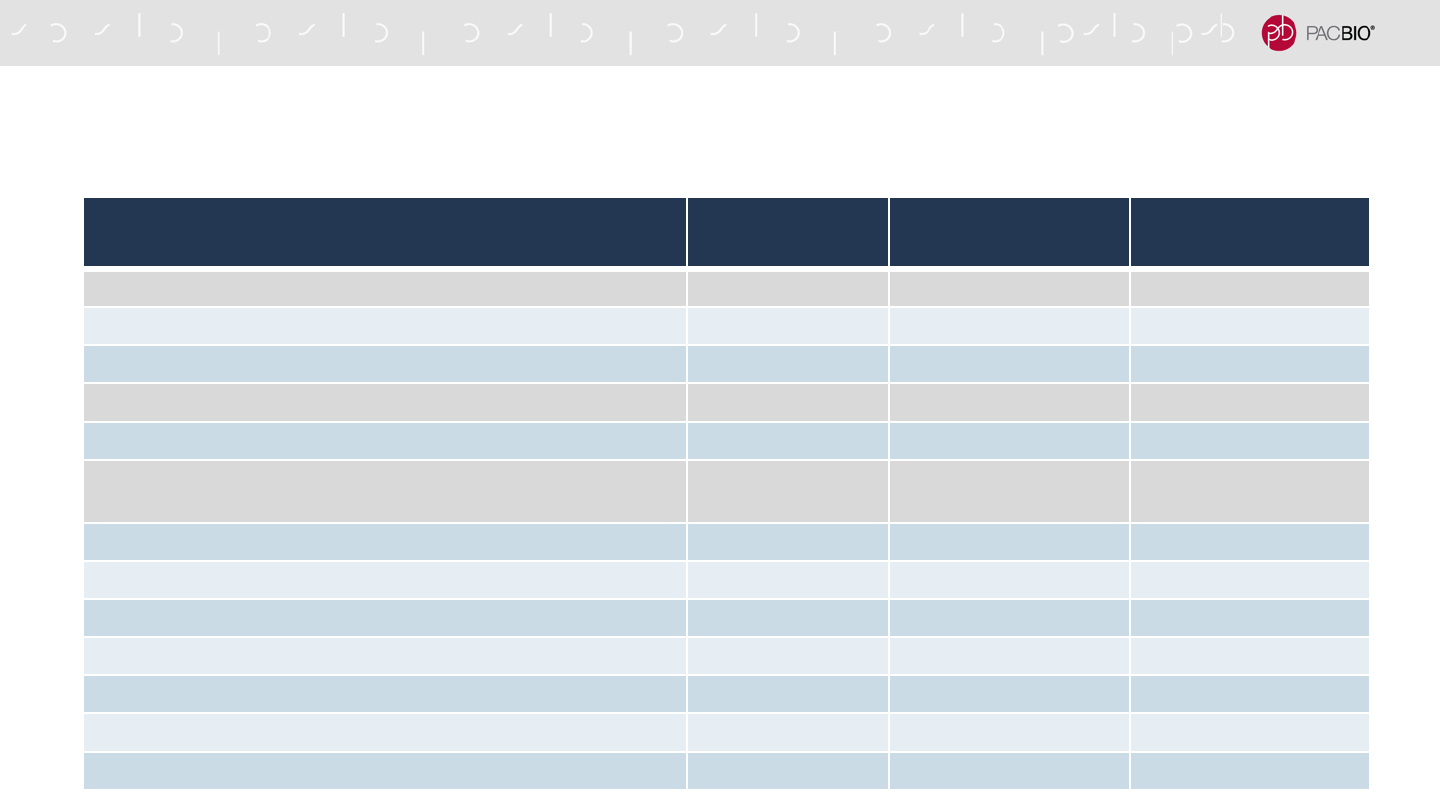
Reagent Kit or Component Part Number Quantity
No. of Reactions
Supported
SMRTbell Enzyme Clean Up Kit 2.0 (NEW) 101-932-600 18
SMRTbell Enzyme Clean Up Mix 1 Tube
SMRTbell Enzyme Clean Up Buffer 1 Tube
Sequencing Primer v5 (NEW) 102-067-400 10
Sequencing Primer v5 1 Tube
Sequel II Binding Kit 2.2 and Internal Control 1.0 (NEW) 102-089-000 24
Sequel II DNA Polymerase 2.2 1 Tube
Adaptive loading Buffer 6 Tubes
Sequel Binding Buffer 2 Tubes
Sequel dNTP 1 Tube
Sequel Complex Dilution Buffer 2 Tubes
Nuclease-Free Water 1 Tube
DNA Internal Control 1.0 1 Tube
NEW SEQUEL II AND IIE SYSTEM CONSUMABLES PRODUCTS
AND REAGENT KIT CONTENTS
12
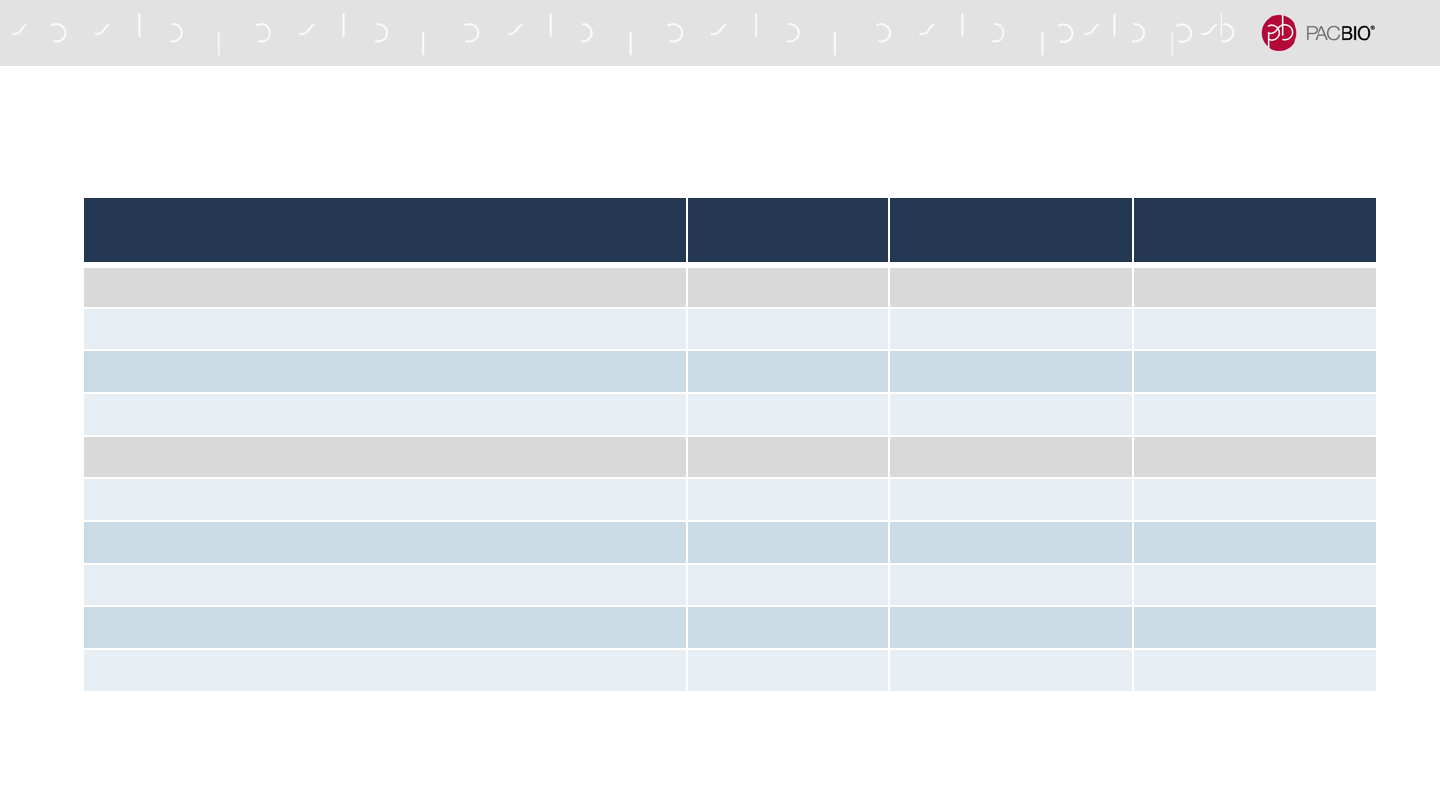
Reagent Kit or Component Part Number Quantity
No. of Reactions
Supported
HiFi Express Template Prep Kit 2.0 Bundle (UPDATED) 102-088-900 18
SMRTbell Express Template Prep Kit 2.0 1 Each
SMRTbell Enzyme Clean Up Kit 2.0 (NEW) 1 Each
Sequencing Primer v5 (NEW) 1 Tube
Sequel II HiFi Bundle-18 2.0 (UPDATED) 102-104-700 18
SMRTbell Express Template Prep Kit 2.0 1 Each
SMRTbell Enzyme Clean Up Kit 2.0 (NEW) 1 Each
Sequencing Primer v5 (NEW) 1 Each
Sequel II Binding Kit 2.2 and Internal Control 1.0 (NEW) 1 Each
AMPure PB, 5 mLs 1 Tube
NEW SEQUEL II AND IIE SYSTEM CONSUMABLES PRODUCTS
AND REAGENT KIT CONTENTS
13

UPDATED HIFI LIBRARY PREPARATION PROTOCOL FOR WGS
DE NOVO ASSEMBLY & VARIANT DETECTION APPLICATIONS
- Procedure & Checklist – Preparing HiFi SMRTbell Libraries using SMRTbell Express
Template Prep Kit 2.0 (PN 101-853-100) protocol document has been updated and
describes a method for constructing SMRTbell libraries (~15 kb - 20 kb) that are suitable for
generating highly accurate long reads on the Sequel II and IIe Systems for WGS de novo
assembly and variant detection applications
- Updated workflow supports high-throughput processing using reduced input genomic DNA
amounts (5 µg per 3 Gb sample genome size)
- Recommend shearing high-quality gDNA using a Megaruptor 3 System (Diagenode)
- Depending on project requirements, SMRTbell libraries can be size-selected using a
PippinHT System (Sage Science), SageELF System (Sage Science), or BluePippin System
(Sage Science)
https://www.pacb.com/support/documentation/
14
APPLICATIONS
WHOLE GENOME SEQUENCING
De Novo Assembly
Variant Detection
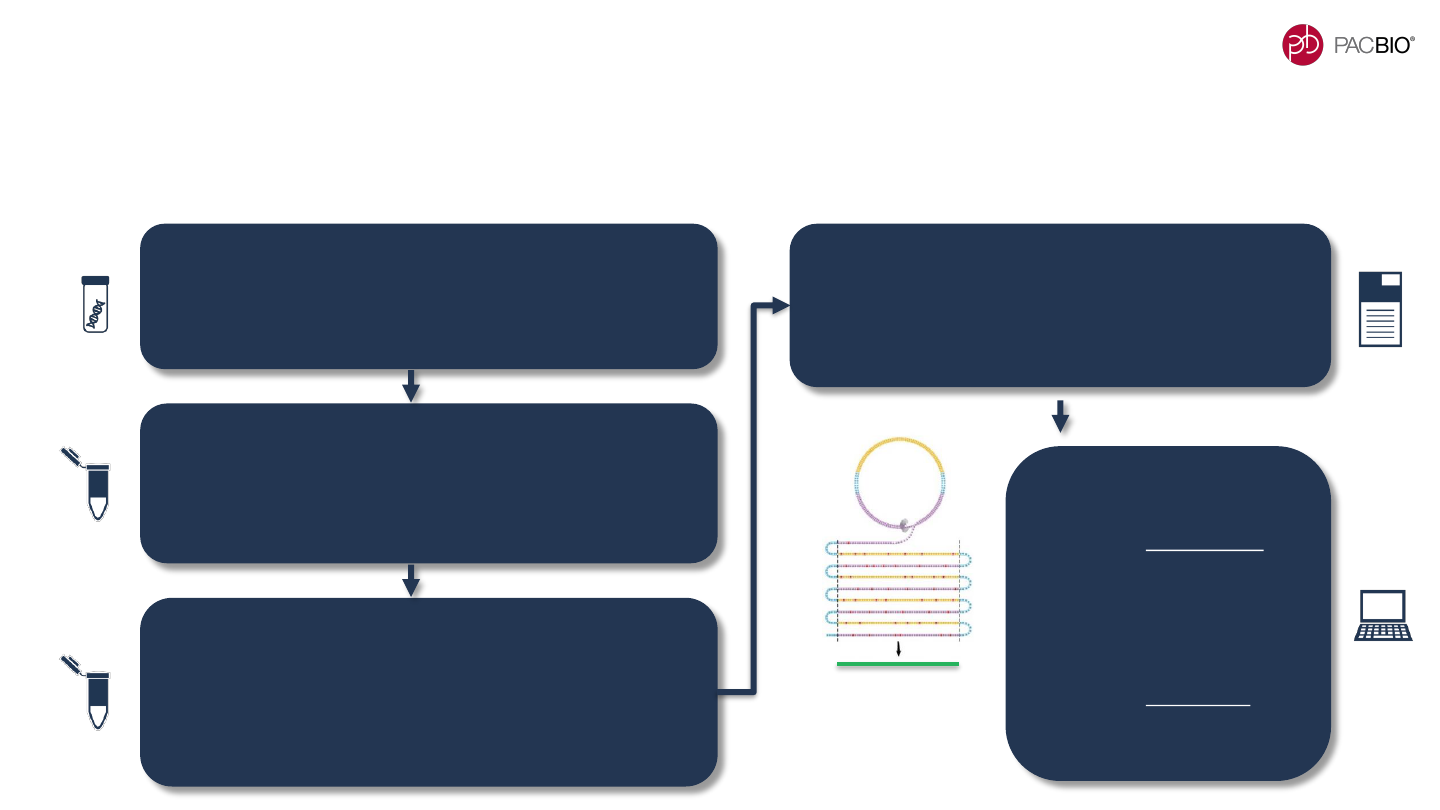
HIFI LIBRARY PREPARATION & SEQUENCING WORKFLOW
IS EFFICIENT AND SCALABLE
Updated HiFi sample preparation workflow provides improved SMRTbell library construction yields and
supports high-throughput processing using reduced input genomic DNA amounts
Genomic DNA QC & Shearing
▪ ≥5 µg of input gDNA for a 3 Gb sample genome size
▪ Shear up to 8 samples in parallel to ~15 kb - 20 kb
target fragment size with the Megaruptor 3 System
SMRTbell Library Construction (5 hrs)
▪ Process up to 16 samples in parallel manually or up
to 96 samples in parallel using automation
▪ Manual workflow supports construction of ≥32 HiFi
libraries per week
SMRTbell Library Size Selection (2 hrs)
▪ Process up to 20 samples in parallel with the
PippinHT System (2 hour run time)
▪ Higher recovery efficiency with PippinHT provides
sufficient SMRTbell library material to run up to ~4 or
more SMRT Cells 8M per 5 µg of starting input gDNA
Sequencing Preparation
(Sequel II and IIe Systems)
▪ Anneal Sequencing Primer v5 (1 hr), bind Sequel
II Polymerase 2.2 (1 hr) and perform complex
cleanup using AMPure PB beads
HiFi Data Analysis
▪ For variant detection,
can use DeepVariant for
small variants <20 bp
and SMRT Link
Structural Variant Calling
for larger variants >20 bp
▪ For de novo assembly,
can use SMRT Link
Genome Assembly or
other third-party software
PacBio HiFi
reads achieve
>99.9% accuracy
HiFi Read
15
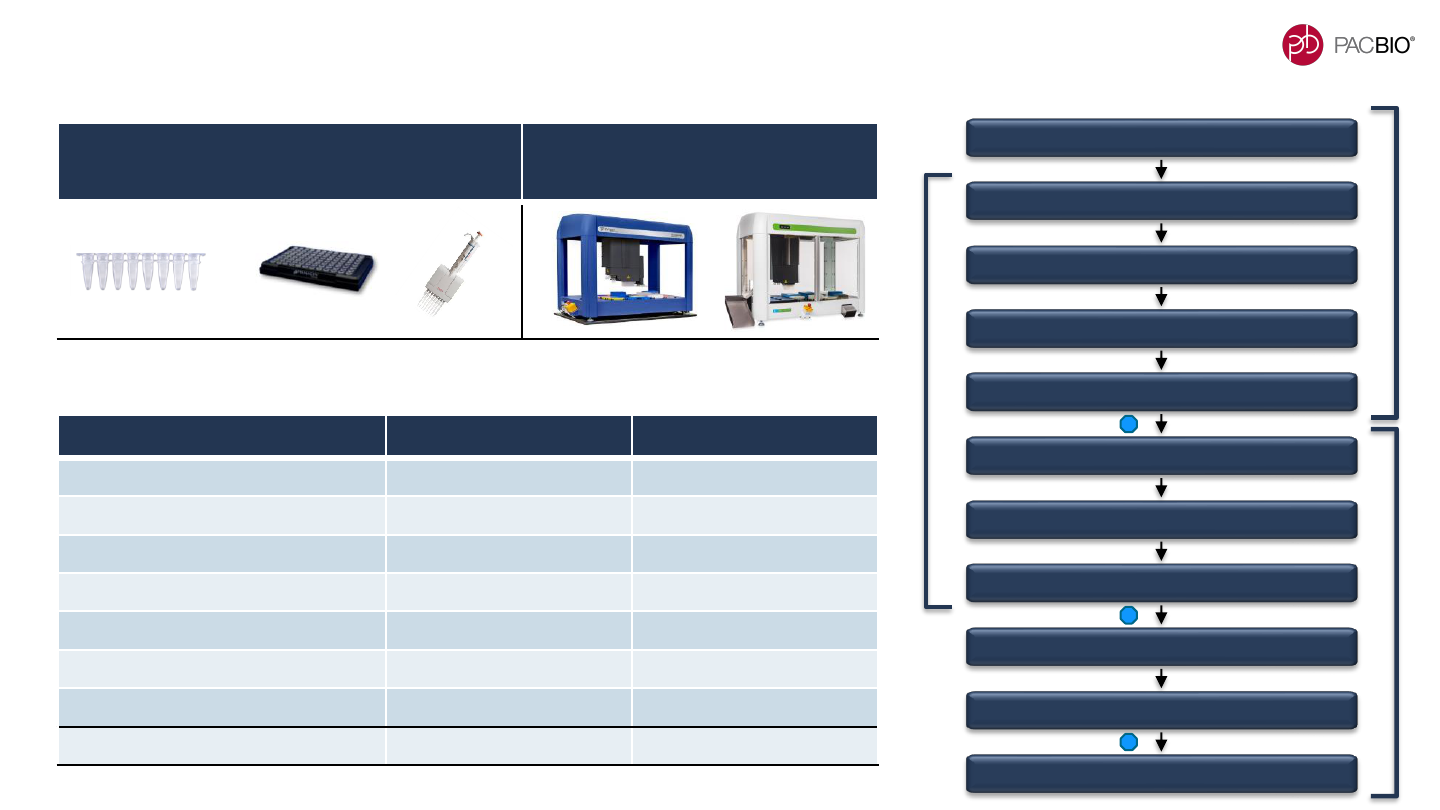
STEP HANDS-ON (MIN) WALK-AWAY (HRS)
Remove SS to A-Tailing 15 1.4
Adapter Ligation** 5 1.0
AMPure PB Bead Purification 5 0.5
Nuclease Treatment 5 0.5
AMPure PB Bead Purification 5 0.5
Size
-
selection (PippinHT System)
10 2.0
AMPure PB Bead Purification 5 0.5
Total 50 6.4
PROCESS UP TO 16 SAMPLES IN PARALLEL
MANUALLY USING 0.2-ML TUBE STRIPS*
PROCESS UP TO 96 SAMPLES IN
PARALLEL USING AUTOMATION*
HIFI LIBRARY CONSTRUCTION WORKFLOW TIMING OVERVIEW
DNA Shearing
DNA Damage Repair
DNA End Repair / A-Tailing
Adapter Ligation
1X AMPure PB Bead Purification
Remove Single-Strand Overhangs
1 X AMPure PB Bead Purification
Nuclease Treatment
Size Selection
1 X AMPure PB Bead Purification
Sequencing Preparation
Enzymatic Reactions ~5 hours
Optional Stop Point
Optional Stop Point
** Adapter Ligation reaction can be performed for 1 hour or left incubating overnight
High-throughput library construction and sequencing preparation in 2 days
Day 1
Day 2
* Reagents can be prepared as Master Mixes
Optional Stop Point
16

THE MEGARUPTOR 3 SYSTEM IS RECOMMENDED FOR HIFI SMRTBELL
LIBRARY CONSTRUCTION
- Megaruptor 3 System (Diagenode) is generally recommended for shearing*
- Allows up to 8 samples to be sheared in parallel for high-throughput applications
- Achieving the same size distribution across multiple samples provides more consistent sequencing
performance
- To maximize HiFi yield per SMRT Cell, PacBio recommends fragmenting the gDNA to a size distribution mode
between 15 kb – 18 kb for human whole genome sequencing
- Note: Libraries with a size distribution mode larger than 20 kb are not recommended for HiFi sequencing.
- Recommended library insert size distributions to use for different WGS applications are summarized below and in
Table 4 in the procedure.
Megaruptor 3 System
17
Application Recommended Library Insert Size (Mode)
Human Variant Detection 15 kb - 18 kb
Human De Novo Assembly 15 kb - 18 kb
Plant / Animal De Novo Assembly 15 kb - 20 kb
* Note: The g-TUBE (Covaris) device generates a broader DNA fragment size-distribution compared to the Megaruptor 3 system. As a result, HiFi read quality and overall HiFi data yield
may be reduced due to the residual presence of very large DNA fragments generated by g-TUBEs. For additional guidance, see Technical Overview: HiFi Library Preparation Using
SMRTbell Express TPK 2.0 for De Novo Assembly and Variant Detection (PN 101-855-400) or contact PacBio Technical Support or your local Field Applications Scientist.

To use the Megaruptor 3 System, perform two cycles of DNA shearing in the same hydropore-syringe device
- Eliminates very large DNA fragments that may not generate sequencing data that meet HiFi read quality requirements
- Example recommended Megaruptor 3 System software settings to achieve a DNA fragment size distribution mode of ~15 kb - 18 kb
(recommended for human whole genome sequencing applications):
- IMPORTANT: Genomic DNA must be in QIAGEN Buffer EB or PacBio Elution Buffer (EB) or an equivalent low-salt buffer (i.e.,
10 mM Tris-Cl, pH 8.5 - 9.0) for shearing
- To minimize sample loss,** the recovered volume (~53 𝜇L) of sheared DNA is used to go directly into the first enzymatic reaction in
SMRTbell library construction (i.e., no intermediate AMPure PB bead purification step is performed)
gDNA TARGET
SHEAR SIZE MODE
MEGARUPTOR 3 SYSTEM
SHEARING CYCLE
MEGARUPTOR 3 SYSTEM
SPEED SETTING*
RUN TIME
PER SHEARING CYCLE
15 – 18 kb
Cycle 1 Speed Setting 31 40 min
Cycle 2 Speed Setting 32 40 min
THE MEGARUPTOR 3 SYSTEM IS RECOMMENDED FOR HIFI SMRTBELL
LIBRARY CONSTRUCTION (CONT.)
* Note: The shearing instructions described in this HiFi sample prep procedure are not compatible with the Megaruptor or Megaruptor 2 systems from Diagenode. If using a Megaruptor or
Megaruptor 2 system, shearing optimization is necessary before proceeding with this procedure. For additional guidance, see Technical Overview: HiFi Library Preparation Using
SMRTbell Express TPK 2.0 for De Novo Assembly and Variant Detection (PN 101-855-400) or contact PacBio Technical Support or your local Field Applications Scientist.
** Losses are mostly due to dead volume in the Megaruptor 3 System [i.e., 5 – 7 𝜇L (<500 ng)]
Because the response of individual gDNA samples may differ, optimization of shearing
conditions is recommended to achieve the desired fragment distribution
18
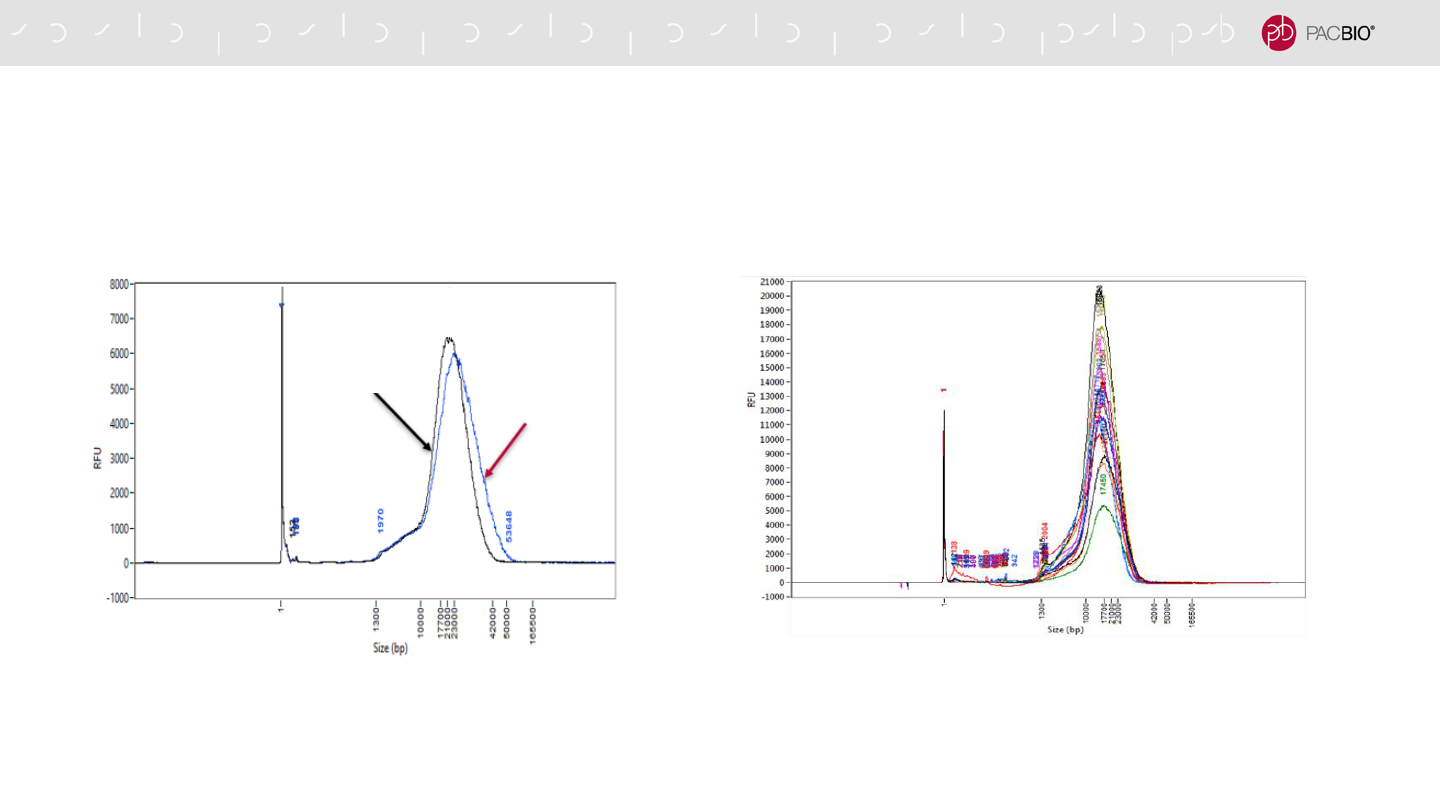
2-CYCLE SHEARING METHOD USING THE MEGARUPTOR 3 SYSTEM IS
RECOMMENDED FOR HIFI SMRTBELL LIBRARY CONSTRUCTION
Femto Pulse DNA sizing QC analysis overlay of 15 human gDNA
samples sheared with 2 cycles of shearing using a Megaruptor 3
System. The size distribution mode of the samples after 2 cycles of
shearing is ~18 kb.
Femto Pulse DNA sizing QC analyses of the same human gDNA
sample sheared with 1 cycle of shearing versus 2 cycles of
shearing using a Megaruptor 3 System. The fragment size
distribution is tighter after performing 2 cycles of shearing compared to
performing 1 cycle of shearing.
1-Cycle Shear
(Speed 32)
2-Cycle Shear
(Speeds 31, 32)
By performing a 2-cycle shear, the resulting DNA fragment size distribution is tighter and more
consistent across multiple samples
2-Cycle Shear
(Speeds 31, 32)
19
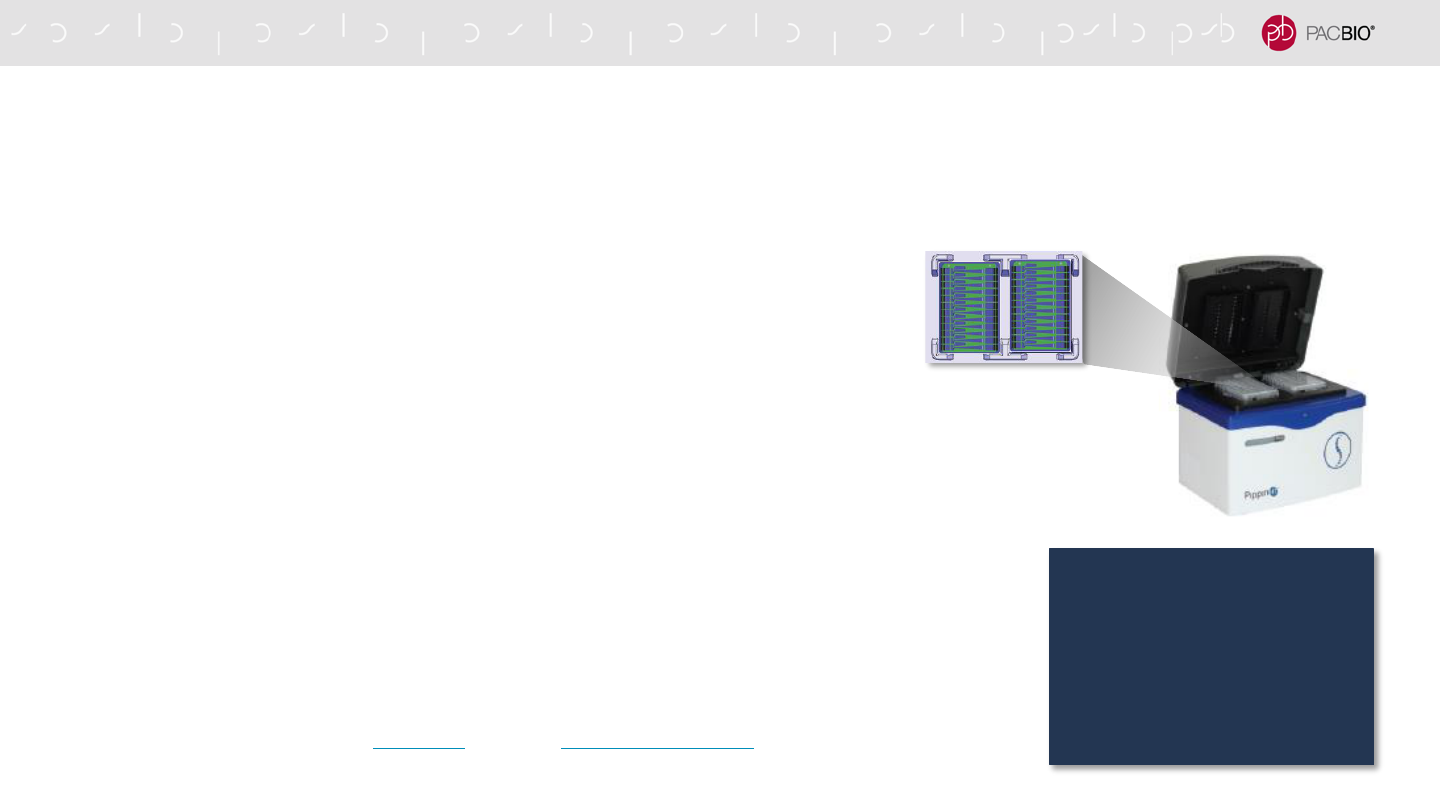
SIZE-SELECTION WITH THE PippinHT SYSTEM IS RECOMMENDED FOR
HIFI SMRTBELL LIBRARY CONSTRUCTION
- PippinHT System (Sage Science) is recommended for size selection
- PippinHT System enables faster run times and higher throughput compared to the
SageELF and BluePippin Systems
- Can process up to 20 samples per instrument run
- 2-hour run time
- SMRTbell templates <10 kb are removed during PippinHT size selection
- PippinHT Cassette Definition File and Run Protocol Setup
- “6-10kb High Pass Marker 75E”
- Using the “Range” selection mode, enter a desired “Start” value of 10000 and a
“End” value of 50000.
- Be sure to assign a marker lane
- PippinHT System shows efficient post-size selection recovery yields (approx. ≥35%
– 50%), which enables reduction of the input gDNA amount required for HiFi
SMRTbell library construction*
* Note: If using BluePippin or SageELF size-selection, library recovery yields may be lower with this HiFi sample prep procedure.
For additional guidance, see Technical Overview: HiFi Library Preparation Using SMRTbell Express TPK 2.0 for De Novo
Assembly and Variant Detection (PN 101-855-400) or contact PacBio Technical Support or your local Field Applications
Scientist.
PippinHT System
With high-quality samples,
higher recovery efficiency with
PippinHT size selection can
provide sufficient SMRTbell
library material to run up to ~4
or more SMRT Cells 8M per
5 µg of starting input gDNA
20
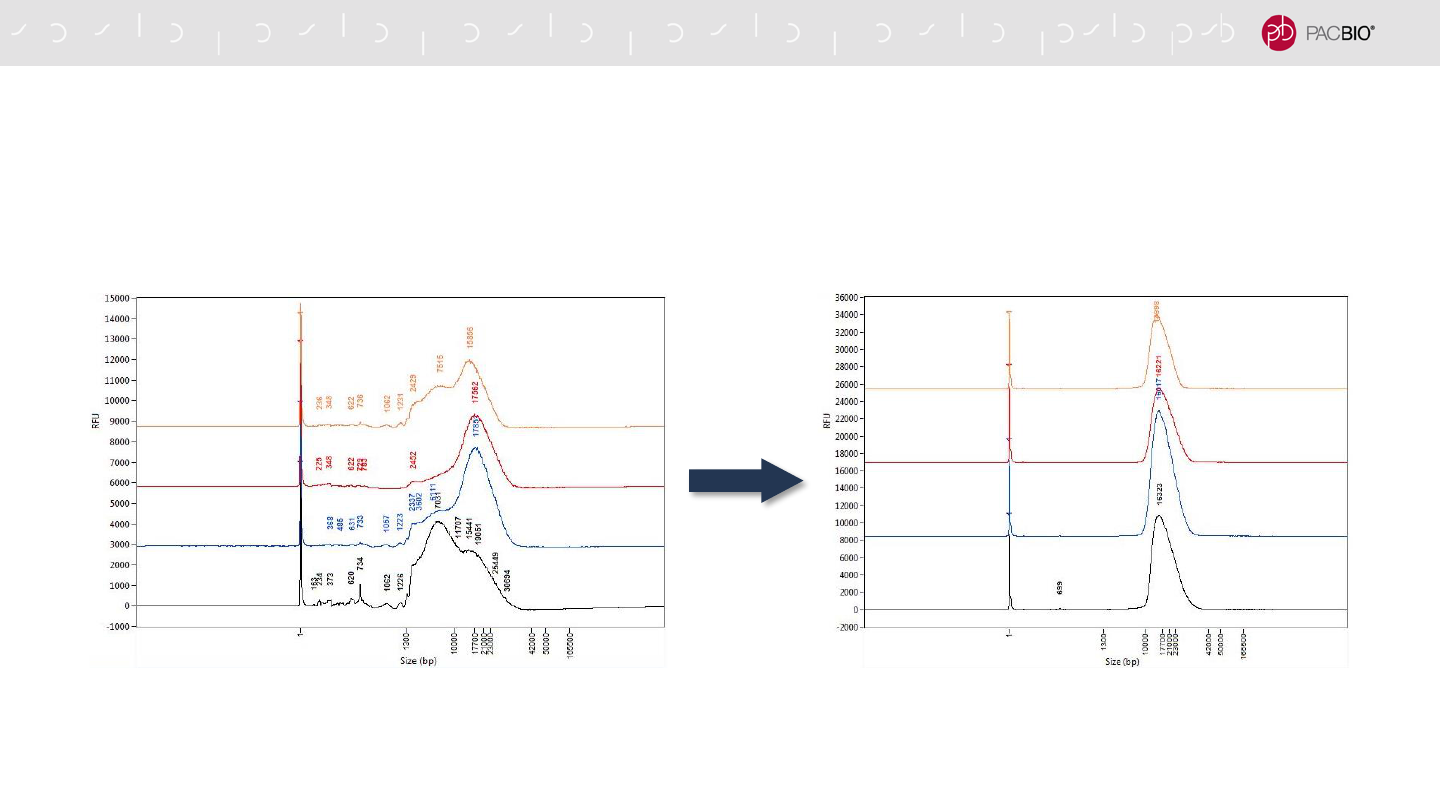
SIZE-SELECTION WITH THE PippinHT SYSTEM IS RECOMMENDED FOR
HIFI SMRTBELL LIBRARY CONSTRUCTION (CONT.)
16.3 kb Mode
18.6 kb Mean
16.2 kb Mode
18.0 kb Mean
16.0 kb Mode
18.0 kb Mean
15.0 kb Mode
16.0 kb Mean
Before PippinHT Size Selection After PippinHT Size Selection
PippinHT size selection is fast (2-hr run time) and efficient (can process up to 20 samples per instrument run)
Femto Pulse DNA sizing QC analyses of the several human
SMRTbell library samples after PippinHT size selection using a 10-
kb lower cutoff setting. The size distribution mode of the size-selected
samples is similar (~18 kb).
Femto Pulse DNA sizing QC analyses of the several human
SMRTbell library samples before size selection. The size distribution
mode ranges from ~7 kb to ~18 kb for the different samples.
21

- Enables greater reproducibility compared with manual-only
methods
- Can process up to 96 samples at a time
HIFI SAMPLE PREPARATION WORKFLOW IS AMENABLE TO AUTOMATION
HiFi SMRTbell library construction can be automated using the Sciclone Liquid Handling
Workstation (Perkin-Elmer)
https://www.perkinelmer.com/category/sciclone-g3-liquid-handling-workstations
Input Requirements:
- Recommend using >6 𝜇g of input sheared gDNA due to liquid dead
volumes
Automated Workflow Steps:
- Enzymatic reactions
- AMPure PB bead purifications
Output:
- SMRTbell libraries ready for downstream size-selection
22
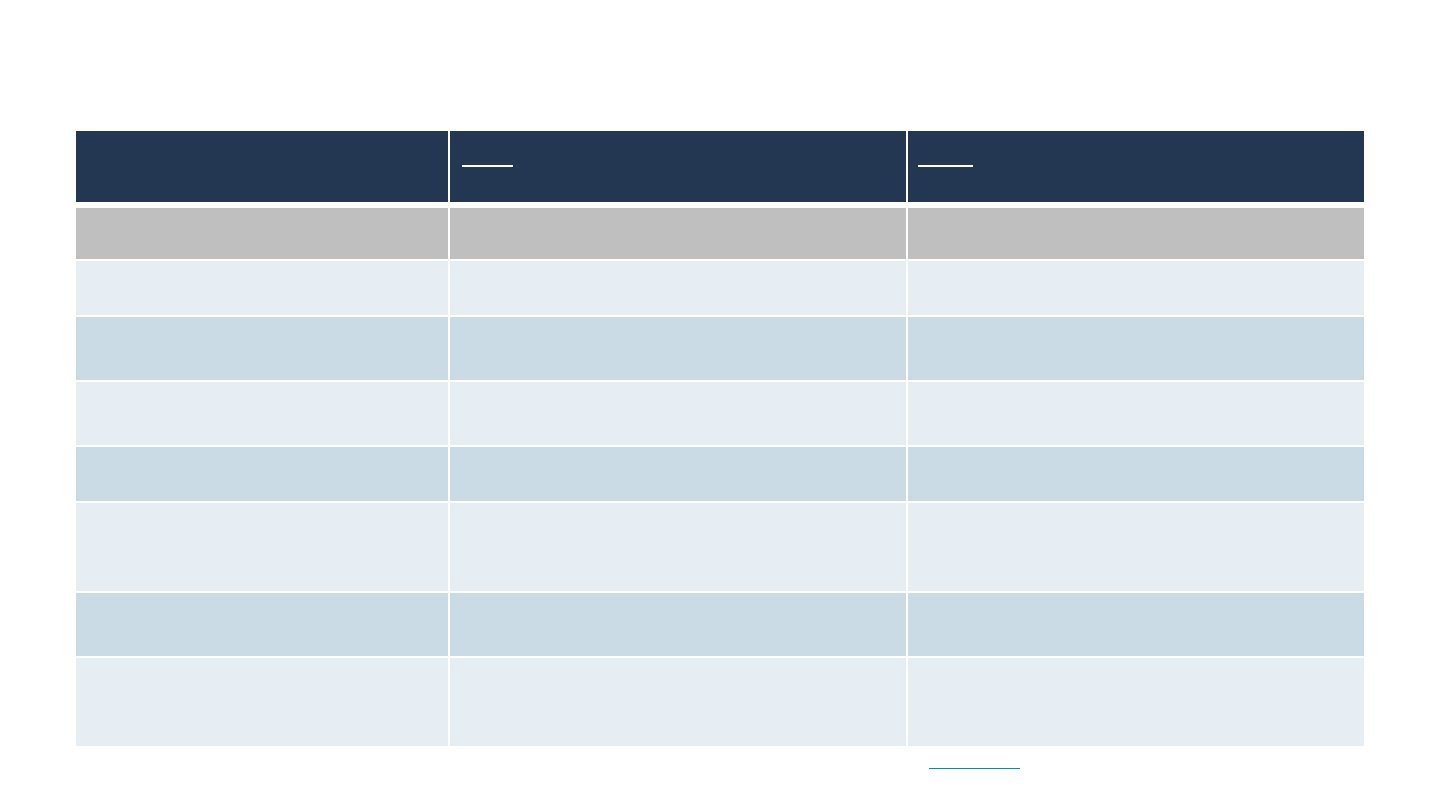
COMPARISON OF HIFI SAMPLE PREPARATION WORKFLOW CHANGES FOR
DE NOVO ASSEMBLY AND VARIANT DETECTION APPLICATIONS
SAMPLE PREP
WORKFLOW STEP
OLD HIFI PROCEDURE & CHECKLIST
(VERSION 3, JAN. 2020)
NEW HIFI PROCEDURE & CHECKLIST
(VERSION 4, APR. 2021)
LIBRARY CONSTRUCTION
Input gDNA Amount ~15 µg ≥5 µg
DNA Shearing
Megaruptor 1/2/3 System; or
g-TUBEs
Megaruptor 3 System (2-cycle shearing)
Post Shearing AMPure PB Bead
Purification
Yes No
Post-ligation Heat Kill Yes No
Buffer Exchange prior to Nuclease
cleanup (AMPure PB beads)
No Yes
SMRTbell Enzyme Clean up Kit
Version (Reaction Time)
1.0 (60 min) 2.0 (30 min)
Size-Selection Options*
SageELF System; or
BluePippin System; or
AMPure PB Bead Size Selection
PippinHT System; or
SageELF System; or
BluePippin System
* AMPure PB bead size selection is under development for the new HiFi sample preparation Procedure & Checklist (PN 101-853-100, Version 4) and specific guidance will be
provided in a future protocol update.
23

SAMPLE PREP
WORKFLOW STEP
OLD HIFI PROCEDURE & CHECKLIST
(VERSION 3, JAN. 2020)
NEW HIFI PROCEDURE & CHECKLIST
(VERSION 4, APR. 2021)
SMRT LINK SAMPLE SETUP
Primer Annealing Sequencing Primer v2 Sequencing Primer v5
Polymerase binding
Sequel II Polymerase 2.0
Binding Time = 4 h
Sequel II Polymerase 2.2
Binding Time = 1 h
Complex Cleanup
Dilute Bound Complex Volume by 3.33-fold and
purify sample using 1.2X AMPure PB beads
If Bound Complex Volume is <100 µL, bring up to
100 µL with Complex Dilution Buffer and purify
sample using 1.2X AMPure PB beads.
SMRT LINK RUN DESIGN SETUP
Pre-Extension Time 2 h (<20 kb) or 4 h (≥20 kb) 0 h (No Pre-extension)
Adaptive Loading (AL) OFF ON
COMPARISON OF HIFI SAMPLE PREPARATION WORKFLOW CHANGES FOR
DE NOVO ASSEMBLY AND VARIANT DETECTION APPLICATIONS (CONT.)
24
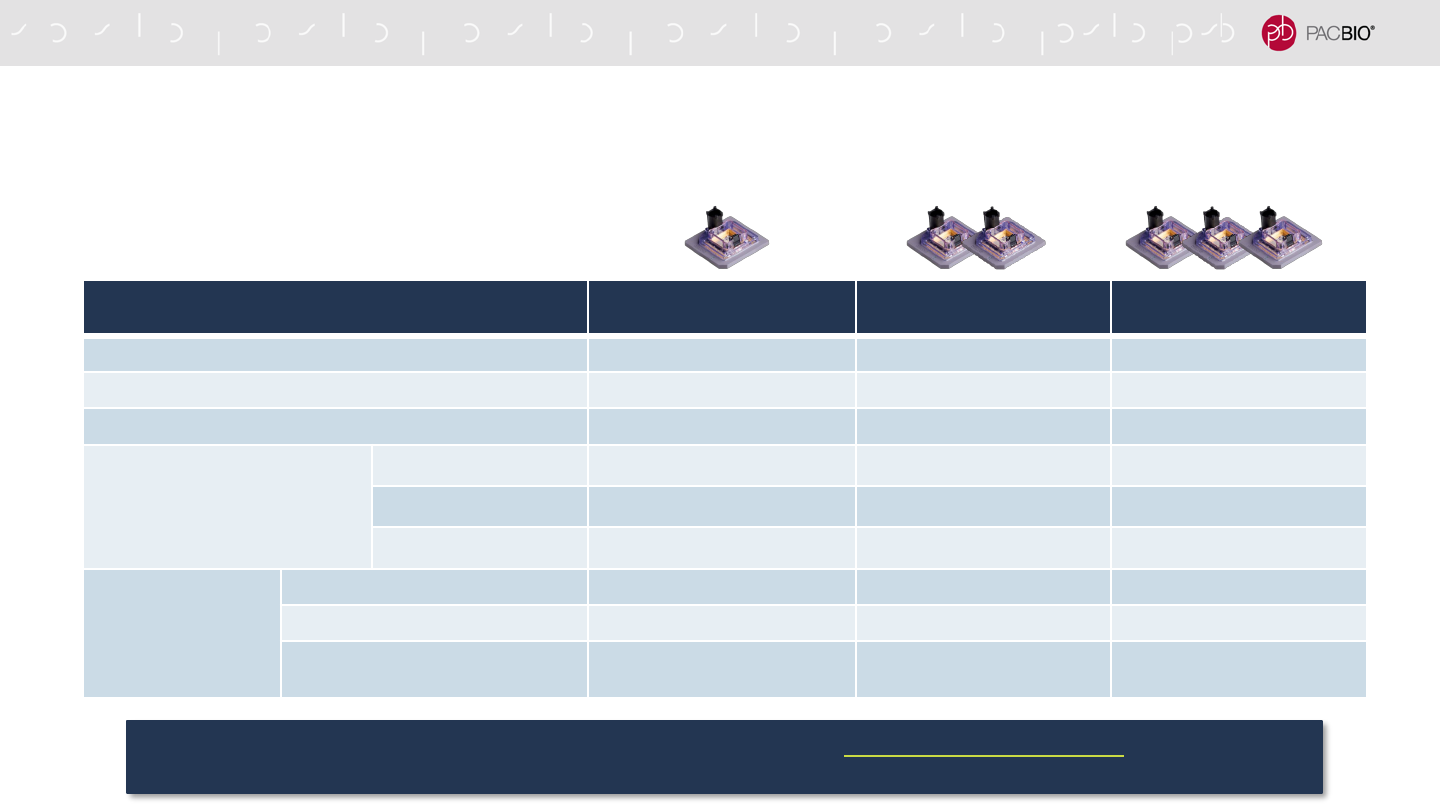
ENABLING PRODUCTION-SCALE THROUGHPUTS FOR HUMAN WHOLE
GENOME SEQUENCING FOR RARE AND INHERITED DISEASE RESEARCH
STRUCTURAL VARIANTS
(SVs)
VARIANT DETECTION
(SNVs, INDELs, SVs)
VARIANT DETECTION
(SNVs, INDELs, SVs)
Number of SMRT Cells 8M/sample 1 2 3
Sequencing time per sample (hrs) 30 60 90
Coverage per human genome sample ~10-fold ~20-fold ~30-fold
Variant Detection
Performance
(% Accuracy, F1)
SNV 99.0% 99.8% 99.9%
Indel 92.6% 96.9% 97.9%
SV 92.6% 95.7% 95.8%
Annual Sample
Throughput
1 Sequel IIe System 256 128 84
6 Sequel IIe Systems 1,536 768 504
12 Sequel IIe Systems 3,072 1,536 1,008
This efficient HiFi sample preparation workflow, developed in collaboration with Children’s Mercy Kansas City, provides a scalable
solution for sequencing 100s to 1000s of whole human genomes per year on the Sequel II and IIe Systems.
25

SMRT Link Sample Setup, Run Design & Run QC
Updates
SMRT LINK V10.1 SAMPLE SETUP & RUN DESIGN
RECOMMENDATIONS FOR SPECIFIC APPLICATIONS
Generally follow SMRT Link Sample Setup & Run Design instructions using the recommendations
provided in the Quick Reference Card – Loading and Pre-Extension Time Recommendations for the
Sequel II and IIe Systems unless specified otherwise in the relevant Procedure & Checklist
27
https://www.pacb.com/support/documentation/
In SMRT Link v10.1, most Sample Setup
and Run Design parameter fields are
auto-filled with the recommended settings for
each application type.
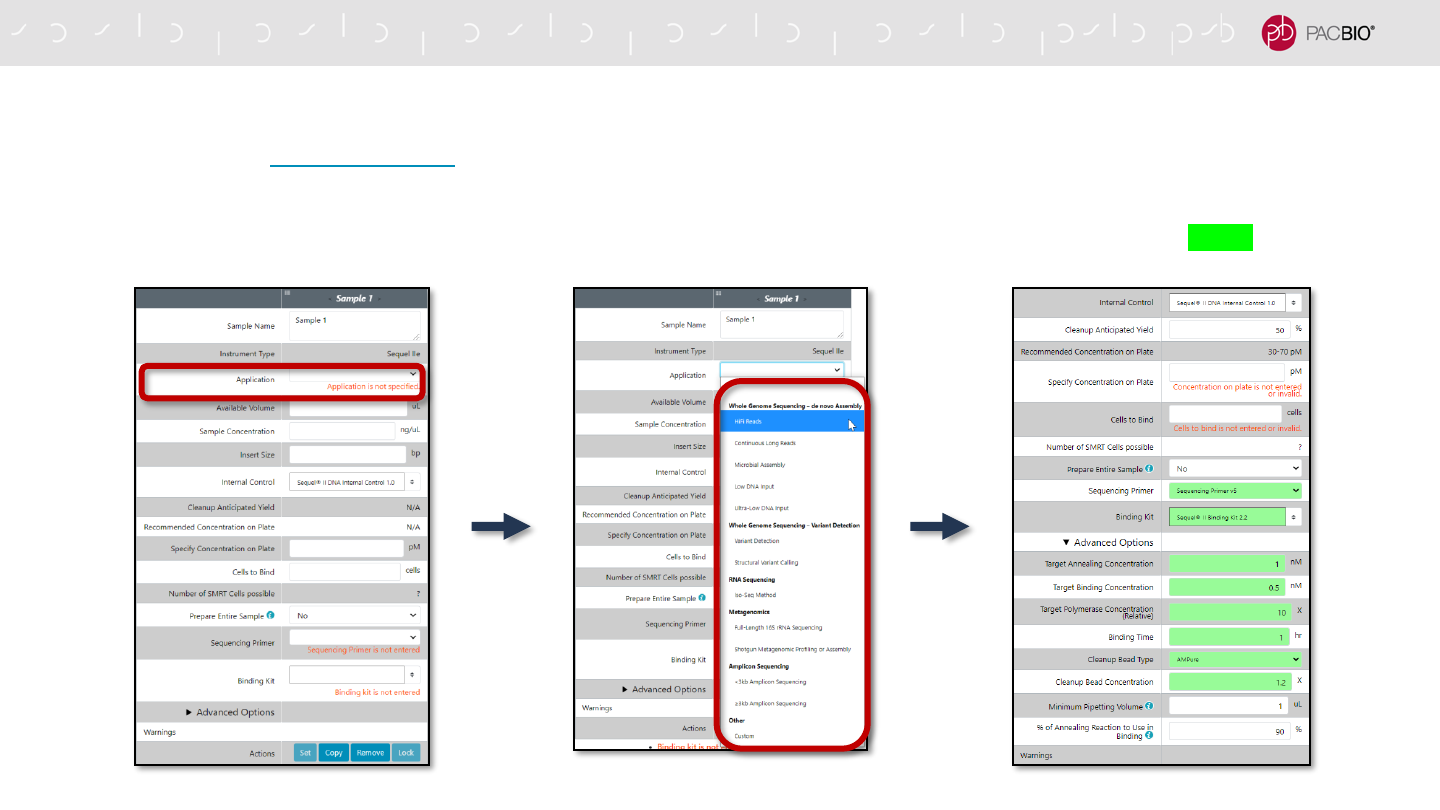
UPDATED SAMPLE SETUP WORKFLOW: SPECIFYING APPLICATION TYPE
28
Sample Setup auto-populates application-specific information for selected fields
- The user starts by first entering the Sample Name and then selecting an Application Type
- Once an application is selected, default values are auto-populated for various fields and highlighted in green
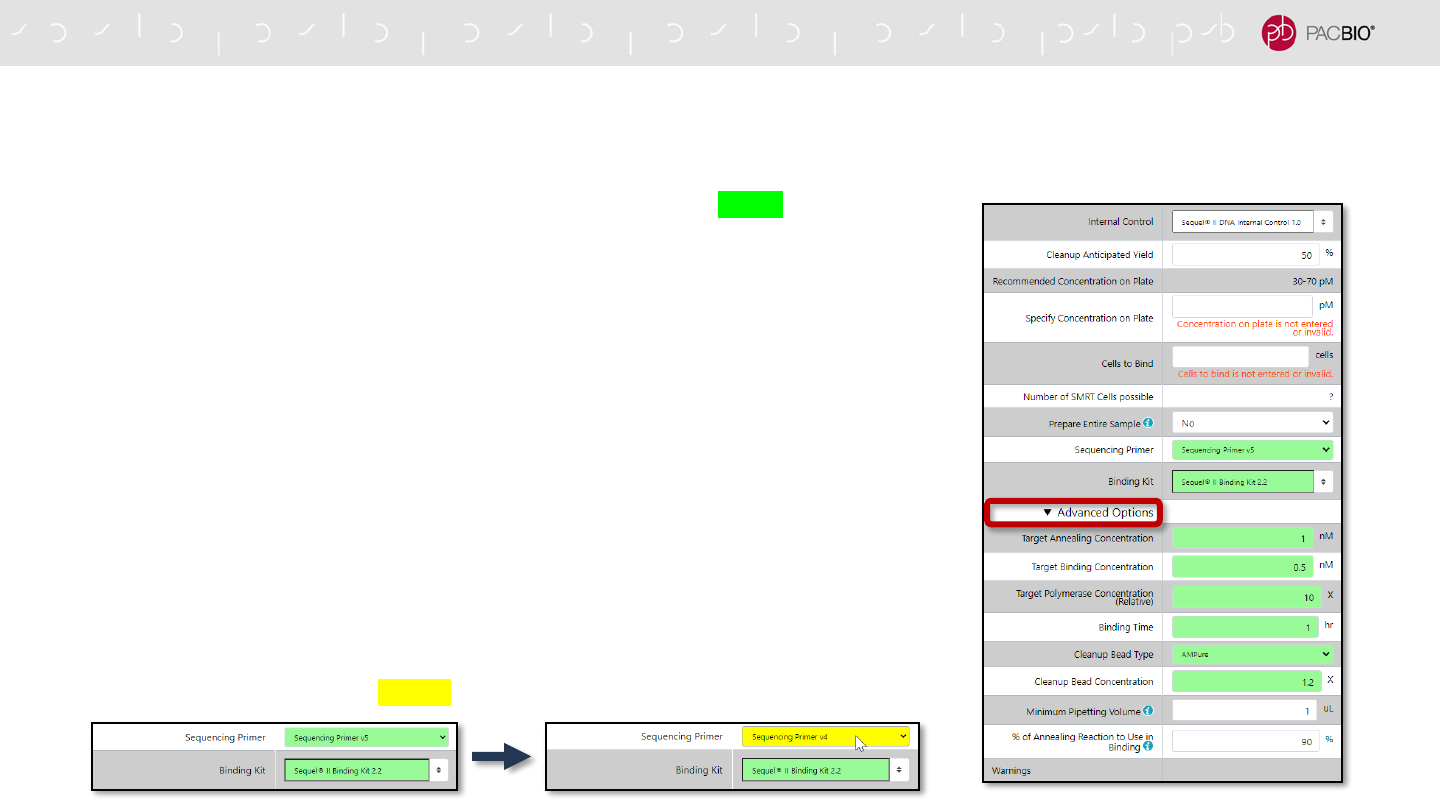
UPDATED SAMPLE SETUP WORKFLOW: SPECIFYING APPLICATION TYPE (CONT.)
29
Auto-populated Sample Setup fields are highlighted in green color
- The following fields are auto-populated and highlighted in green:
- Sequencing Primer
- Binding Kit
- Note: The following auto-populated fields are located in Advanced Options:
- Target Annealing Concentration
- Target Binding Concentration
- Target Polymerase Concentration (Relative)
- Binding Time
- Cleanup Bead Type
- Cleanup Bead Concentration
- If any auto-populated entry is manually changed to a different value, then the
field will be highlighted in yellow color
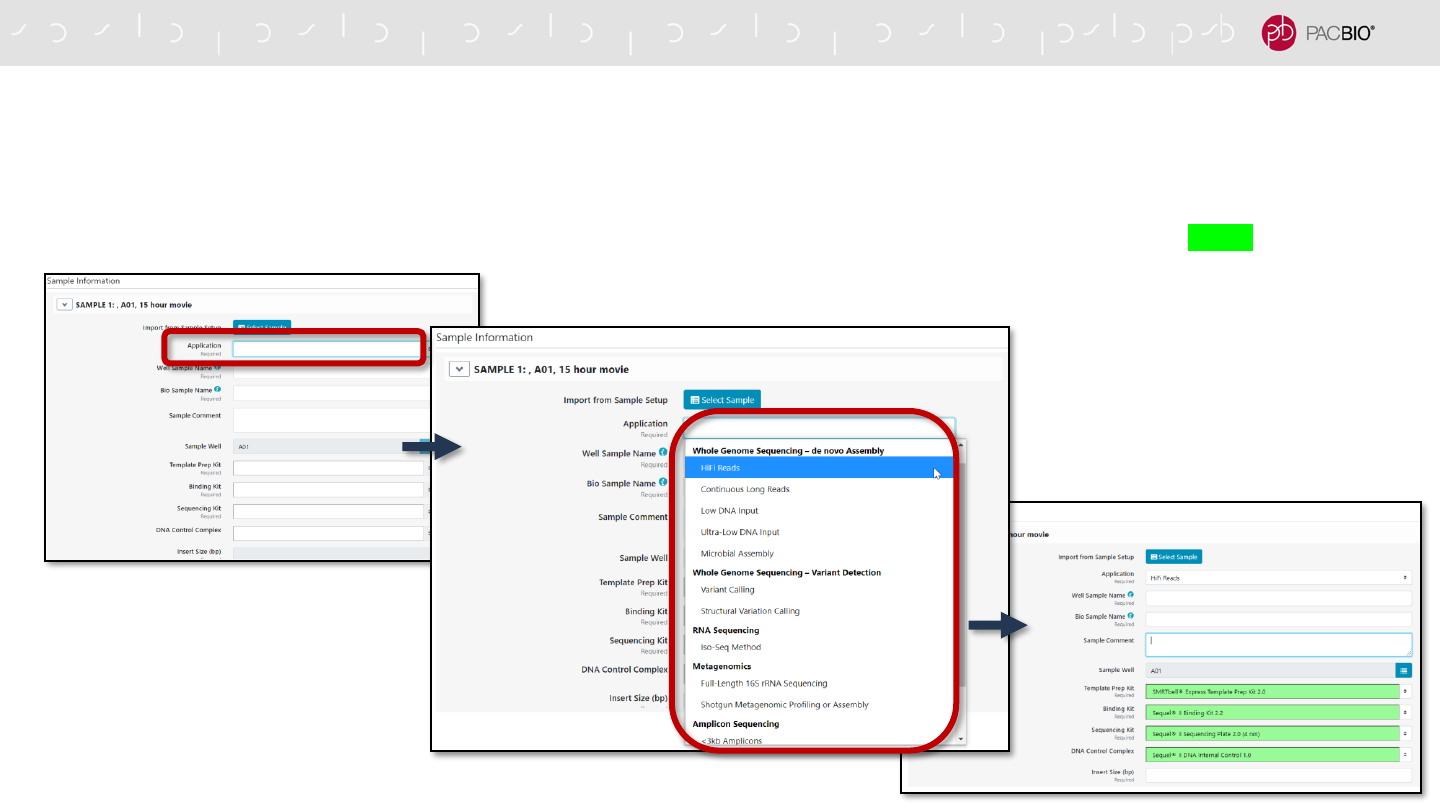
UPDATED RUN DESIGN WORKFLOW: SPECIFYING APPLICATION TYPE
30
Run Design auto-populates application-specific information for selected fields
- In not importing sample information from Sample Setup, the user can start by first selecting an Application Type
- Once an application is selected, default values are auto-populated for various fields and highlighted in green

UPDATED RUN DESIGN WORKFLOW: SPECIFYING APPLICATION TYPE (CONT.)
31
Auto-populated fields are highlighted in green color
- The following fields are auto-populated and highlighted in green:
- Template Prep Kit
- Binding Kit
- Sequencing Kit
- DNA Control Complex
- Movie Time Per SMRT Cell
- Pre-Extension Time (If applicable)
- If any auto-populated entry is manually changed to a different
value, then the field will be highlighted in yellow color
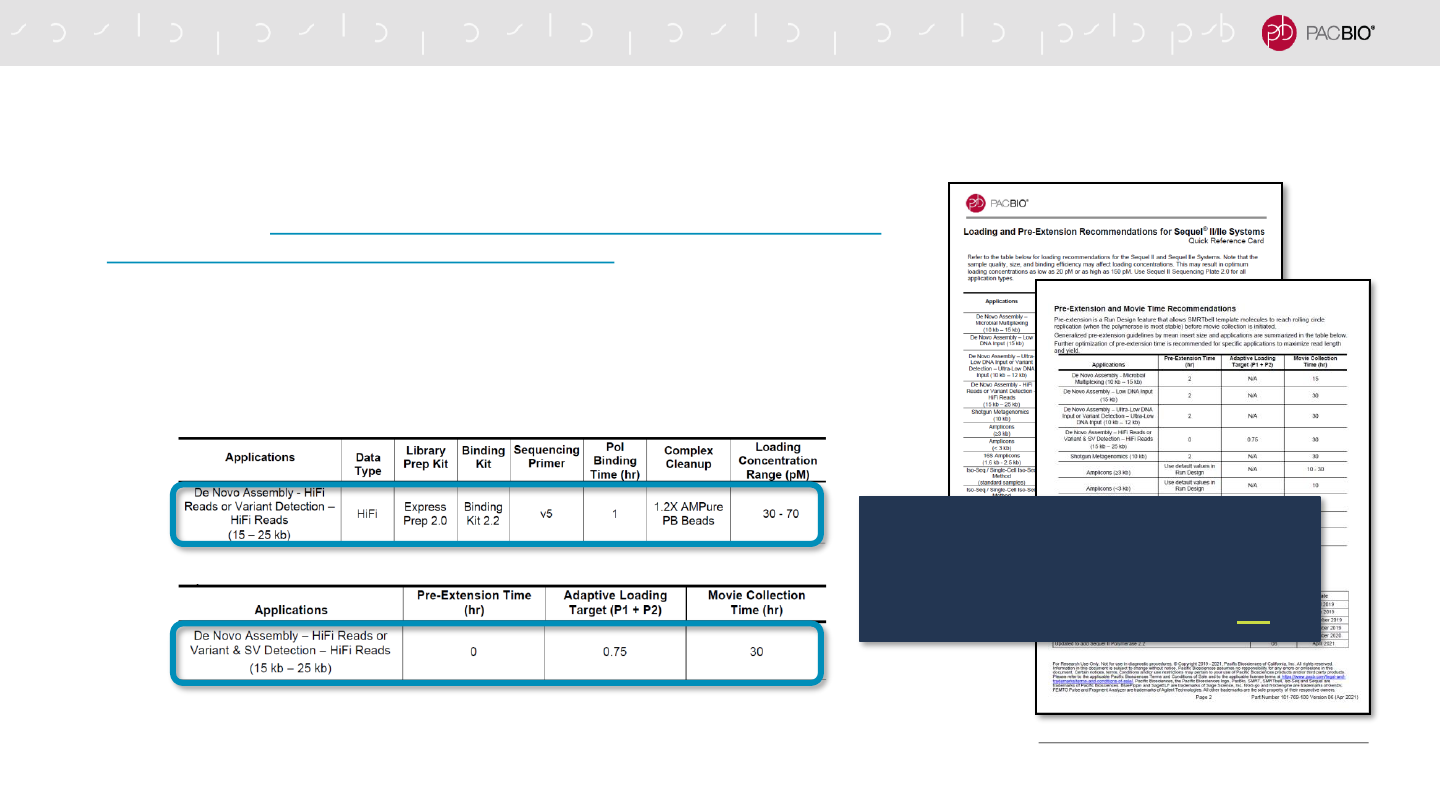
SAMPLE SETUP RECOMMENDATIONS FOR HIFI DE NOVO ASSEMBLY AND
VARIANT DETECTION APPLICATIONS (SEQUEL II AND IIe SYSTEM V10.1 RELEASE)
32
https://www.pacb.com/support/documentation/
- Follow SMRT Link Sample Setup instructions using the recommendations
provided in the Quick Reference Card – Loading and Pre-Extension Time
Recommendations for the Sequel II/IIe Systems for sequencing HiFi library
samples for De Novo Assembly or Variant Detection applications
→For SMRT Link v10.1 (or higher): Select ‘WGS – De Novo Assembly / HiFi
Reads’ or ‘WGS – Variant Detection / Variant Calling’ from the Application
field drop-down menu in the SMRT Link Sample Setup and SMRT Link Run
Design user interface
* Recommendations for using Sequel II Binding Kit 2.2 with other application use cases aside from WGS de novo assembly and variant detection will be provided in future protocol
documentation releases.
For HiFi WGS de novo assembly and
variant detection applications, we
recommend enabling Adaptive Loading
and using Sequel II Binding Kit 2.2.*

NEW ADAPTIVE LOADING FEATURE FOR SEQUEL II AND IIe
SYSTEMS
Adaptive Loading reduces sample overloading, allowing users to load higher with
confidence
Adaptive Loading (AL) uses active monitoring of polymerase binding
to the bottom of the ZMW during loading to reduce variability and the
risk of overloading with high-concentration samples
- Adaptive loading technology actively monitors
polymerase complex binding to the bottom of
ZMWs during the sample immobilization step.
- Detection of these active polymerase complexes
allows the system to terminate the immobilization
step when the desired loading target has been
achieved.
→ This approach can help reduce sample overloading
and run-to-run yield variability
33
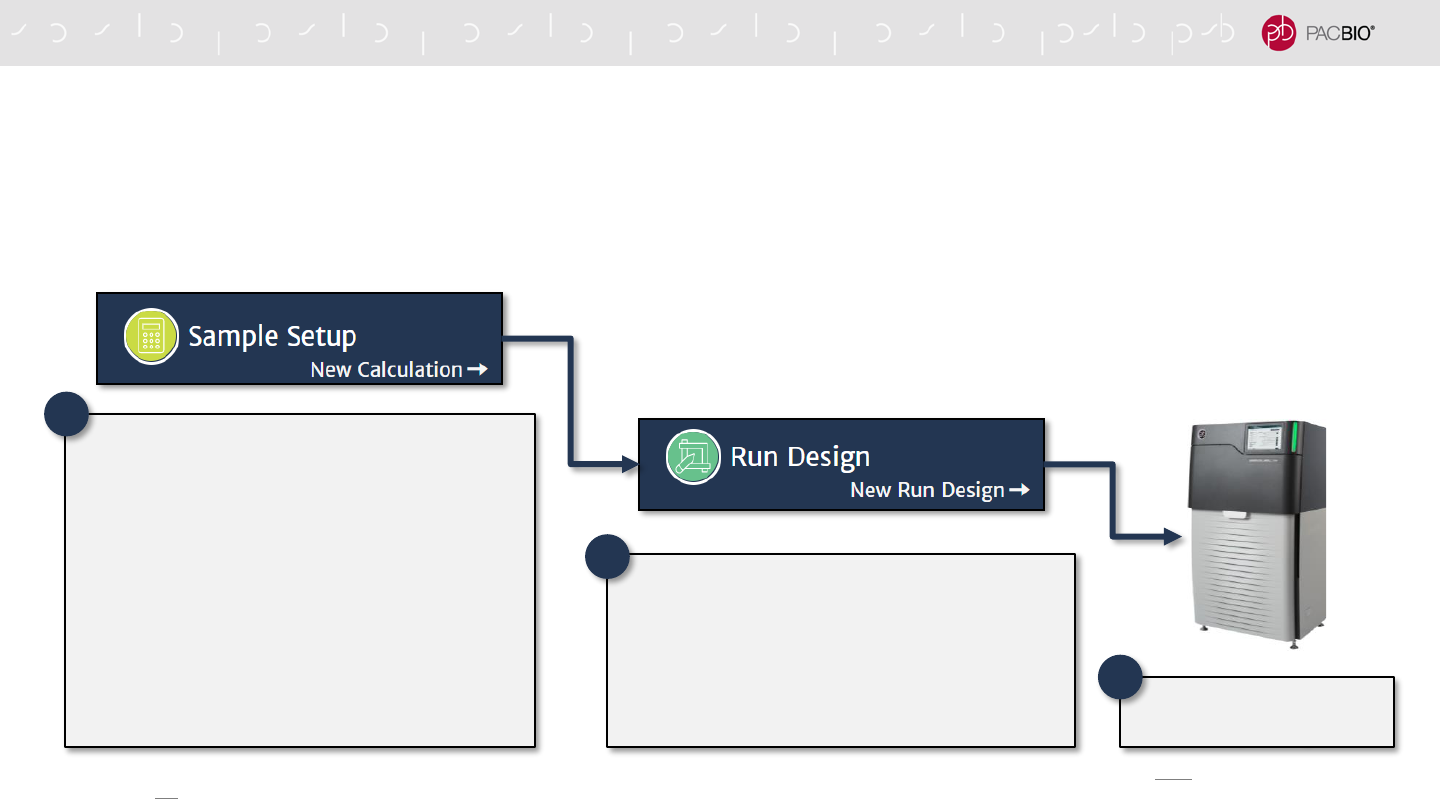
OVERVIEW OF SMRT LINK V10.1 SAMPLE SETUP AND RUN DESIGN
WORKFLOW TO ENABLE ADAPTIVE LOADING
Adaptive Loading is automatically enabled by default in SMRT Link v10.1 Sample Setup
and Run Design for sequencing applications using Sequel II Binding Kit 2.2
1. Binding Kit Version Selection
- Specify Sequel II Binding Kit 2.2*
2. Complex Cleanup Step
- Use Adaptive Loading (AL) Buffer in place of
Complex Dilution Buffer (CDB) to elute purified
polymerase-bound sample from AMPure PB
beads
3. Final Loading Dilution Step
- Use AL Buffer in place of CDB to dilute the
purified polymerase-bounds sample to the final
on-plate loading concentration
Advanced Options
- Specify Use Adaptive Loading = YES
- Specify Loading Target (P1 + P2)
- Specify Maximum Loading Time (hours)
Load Sample Plate
- Start sequencing run
1
2
3
34
* In SMRT Link v10.1 Sample Setup, the Adaptive Loading sample setup procedure is only enabled by selecting Sequel II Binding Kit 2.2. Selection of other Sequel II Binding Kit
versions will not enable the Adaptive Loading sample setup procedure.
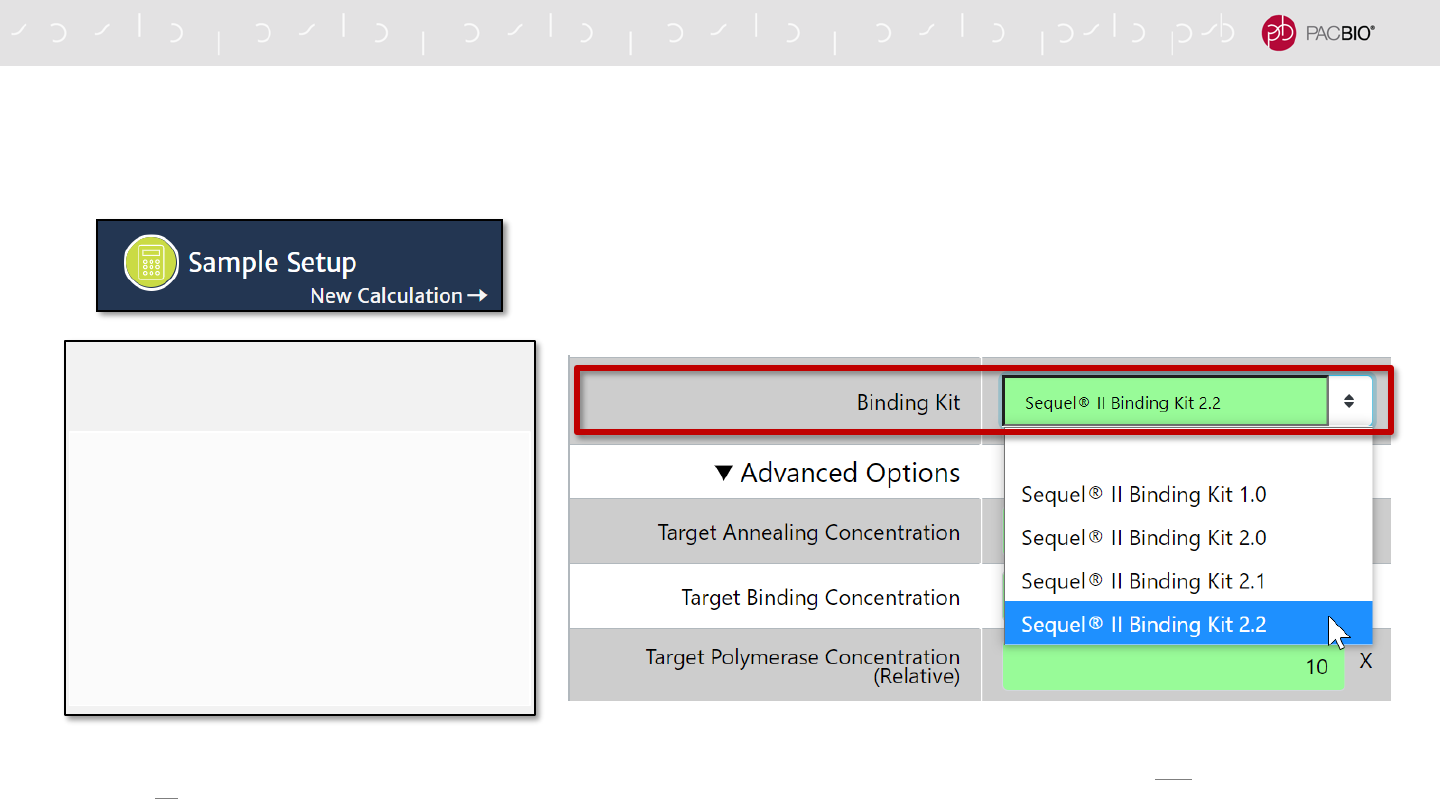
Binding Kit Version Selection
- Specify Sequel II Binding Kit 2.2*
Complex Cleanup Step
- Use Adaptive Loading (AL) Buffer in
place of Complex Dilution Buffer (CDB) to
elute purified polymerase-bound sample
from AMPure PB beads
Final Loading Dilution Step
- Use AL Buffer in place of CDB to dilute the
purified polymerase-bounds sample to the
final on-plate loading concentration
SMRT LINK V10.1 SAMPLE SETUP WORKFLOW TO ENABLE
ADAPTIVE LOADING
1. Binding Kit Version Selection
35
* In SMRT Link v10.1 Sample Setup, the Adaptive Loading sample setup procedure is only enabled by selecting Sequel II Binding Kit 2.2. Selection of other Sequel II Binding Kit
versions will not enable the Adaptive Loading sample setup procedure.
SMRT LINK V10.1 SAMPLE SETUP WORKFLOW TO ENABLE
ADAPTIVE LOADING (CONT.)
Binding Kit Version Selection
- Specify Sequel II Binding Kit 2.2
Complex Cleanup Step
- Use Adaptive Loading (AL) Buffer in
place of Complex Dilution Buffer (CDB) to
elute purified polymerase-bound sample
from AMPure PB beads
Final Loading Dilution Step
- Use AL Buffer in place of CDB to dilute the
purified polymerase-bounds sample to the
final on-plate loading concentration
2. Complex Cleanup Step
36

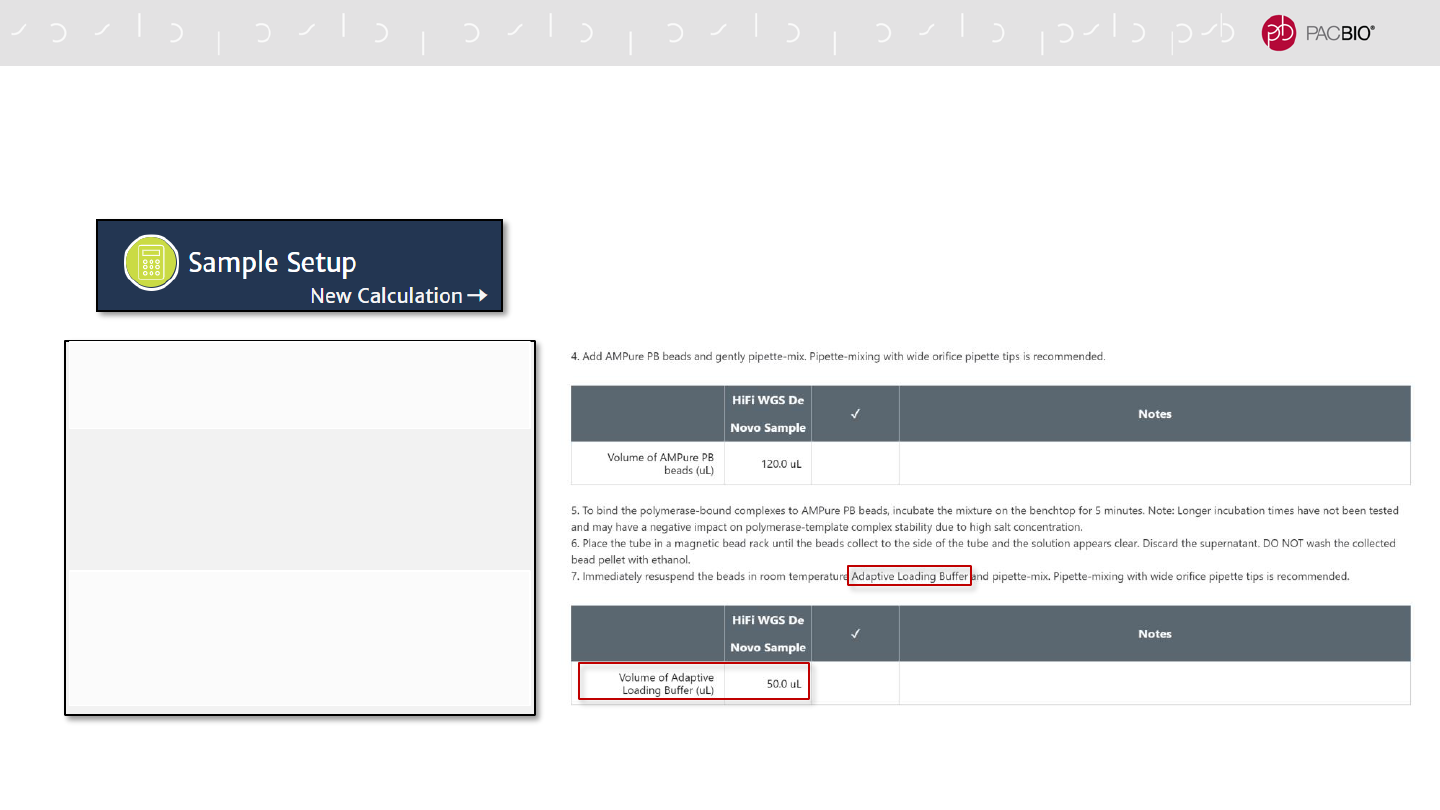
SMRT LINK V10.1 SAMPLE SETUP WORKFLOW TO ENABLE
ADAPTIVE LOADING (CONT.)
3. Final Loading Dilution Step*
Binding Kit Version Selection
- Specify Sequel II Binding Kit 2.2
Complex Cleanup Step
- Use Adaptive Loading (AL) Buffer in
place of Complex Dilution Buffer (CDB) to
elute purified polymerase-bound sample
from AMPure PB beads
Final Loading Dilution Step
- Use AL Buffer in place of CDB to dilute the
purified polymerase-bounds sample to the
final on-plate loading concentration
37
* In SMRT Link v10.1 Sample Setup, no DTT or Sequel Additive is added during the Final Loading Dilution step for sample complexes prepared with Sequel II Binding Kit 2.2.
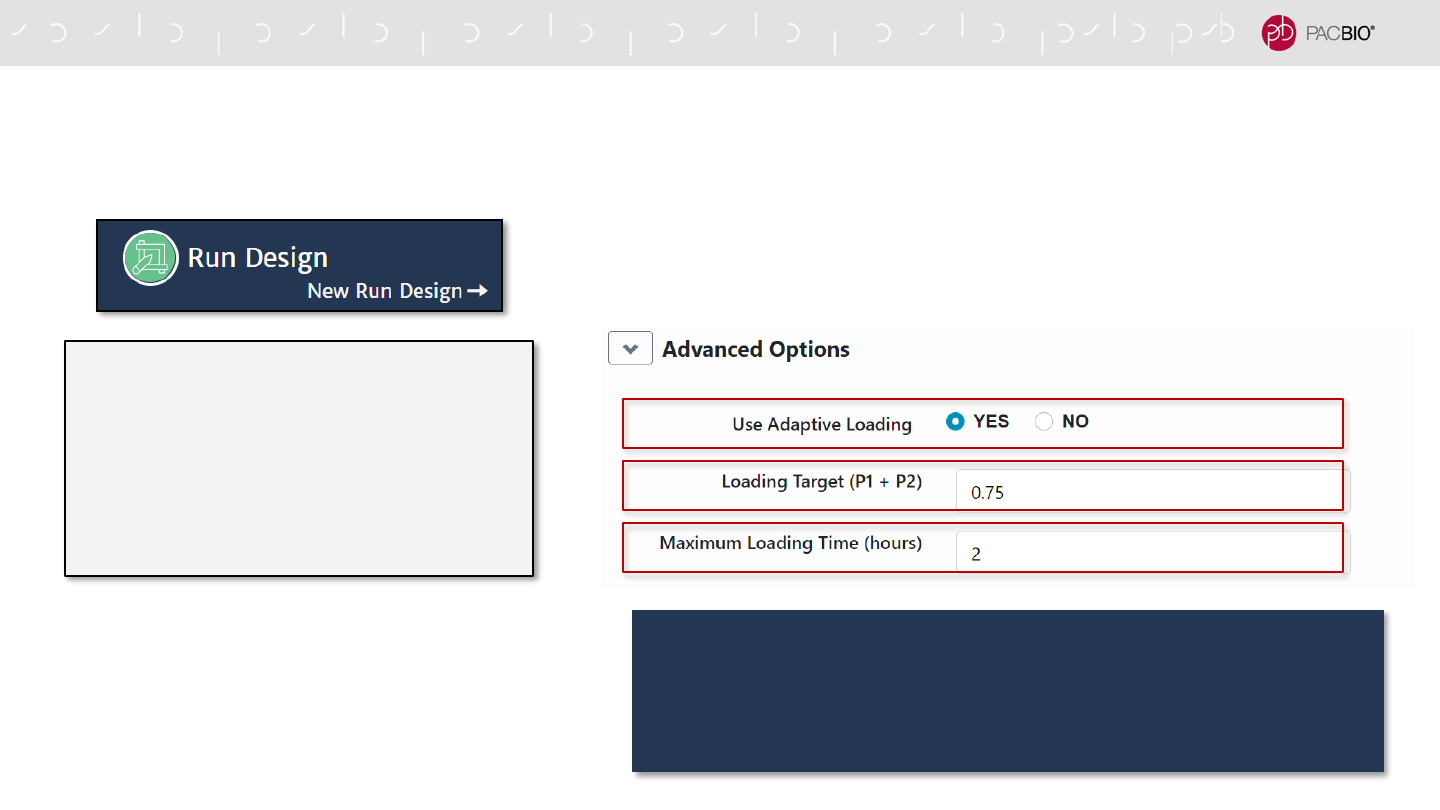
SMRT LINK V10.1 RUN DESIGN WORKFLOW TO ENABLE
ADAPTIVE LOADING
Advanced Options
- Specify Use Adaptive Loading = YES
- Specify Loading Target (P1 + P2)
- Specify Maximum Loading Time (hours)
For HiFi WGS de novo assembly and variant detection applications
using Sequel II Binding Kit 2.2, we highly recommend using the
Adaptive Loading default values of 0.75 for the Loading
Target and 2 hours for Maximum Loading Time.
Run Design Advanced Options
38
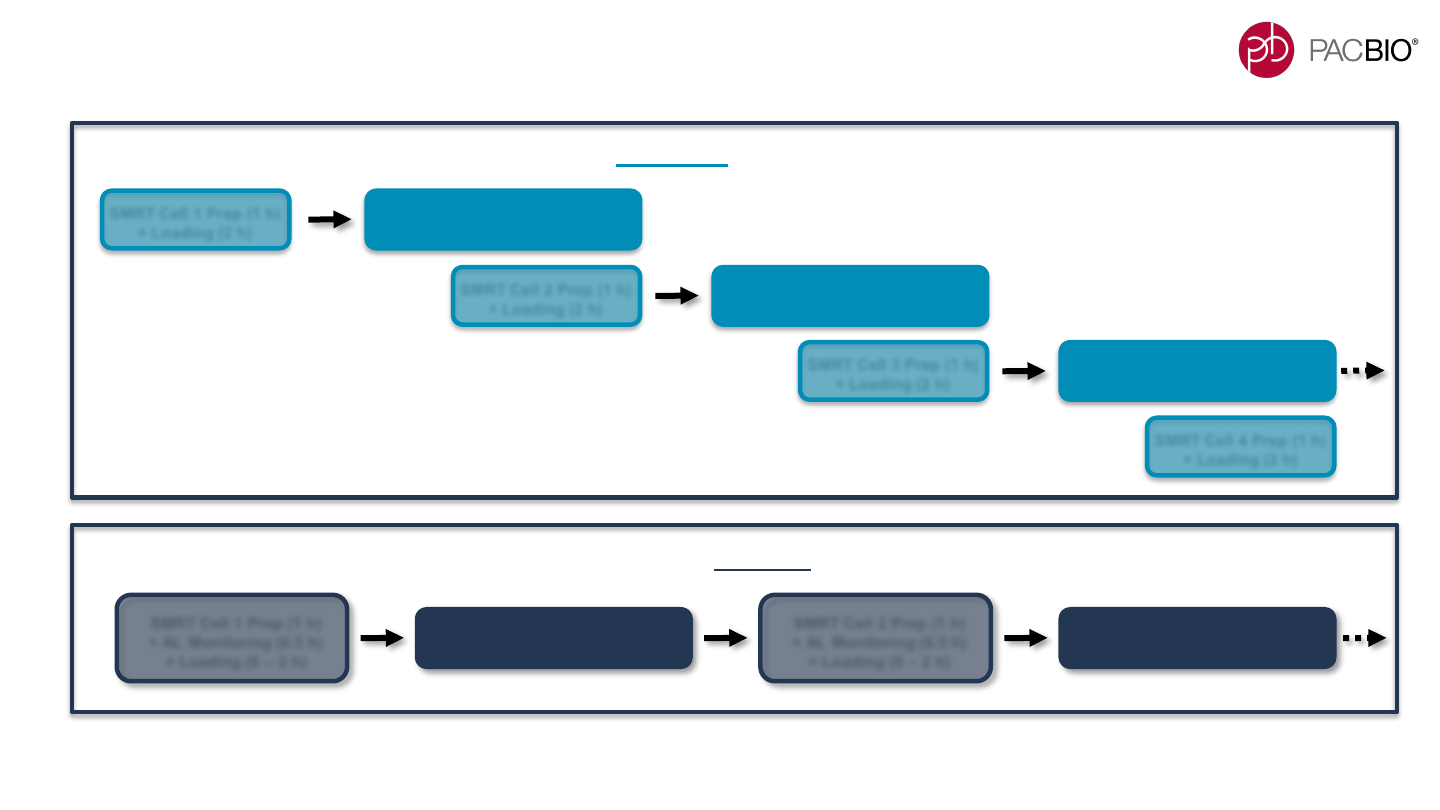
ADAPTIVE LOADING REQUIRES SERIALIZATION OF THE
SEQUEL II/IIe SYSTEM INSTRUMENT ROBOTIC WORKFLOW
Adaptive Loading Workflow: SMRT Cell prep occurs in series with data acquisitions
SMRT Cell 1 Prep (1 h)
+ AL Monitoring (0.5 h)
+ Loading (0 – 2 h)
SMRT Cell 1 Prep (1 h)
+ Loading (2 h)
SMRT Cell 1 Data Acquisition
SMRT Cell 2 Prep (1 h)
+ Loading (2 h)
SMRT Cell 3 Prep (1 h)
+ Loading (2 h)
SMRT Cell 2 Data Acquisition
SMRT Cell 4 Prep (1 h)
+ Loading (2 h)
SMRT Cell 3 Data Acquisition
Standard Workflow: SMRT Cell prep occurs in parallel with data acquisitions
SMRT Cell 1 Data Acquisition
SMRT Cell 2 Prep (1 h)
+ AL Monitoring (0.5 h)
+ Loading (0 – 2 h)
SMRT Cell 2 Data Acquisition
- With the Adaptive Loading feature enabled, instrument run times will be longer compared to non-AL runs depending on
the actual duration of the AL monitoring + immobilization (loading) time period (up to ~2.5 hours) per SMRT Cell 8M.
39

NEW RUN QC VISUALIZATION PLOTS IN SMRT LINK V10.1
The following plots below are generated for any run where CCS processing is enabled on-instrument
(Sequel IIe System) or through CCS Pre-Analysis (Sequel or Sequel II Systems)
40
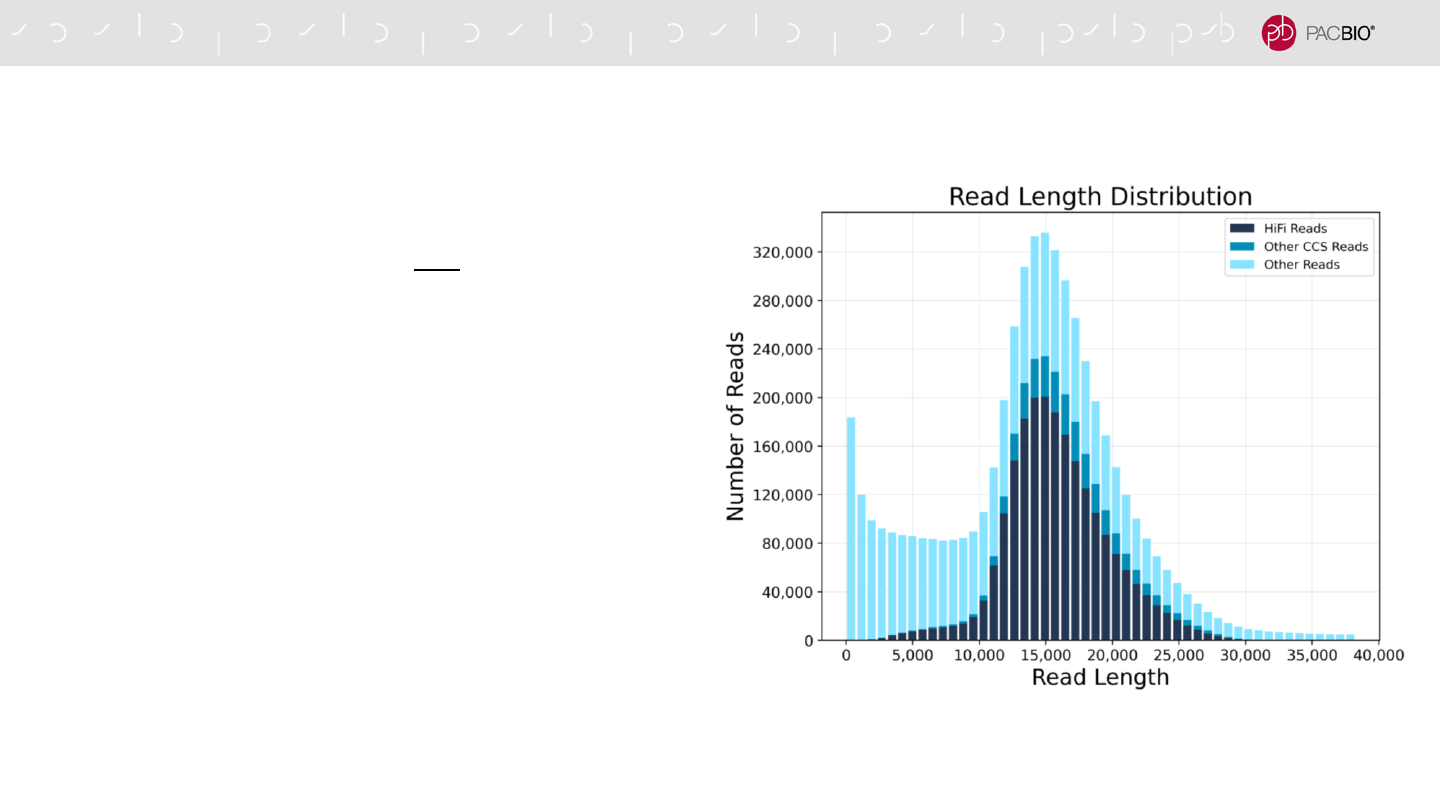
Displays a histogram distribution of
HiFi Reads (QV ≥20), other CCS
Reads (three or more passes, but QV
<20), and other reads, by read
length.
Displays a histogram distribution of
HiFi Reads (QV ≥20) and other CCS
Reads by read quality.
Displays a heat map of CCS Read
lengths and predicted accuracies.
The boundary between HiFi Reads
and other CCS Reads is shown as a
dashed line at QV 20.
HiFi Reads
(≥Q20)
Q20

Example HiFi Library Sequencing Performance
Data

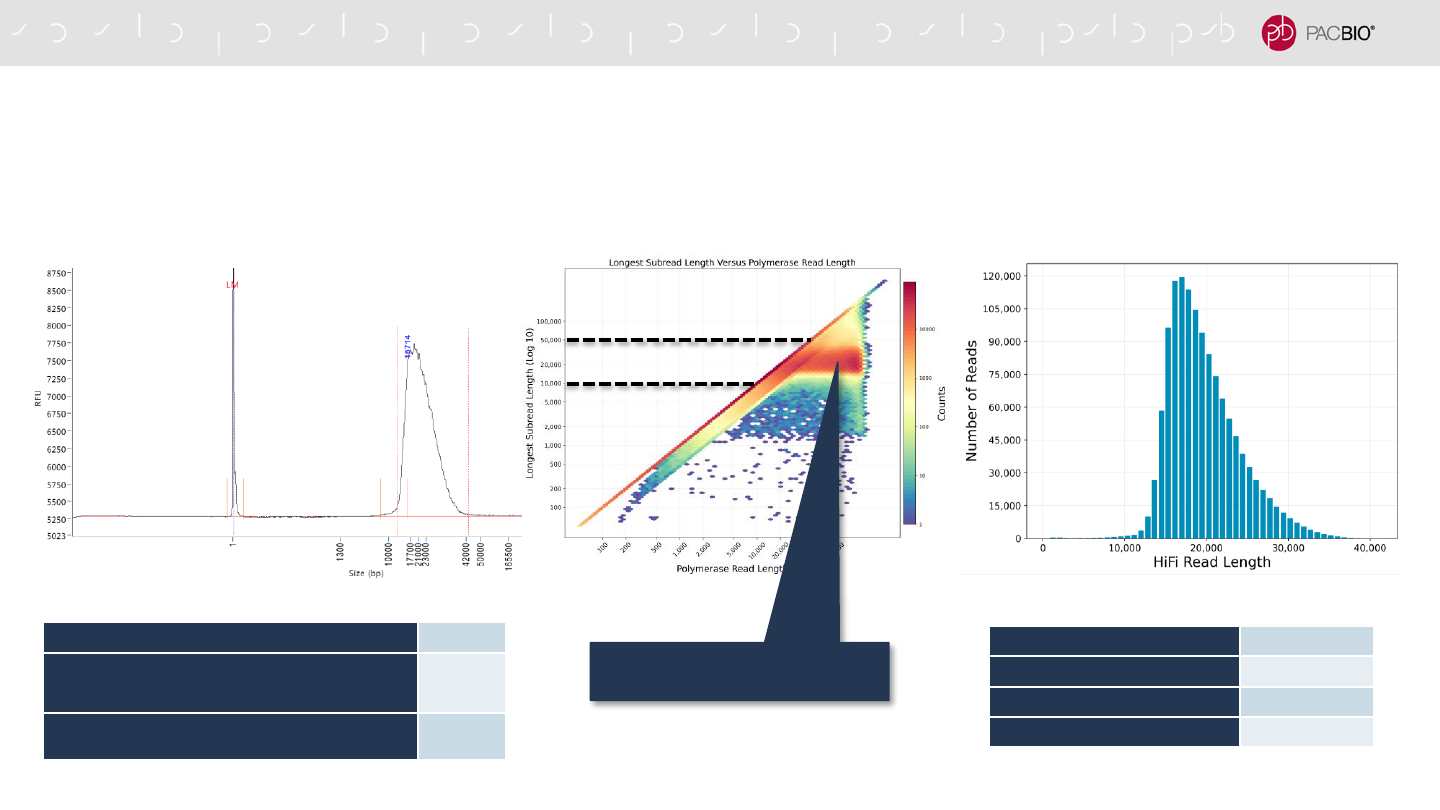
EXAMPLE SEQUENCING PERFORMANCE OF A 18-KB HUMAN HIFI LIBRARY
FOR WGS VARIANT DETECTION APPLICATIONS
HiFi Reads 2.0 M
HiFi Base Yield** 34.7 Gb
Mean HiFi Read Length 17,212 bp
Median HiFi Read Quality Q30
Size-Selected HiFi Library QC Insert Read Length Density Plot HiFi Read Length Distribution
* 40 pM on-plate loading concentration
generated P1 = 70% using a 30-hour
movie collection time (Sequel IIe System)
50 kb
10 kb
Mean HiFi
Read Length
= 17.2 kb
IRLD plot shows most HiFi read
lengths are ~10 – 30 kb*
** For this human library data set, typical HiFi base
yields were ~25 Gb – 35 Gb per SMRT Cell 8M.
Input gDNA for Megaruptor 3 Shearing 5 µg
Post-Library Construction Recovery (%)
[Pre-Nuclease Treatment]
3540 ng
(71%)
Post-Library Construction Recovery (%)
[Post-Nuclease Treatment]
1368 ng
(27%)
Post-PippinHT Size Selection Recovery (%)
640 ng
(13%)
~17.5 kb Mode
(Pre-Size Selection)
~18 kb Mode
(Post-Size Selection
with PippinHT System)
42
EXAMPLE SEQUENCING PERFORMANCE OF A 20-KB PLANT HIFI LIBRARY
FOR WGS DE NOVO ASSEMBLY APPLICATIONS
HiFi Reads 1.3 M
HiFi Base Yield 25.1 Gb
Mean HiFi Read Length 19,696 bp
Median HiFi Read Quality Q28
Size-Selected HiFi Library QC Insert Read Length Density Plot HiFi Read Length Distribution
* 60 pM on-plate loading concentration
generated P1 = 85% [Adaptive Loading
Target (P1 + P2) = 0.85] using a 30-hour
movie collection time (Sequel IIe System)
50 kb
10 kb
Mean HiFi
Read Length
= 19.7 kb
IRLD plot shows most HiFi read
lengths are ~10 – 30 kb*
Input gDNA for Megaruptor 3 Shearing 5 µg
Post-Library Construction Recovery (%)
[Post-Nuclease Treatment]
1900 ng
(38%)
Post-PippinHT Size Selection Recovery (%)
850 ng
(17%)
~20 kb Mode
(Post-Size Selection
with PippinHT System)
43

SEQUEL II DNA INTERNAL CONTROL PERFORMANCE UPDATE
- Sequel II Binding Kits 2.0, 2.1 and 2.2 include Sequel II DNA Internal Control Complex
pre-bound with Sequel II Polymerase 1.0
- Sequel II Polymerase 2.2 is bound (tethered) slightly higher above the surface of the
ZMW compared to Sequel II Polymerases 2.0 and 2.1
- A higher laser power setting is required to illuminate the ZMW when sequencing samples
bound to Sequel II Polymerase 2.2
→ Sequel II DNA Internal Control Complex 1.0 shows reduced mean read length
performance when used with samples bound to Sequel II Polymerase 2.2
compared to samples bound with Sequel II Polymerase 2.0 or 2.1
Sequel II DNA Internal Control Complex 1.0 is included with Sequel II Binding Kit 2.0 / 2.1 / 2.2
Polymerase Version
Bound to Sample
Estimated DNA Internal Control Complex 1.0
Mean Polymerase Read Length (30-h Movie)
Sequel II Polymerase 2.2 ~30 kb
Sequel II Polymerase 2.0
Sequel II Polymerase 2.1
~50 kb
Comparison of Sequel II Polymerase 2.0-
DNA Template complex (A) vs. Sequel II
Polymerase 2.2-DNA Template complex (B)
immobilized to a ZMW.
A
B
44
The higher laser power setting required for sequencing samples bound to Sequel II
Polymerase 2.2 results in increased photodamage to the Sequel II DNA internal
Control Complex 1.0 and hence shorter control polymerase read lengths

For Research Use Only. Not for use in diagnostic procedures. © Copyright 2021 by Pacific Biosciences of California, Inc. All rights reserved.
SMRT Link v10.1 Overview
Subhead should be no longer than 1 line

SMRT LINK V10.1 SOFTWARE RELEASE: KEY FEATURES &
IMPROVEMENTS
▪ Supports complete SMRT Link analysis
workflow on Amazon Cloud (SMRT Link
Cloud)
▪ Features several new and improved
analysis applications for HiFi de novo
assembly, SV calling, multiplexed Iso-Seq
analysis and SARS-CoV-2 full-viral genome
sequencing
▪ Reduces HPC requirements to enable
lower-cost data analysis & storage
configurations
▪ Provides a simplified user experience for
run setup and includes usability
improvements to support high-throughput
sequencing
46

Supports Complete SMRT Link Workflow on Amazon Cloud*
- Flexible data analysis optimized for speed or cost based on user
preferences
- No need for internal HPC
Features New and Improved Analysis Applications
- New Genome Assembly application for HiFi data*
- New SARS-CoV-2 application for COVID-19 surveillance
- Updated Iso-Seq application for improved multiplexed sample analysis
- Updated SV Calling application for improved precision
- Enhanced alignment concordance in mapping applications
- Bioconda: New HiFi Amplicon Analysis application*
- Bioconda: New Single-cell Iso-Seq application*
Provides Simplified User Experience & Usability Improvements
- New application-centric Sample Setup and Run Design*
- New HiFi sequencing metrics and data visualizations in Run QC*
- Reduced HPC requirements to enable lower-cost configurations*
- New Sample Setup import feature to support for high-throughput
production environments
SMRT LINK V10.1 SOFTWARE RELEASE: KEY FEATURES &
IMPROVEMENTS
▪ Supports complete SMRT Link analysis
workflow on Amazon Cloud (SMRT Link
Cloud)
▪ Features several new and improved
analysis applications for HiFi de novo
assembly, SV calling, multiplexed Iso-Seq
analysis and SARS-CoV-2 full-viral genome
sequencing
▪ Reduces HPC requirements to enable
lower-cost data analysis & storage
configurations
▪ Provides a simplified user experience for
run setup and includes usability
improvements to support high-throughput
sequencing
SMRT Link
Cloud
SMRT
Analysis
* Feature first introduced with SMRT Link v10.0 limited release
47
Head Node
Cores
32
RAM
64 GB
Local Storage
1 TB SSD/Flash storage
db_datadir
(Local Storage)
250 GB
Compute Nodes
Cores (Total)
64
Minimum RAM per slot (1 slot = 1 core)
>4 GB
Local Storage
100 GB
Shared Data Storage
Sequencing Data
20 TB
a
Analysis Data
40 TB
a
Network
10 GbE strongly recommended, 1GbE required
b
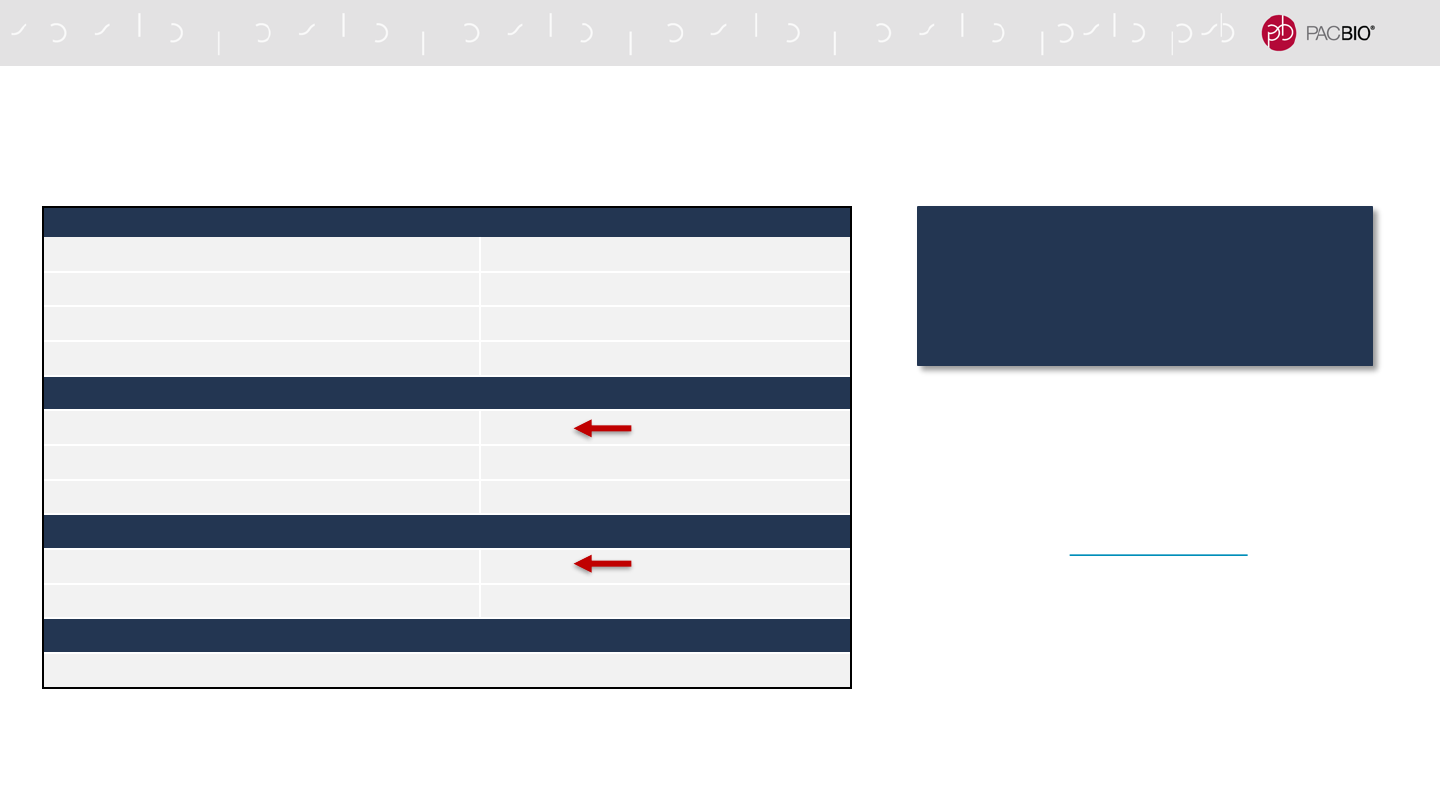
SEQUEL IIe SYSTEM ENABLES REDUCED COMPUTE
REQUIREMENTS FOR SMRT LINK INSTALLATIONS
a
Storage is calculated for one Sequel IIe System, assuming 100 human genomes per year at
30-fold HiFi coverage, de novo assembly
b
Connection between the Head Node and Sequel IIe System
48
Reduced HPC requirements for the
Sequel IIe System enable lower-cost
compute configurations
compared to the Sequel II System
- Approx. 5-fold lower HPC costs: $20K (Sequel IIe
System) vs. $100K (Sequel II System)
- Note: For a single Sequel IIe System deployment, a
Single System Compute configuration is available
– contact PacBio Technical Support or your local
Bioinformatics FAS for details. (Supports ONE
Sequel IIe system ONLY. Not suggested for sites
with multiple instruments.)
Previously 384
(Sequel II System)
Previously >100 TB
(Sequel II System)

CCS Analysis Application Features &
Reports
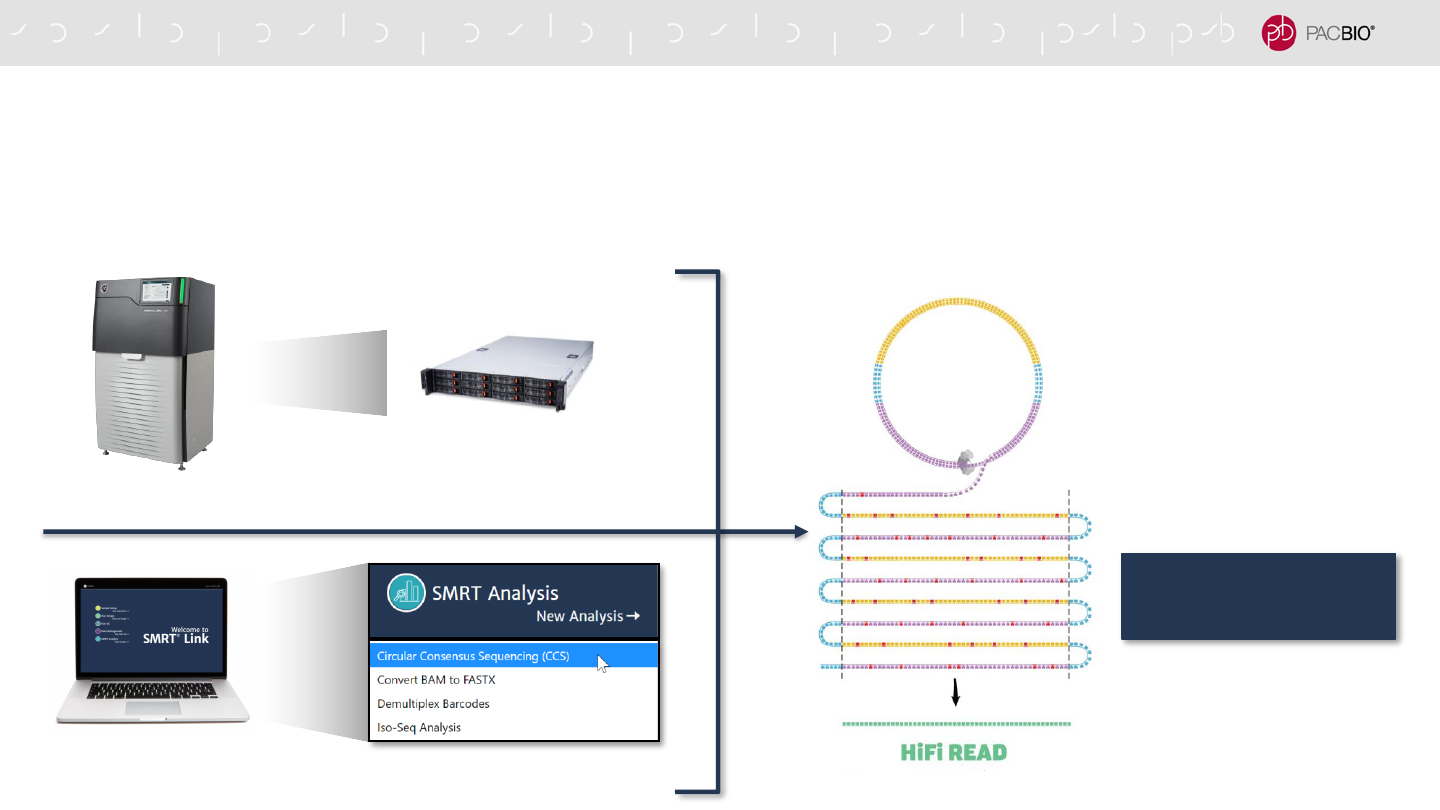
CCS ANALYSIS ALGORITHM AND DATA OUTPUTS
CCS analysis and output are unified for Sequel IIe System and SMRT Link
- CCS algorithm and output files (metrics, reports) are the same for Sequel IIe System and SMRT Link
50
On-Instrument CCS
CCS Analysis Application
Sequel IIe System
SMRT Link
HiFi reads achieve
≥99.9% accuracy
CCS ANALYSIS reads.ba m DATA FILE FORMAT
- Sequel IIe System on-instrument CCS (OICCS)* and
SMRT Link CCS Analysis application outputs
a reads.bam file containing one read per
productive ZMW.
- This format is far more compact than subread data.
- There are three classes of reads in this reads.bam
file:
▪ HiFi Reads: CCS reads with QV ≥20
▪ Other CCS Reads: CCS reads with lower quality (<Q20)
▪ Other Reads: Single-pass reads or reads not meeting
minimum CCS requirements
- For OICCS, when the reads.bam file is imported into
SMRT Link, a filtered file containing only HiFi reads
is automatically generated
51
* Note: Users can optionally specify in SMRT Link Run Design to include polymerase kinetics information (for secondary epigenetics analysis) in the
reads.bam file produced through either on-instrument CCS or SMRT Link – however, BAM file size is 5X larger if kinetics information is included.

HIFI DATA-SPECIFIC FILES ARE AUTOMATICALLY GENERATED IN
SMRT LINK WHEN CCS ANALYSIS IS PERFORMED
HiFi Data File Generation With On-instrument CCS Analysis (Sequel IIe System ICS v10.1+)
- An on-instrument CCS analysis generates a reads.bam file and transfers it to the
network server.
- The reads.bam file contains HiFi Reads and non-HiFi Reads, and should not be
used unfiltered as input for third-party tools that expect ≥QV20 sequencing data .
- SMRT Link automatically launches an Export Reads analysis on the reads.bam to
filter out the HiFi Reads, and generates the following three HiFi data files by default*:
▪ <Movie_Name>.hifi_reads.fastq.gz → FASTQ file with HiFi reads
▪ <Movie_Name>.hifi_reads.fasta.gz → FASTA file with HiFi reads
▪ hifi_reads.bam → BAM file with HiFi reads
- Refer to Sequel IIe System: Location of HiFi Reads Files (PN 102-110-200) to
locate the hifi_reads files generated by SMRT Link when you perform an on-
instrument CCS analysis on the Sequel IIe System.
52
* If not using SMRT Link for subsequent analysis, please use these three files as input with any third-party analysis tools
HIFI DATA-SPECIFIC FILES ARE AUTOMATICALLY GENERATED IN
SMRT LINK WHEN CCS ANALYSIS IS PERFORMED (CONT.)
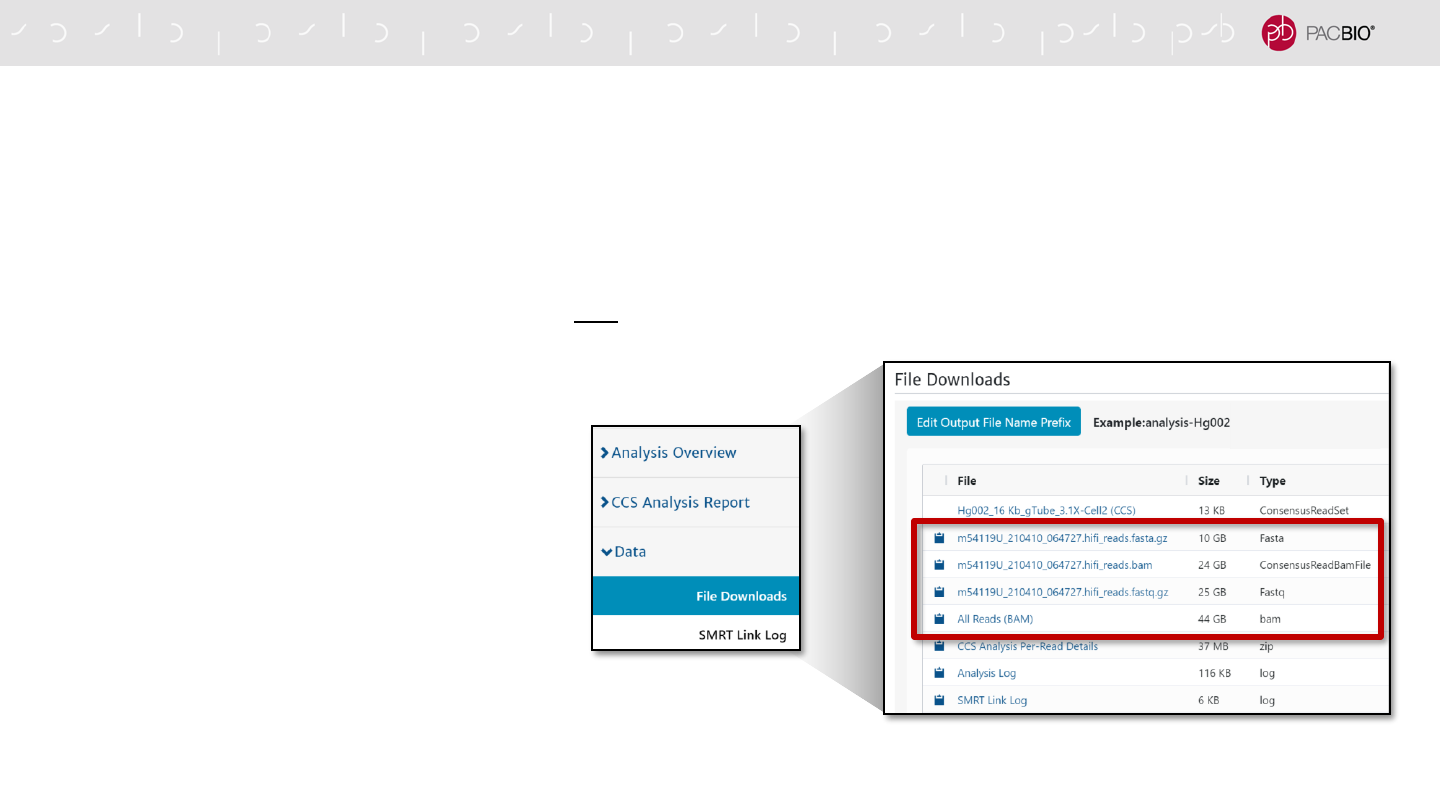
HiFi Data File Generation With SMRT Link CCS Analysis Application (SMRT Link v10.1+)
- The Circular Consensus Sequencing (CCS) analysis application in SMRT Link generates one reads.bam file (labeled
‘All Reads (BAM)’ in the File Downloads tab) plus the following three HiFi data-specific files below by default*:
53
▪ <Movie_Name>.hifi_reads.fastq.gz
→ FASTQ file with HiFi reads
▪ <Movie_Name>.hifi_reads.fasta.gz
→ FASTA file with HiFi reads
▪ hifi_reads.BAM
→ BAM file with HiFi reads
- To download the above files in SMRT Link, go
to the File Downloads tab for the CCS
analysis job

SMRT LINK ANALYSIS APPLICATIONS AND DATA FILTERING
CCS-based analysis applications require a reads.bam file as input and use built-in default
read quality filter settings
- All SMRT Link CCS-based applications* use reads.bam dataset as
input
▪ Built-in default filtering applied prior to analysis execution for each application
▪ All applications except Iso-Seq analysis use default HiFi reads (Q20 or higher)
▪ Iso-Seq application uses reads with Q10 or higher
The following analysis parameters
are deprecated as they no longer
need to be specified by the user:
▪ Minimum Number of Passes
▪ Minimum Predicted Accuracy
* Note: Microbial Assembly, Base Modification Analysis and HGAP4 Assembly remain CLR-based analysis applications
- Custom/non-HiFi data filtering:
▪ Use SMRT Link “Export Reads” application to specify a custom QV value in Advanced Parameters to create
FASTX and/or BAM files containing reads with a specified minimum CCS Predicted Accuracy
▪ Use SMRT Link Data Management to create a Data Set with custom read quality filtering
54
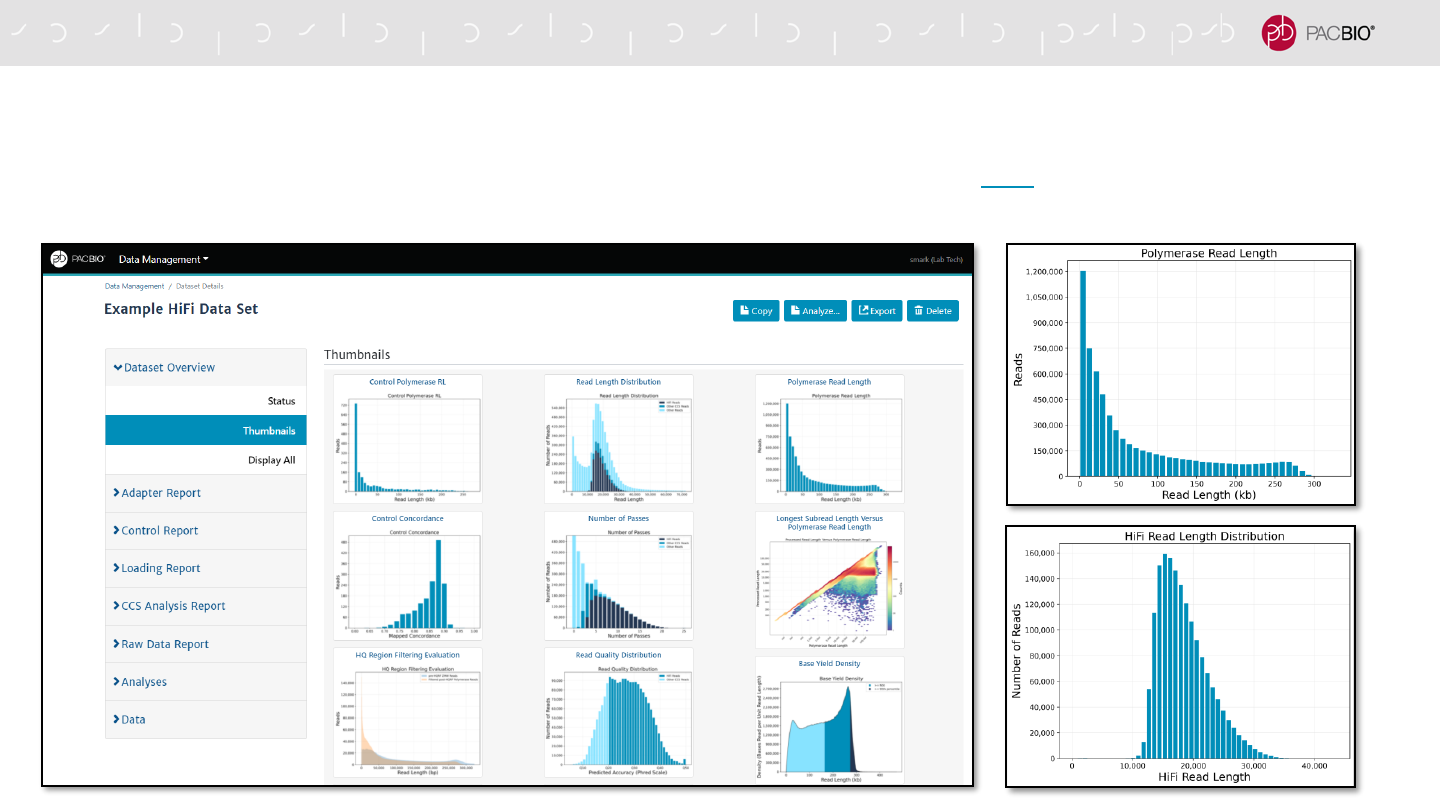
SMRT LINK DATA MANAGEMENT DATA SET REPORTS
Data Set Overview tab displays summary information for all reads and HiFi reads
55
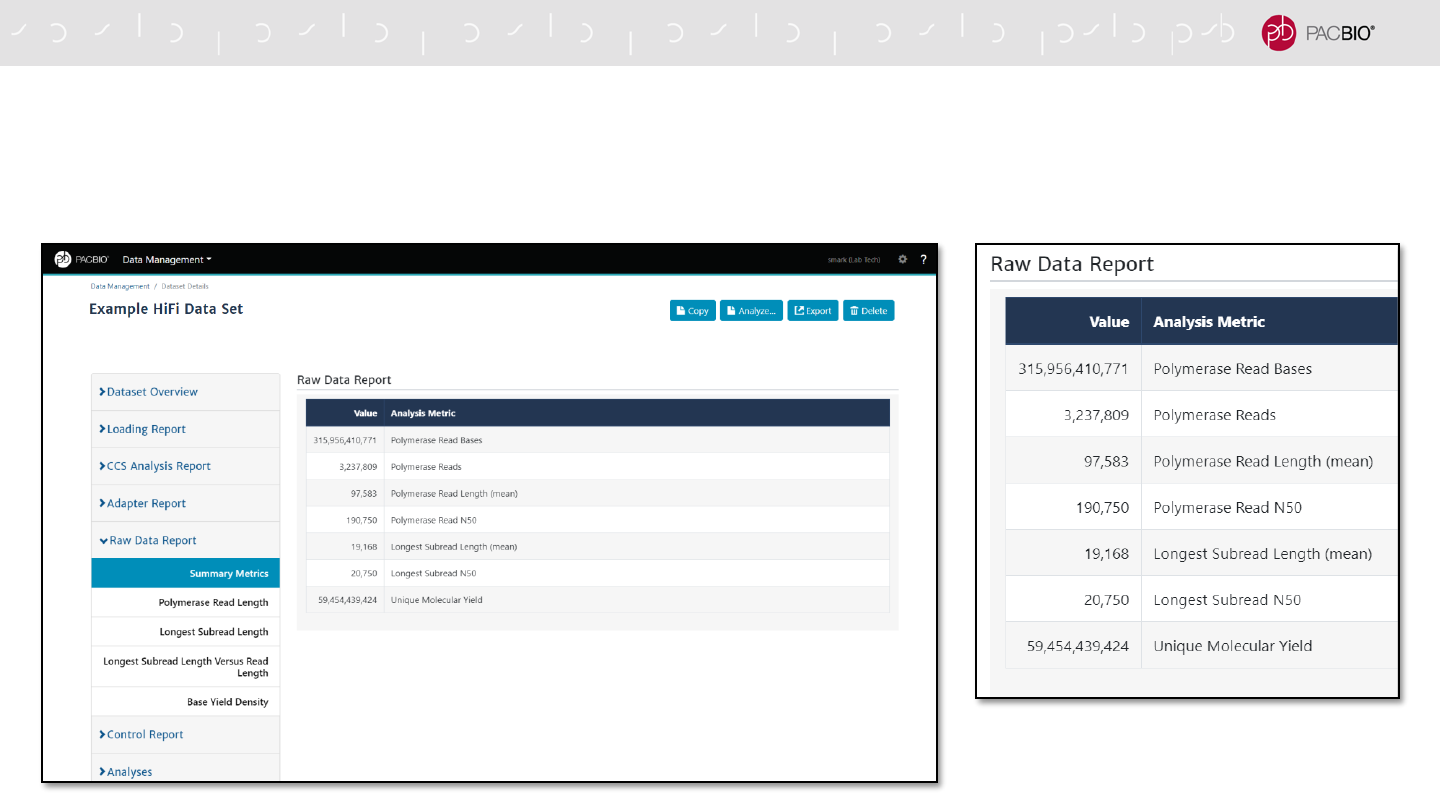
SMRT LINK DATA MANAGEMENT DATA SET REPORTS (CONT.)
Raw Data Report tab displays summary metrics for all reads
56
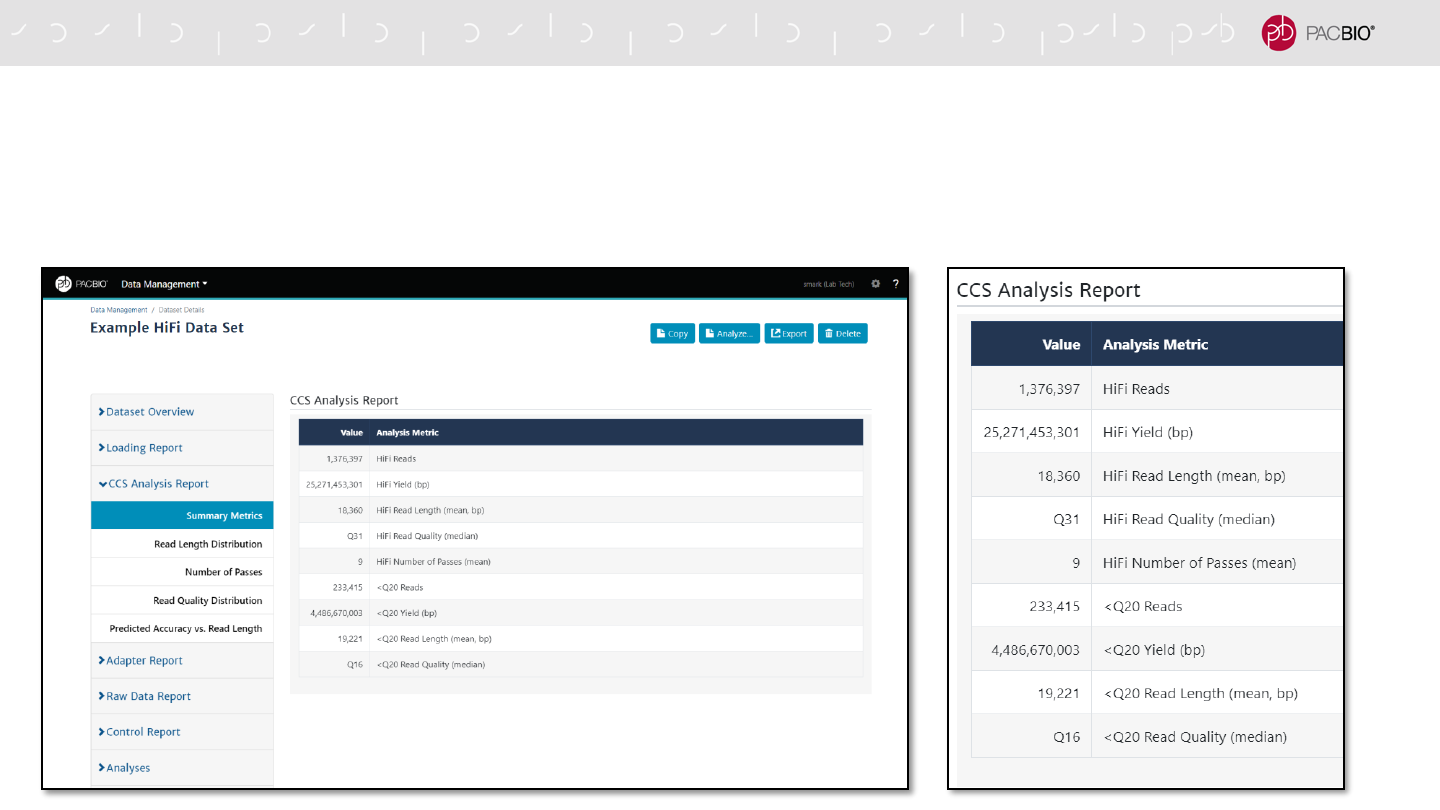
SMRT LINK DATA MANAGEMENT DATA SET REPORTS (CONT.)
CCS Analysis Report tab includes summary metrics for HiFi Reads (≥Q20) and Other CCS
Reads (<Q20)
57

SMRT Link v10.1 Cloud

SMRT LINK CLOUD INTEGRATION
A cloud-based end-to-end analysis workflow enabled on Amazon Web Services
- Complete SMRT Link v10.1 functionality is now available on the cloud
- Cloud-agnostic solution – Amazon Web Services (AWS) support is being offered first
- Enabled for all Sequel Systems – Sequel, Sequel II and Sequel IIe Systems
- SMRT Link Cloud Advantages:
- No dependency on having internal compute hardware infrastructure
- Flexible data analysis options to optimize for speed or cost based on user preferences
- Ability to easily share data with collaborators
59

DATA STREAMING TO THE CLOUD
SMRT Link v10.1 enables data streaming to AWS
- Seamless automated data streaming from the sequencing instrument to AWS
- Once the Sequel / Sequel II / Sequel IIe System, SMRT Link and local network storage are configured,
automated data streaming is enabled through Amazon tools and utilities included with SMRT Link
- Fail-safe data streaming is enabled through use of a local fail-safe network storage server
- The sequencing data for each run is transferred to a local network storage server and then streamed to
AWS
- Network storage server requirements (provided by customer)
▪ Disk space requirements are based on a typical run for Sequel II/IIe Systems
▪ Disk space is managed by the customer – availability, data back up, etc.
60
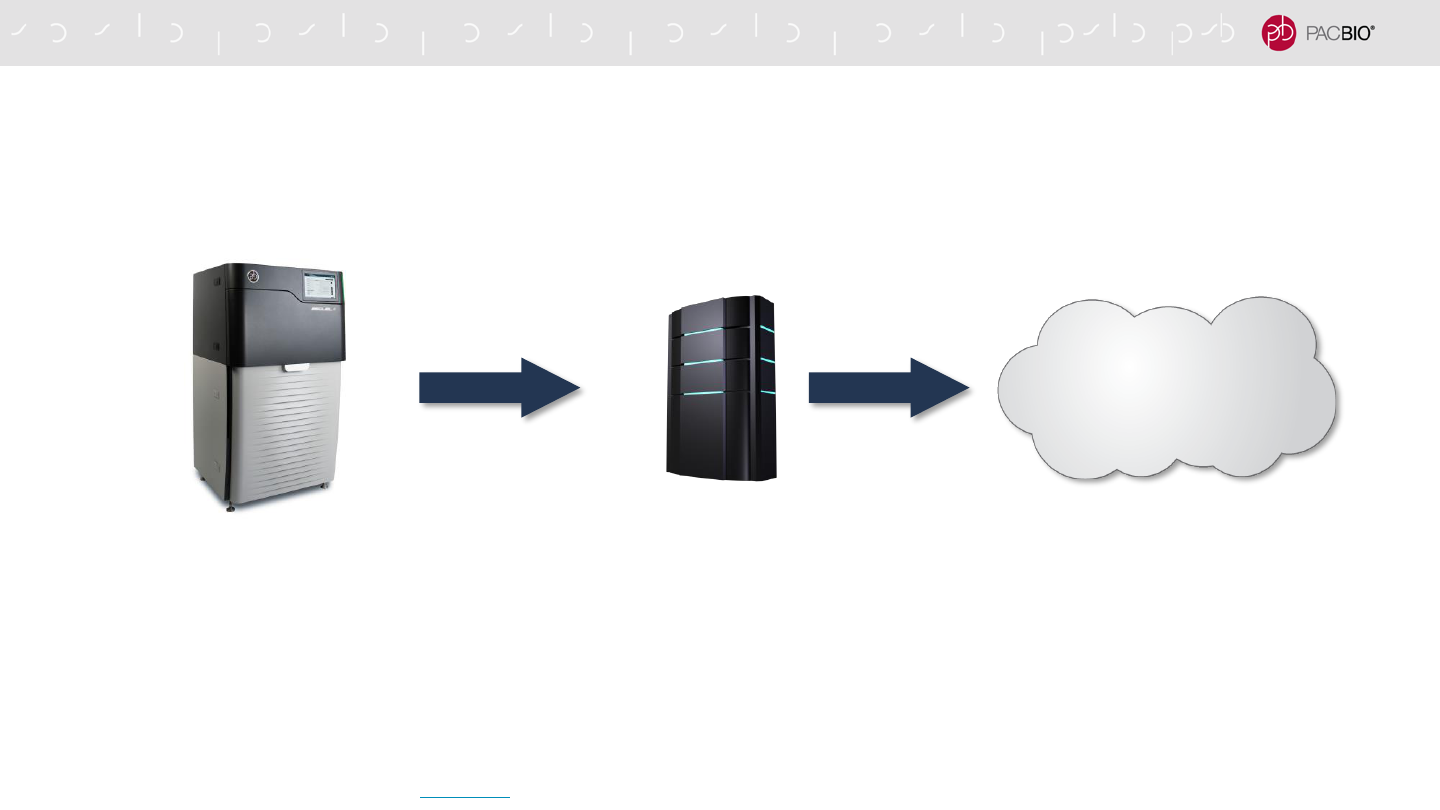
* Optionally, a customer-preferred transfer mechanism can be used
DATA STREAMING TO THE CLOUD (CONT.)
SMRT Link provides a solution for streaming data from the network server to AWS
Cloud
(AWS)
Network ServerSequel Instrument Amazon Web Services
Generate Sequencing Data Transfer Data to a Network Server Stream data to AWS*
61
See SMRT Link Cloud Reference Guide (v10.1) (PN 101-985-900)
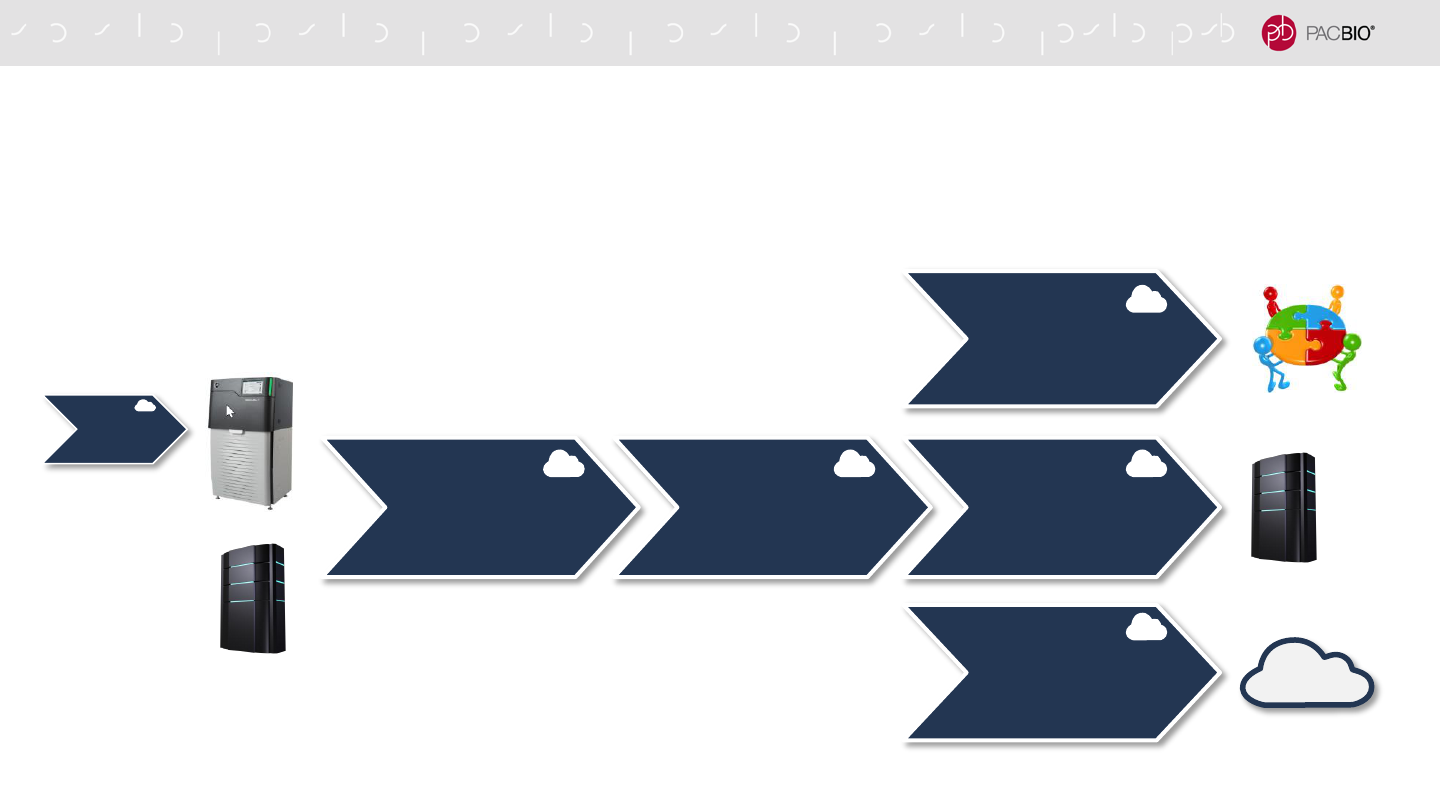
SMRT LINK CLOUD WORKFLOW
Versatile post-analysis options
Cloud
(AWS)
SMRT
Sequencing
Data on a
Network Server
SMRT Link
Data Transfer
Network
Server
SMRT Link
SMRT Analysis
Design a
Run
Stream
Sequencing
Data to AWS
Analyze
Sequencing
Data
Share
Analysis
Results
Archive
Data on the
Cloud
Download
Analysis
Results
Cloud Benefits
62

SMRT LINK CLOUD DATA SAFETY AND SECURITY
Compliance and safety in place
- Data safety and security mechanisms
- Utilizing existing AWS and SMRT Link safety features
- Compliance with data localization requirements
- Data localization provided by AWS data locality
- For information regarding specific security questions and concerns, see the SMRT Link
Cloud Reference Guide (v10.1) (PN 102-043-900)
63
SMRT LINK CLOUD USAGE
Same functionality and usage as a local SMRT Link instance
- All SMRT Link features
available
- SMRT Link access URL
points to the Cloud instance
- All available User Documents
are applicable for SMRT Link
Cloud
64
SMRT LINK CLOUD – TRANSFER SERVER REQUIREMENTS
AWS DATASYNC VM SPECIFICATIONS (MINIMUM REQUIREMENTS)
Virtual processors – Four (4) virtual processors assigned to the VM
Disk space – 80 GB of disk space for installation of VM image and system data
RAM – Depending on your configuration, one of the following:
• 32 GB of RAM assigned to the VM, for tasks to transfer EC2 instance types with up to 20 million files.
• 64 GB of RAM assigned to the VM, for tasks to transfer more than 20 million files
STORAGE REQUIREMENTS
Sequel
IIe System 1.5 – 2 TB (no kinetics info), 9 – 12 TB (with kinetics info)
Sequel II
System 12 TB
Sequel
System 1.5 – 2 TB
NETWORK
10 GBE highly recommended, 1 GBE required
Time for data transfer to AWS is highly dependent on network speed and load
65
COST SAVINGS: HUMAN TRIO ANALYSIS (20-FOLD HIFI COVERAGE)
WITH CLOUD COMPUTE
Sequel II System Sequel IIe System Sequel IIe System Savings
Instrument data transfer (data type)
6,000 GB (Subreads) 320 GB (HiFi Reads) 95%
HiFi read generation
12,000 CPU-hours - 100%
Application compute
2,000 CPU-hours 2,000 CPU-hours -
Total compute
14,000 CPU-hours 2,000 CPU-hours 85%
Total data storage
6,000 GB 320 GB 95%
CPU cost (AWS)*
$602 $85 $517
Annual data storage cost (AWS)**
$1,671 $85 $1,586
Compute cost (AWS)***
$2,273 $170 $2,103
* m5a.12xlarge (48 vCPUs, 192 GiB RAM, $2.064/hr), https://aws.amazon.com/ec2/pricing/on-demand/
** Assumes 1 trio for 1 year at $0.023 per month, https://aws.amazon.com/s3/pricing/
66
*** All prices are listed in USD and costs may
vary by region.

SMRT Link Applications Updates
ANALYSIS APPLICATION UPDATES
Genome Assembly – De Novo Assembly Using HiFi Reads (SMRT Link v10.0 Release) [NEW]
- Generate highly accurate polished contiguous assemblies and fully phased haplotigs
- Fast and easy to use
68
Amplicon Analysis – HiFi Reads (Bioconda Release) [NEW]
- Clustering and allele detection using HiFi reads
Iso-Seq Analysis (SMRT Link v10.1 Release) [UPDATED]
- Features improved graphical and tabular report outputs for analysis of multiplexed Iso-Seq samples
SARS-CoV-2 Analysis (SMRT Link v10.1 Release) [NEW]
- Analyze multiplexed SARS-CoV-2 viral amplicon samples to identify variants and call a single consensus
sequence per sample using HiFi reads
Single-Cell Iso-Seq Analysis (Bioconda Release) [NEW]
- Supports analysis of Unique Molecular Identifier (UMI) sequence tags in single-cell Iso-Seq samples
Contiguity Correctness
Completeness Compute
-Resolve Repetitive
Regions
-High Contig N50
-Gene Space
-Repetitive
Regions
-Base QV
-Phasing accuracy
AGTTTCGATAGA
AGTT-CGAAAGA
-CPU / Wall Time
-RAM
-Disk Storage
GENOME ASSEMBLY ANALYSIS APPLICATION [NEW]
SMRT Link Genome Assembly analysis application uses HiFi reads for improved de novo
assemblies
69

GENOME ASSEMBLY ANALYSIS APPLICATION ALGORITHM
Powered by IPA (Improved and Phased Assembly) algorithm
- Fast and efficient assembly – 5.5 hours* for a human assembly with 20-fold HiFi read
coverage
- High contiguity
- Fully phased haplotigs
- High per-base quality of polished assemblies
- Easy to use
* Compute environment:
Head Node - Cores: 32, RAM: 64 GB, 1 TB local tmp, 256 GB local db_datadir
Compute Nodes – Cores 64, RAM: 4GB per core, 1 TB local tmp, 256 GB local db_datadir
70
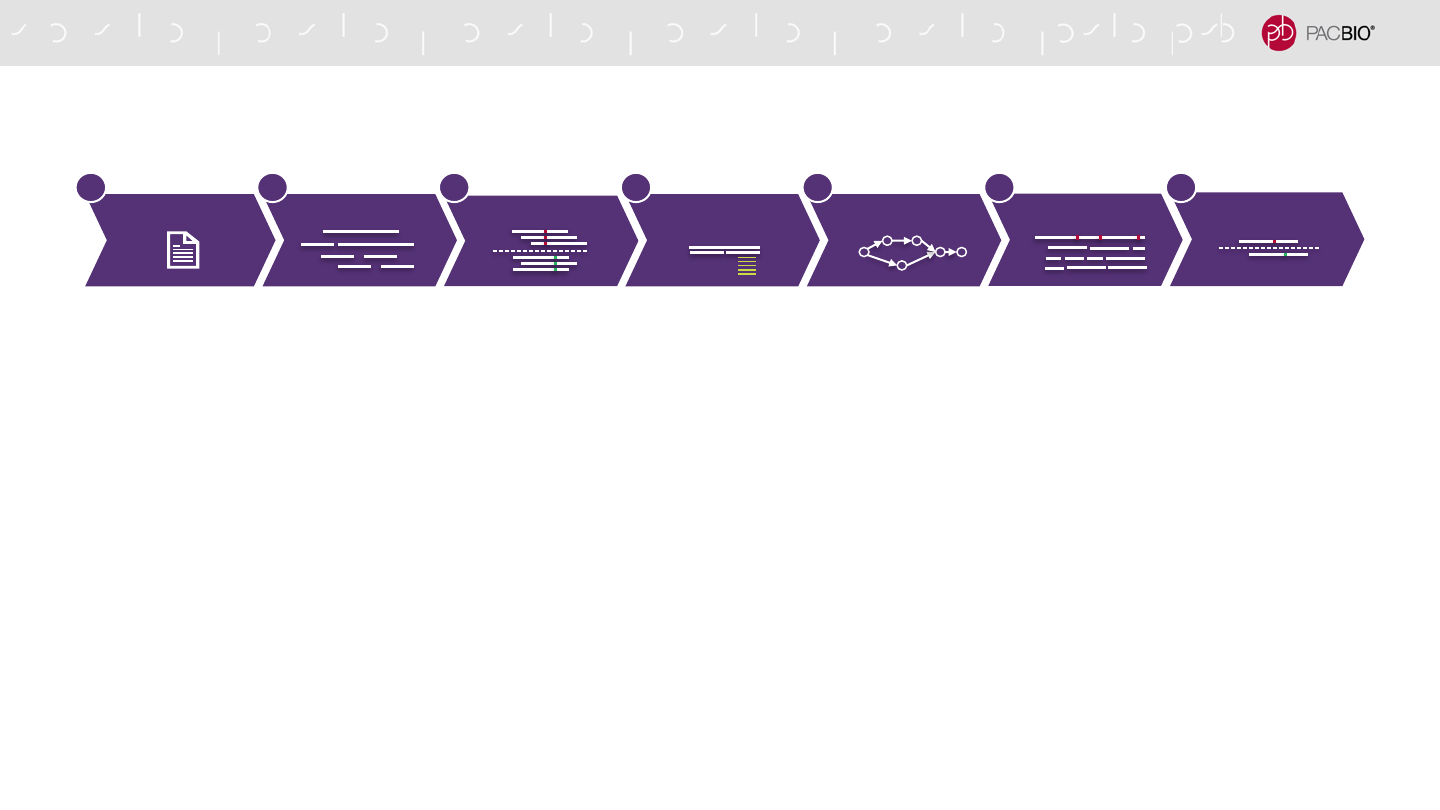
GENOME ASSEMBLY ANALYSIS APPLICATION WORKFLOW
1. Convert inputs to compressed database format for fast and easy retrieval
2. Overlap reads to form read piles using local alignment initialization and extension
3. Separate overlapped reads by phase using de Bruijn graph
4. Remove chimeras and repeats to improve contiguity and reduce missasembly
5. Build a string graph with primary and associate contigs, assign reads to contigs based on phased overlaps
6. Polish using phased-aware assignment-based mapping
7. Purge duplicates
Building SEQ. Db
Phase Separation
Chimera & Repeat
Filtering
Fast Overlap Layout
Polishing
Purge Duplicates
2 3 4 5 71 6
71

SARS-CoV-2 ANALYSIS APPLICATION [NEW]
Use Case
- Analysis support for HiFiViral for SARS-CoV-2 Workflow (See Procedure & Checklist – Multiplexing 1.2 kb
Amplicons for Full-Viral Genome Sequencing [PN 102-075-000 (High-Throughput) / PN 102-082-500 (Low-
Throughput)]
- For each sample, identifies a single SARS-CoV-2 species and consensus sequence
- Input Data: HiFi sequencing data for multiplexed SARS-CoV-2 amplicon samples*
- Amplicon sizes supported: From a few hundred bases to kilobases, tiled across the entire 30 kb SARS-CoV-2
viral genome
- Sample multiplexing level supported: 10- to 1000-plex
Analysis output per sample
- Amplicon coverage (CSV)
- Variant calls (VCF)
- Consensus sequence (FASTA)
- Aligned reads (BAM)
* SARS-CoV-2 analysis application does not support non-amplicon SARS-CoV-2 data (capture-based data, WGS or transcriptome) and non-SARS-CoV-2 viral data.
72
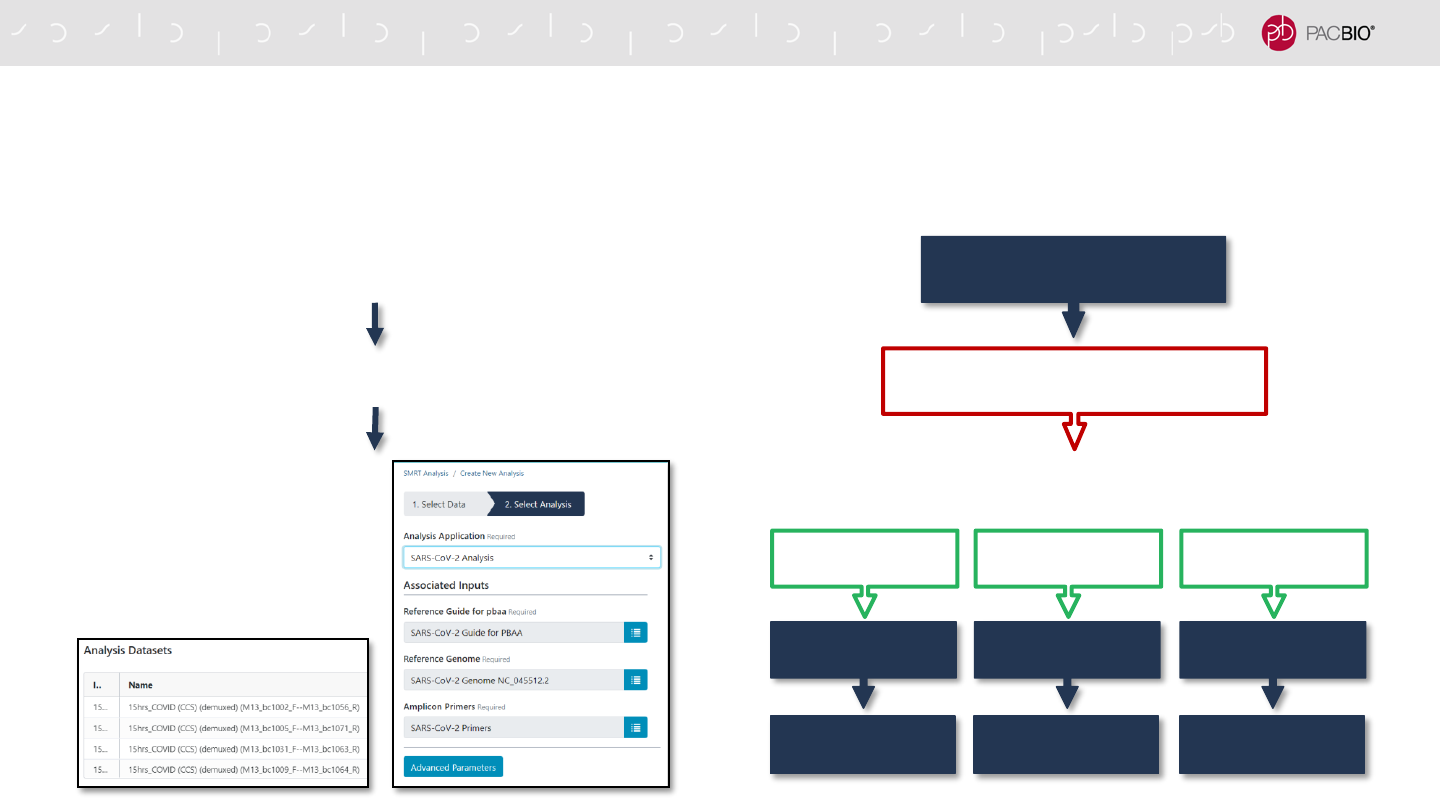
SARS-CoV-2 ANALYSIS APPLICATION WORKFLOW
HiFi Reads
Demultiplex Barcodes
SARS-CoV-2
Analysis Pipeline
(Select all samples to analyze)
HiFi Reads
Demultiplex to remove barcoded
M13 primers from PCR 2
Trim target-
specific primers
Variant Calling
Consensus
Sequence
Trim target-
specific primers
Variant Calling
Consensus
Sequence
Trim target-
specific primers
Variant Calling
Consensus
Sequence
Sample 1 Sample 2 ….Sample N
Use the SARS-CoV-2 analysis application in SMRT Link to analyze multiplexed viral surveillance
samples for SARS-CoV-2
73
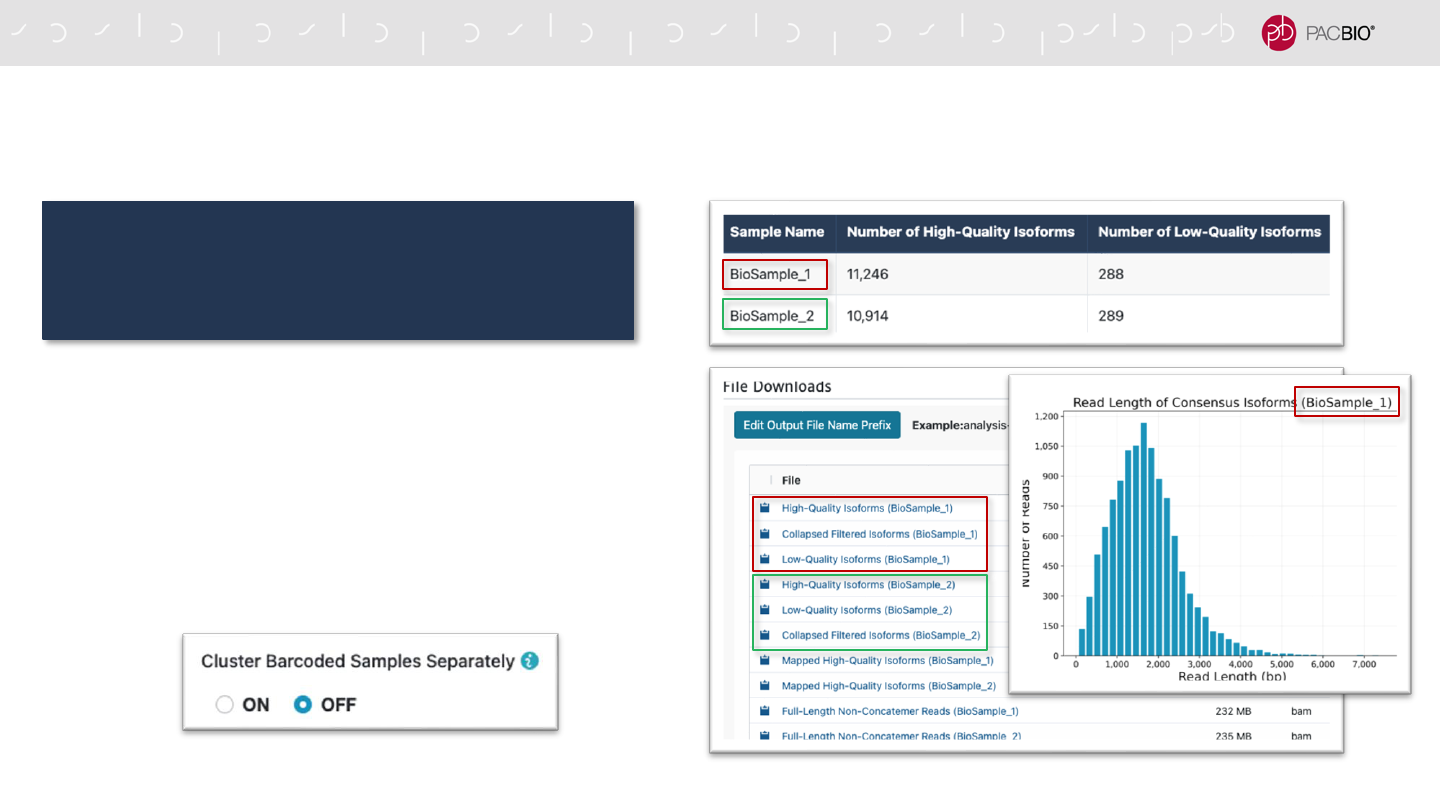
IMPROVED ISO-SEQ ANALYSIS APPLICATION FOR MULTIPLEXED
SAMPLES
- For multiplexed datasets, Iso-Seq Analysis reports
and graphs now include isoforms per barcode in
addition to the total number of isoforms across all
barcodes
- Per-barcode summary metrics, plots, and file
downloads are generated
- Iso-Seq Analysis now supports both separate and
joint clustering of barcoded samples
In SMRT Link v10.1, use the Iso-Seq Analysis
application to analyze multiplexed Iso-Seq
samples* – do not run Demultiplex Barcodes first
* In Iso-Seq Analysis, select primer set IsoSeq_Primers_12_Barcodes_v1 or another custom set containing multiple barcodes Do not use the default of IsoSeqPrimers_v2
74

PACBIO AMPLICON ANALYSIS APPLICATION (pbAA) [BIOCONDA
RELEASE]
PacBio Amplicon Analysis Application for HiFi reads
- Functionality:
▪ A reference-guided application for clustering and generation of high-quality phased consensus
sequences using HiFi data
- Benefits:
▪ Accurate - base level resolution
▪ Sensitive – no missed alleles, favoring false positive over false negatives
▪ Fast – results in less than five minutes for samples with high read depth (>500-fold)
- Optimized performance with low computational complexity
▪ Flexible – general amplicon analysis application with tunable parameters
▪ Bonus feature: Visualization sub-tool for coloring aligned reads by cluster (helpful for interpreting
and troubleshooting analysis results)
75
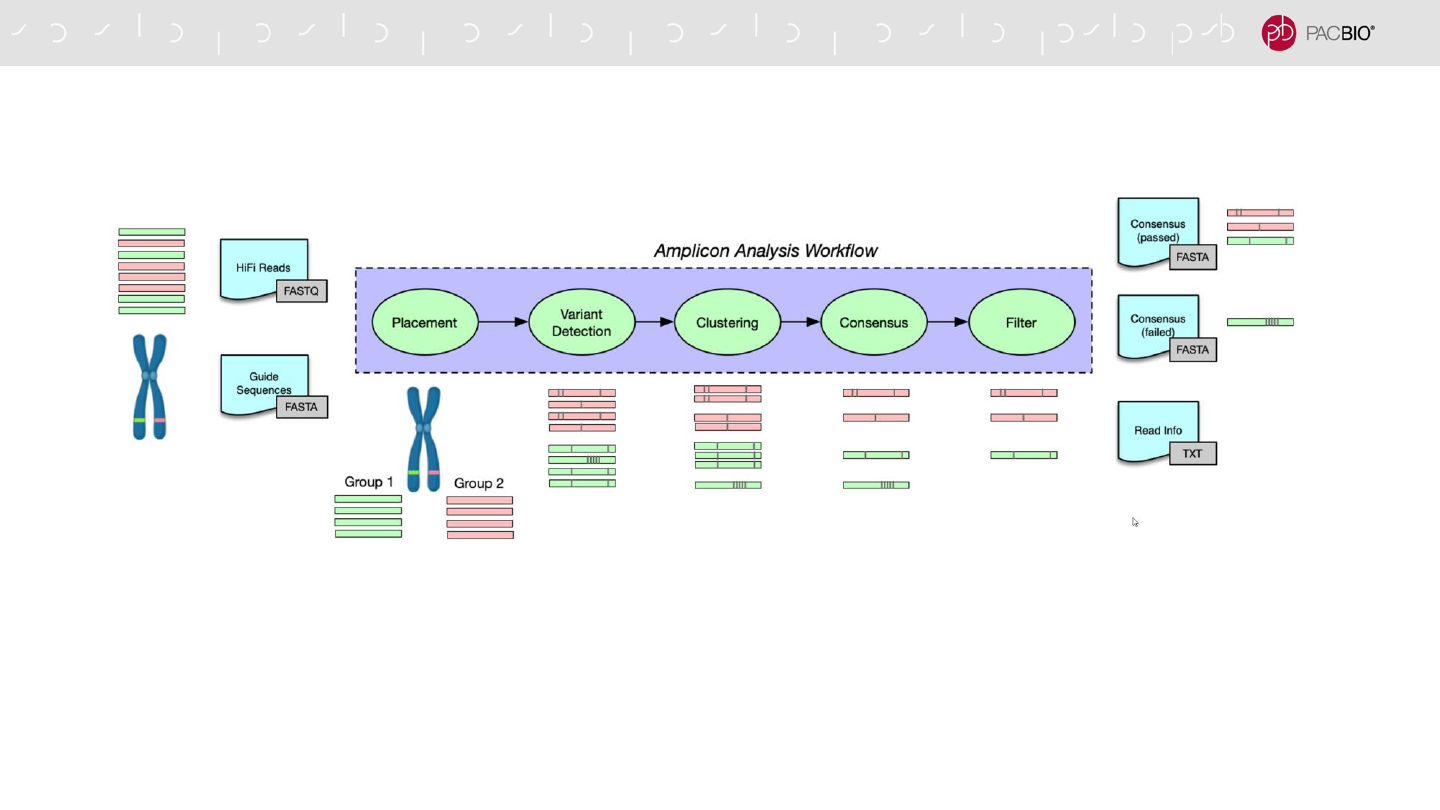
PACBIO AMPLICON ANALYSIS APPLICATION (pbAA) WORKFLOW
AND OUTPUT
1. Generate guide information
2. Assign reads to a loci
3. Detect variants
4. Cluster using a custom model
5. Generate consensus
6. Filter results
Analysis Workflow
- Two cluster consensus files – reads passing
and failing filtering
- Reads information file – details on read
classification
Output
76

SINGLE-CELL ISO-SEQ METHOD SUMMARY
77
- Procedure & Checklist – Preparing Single-Cell Iso-Seq Libraries
Using SMRTbell Express Template Prep Kit 2.0 protocol (PN 101-
892-000) provides detailed workflow guidance
- Uses standard Iso-Seq Express library preparation & sequencing
workflow
- Generating matching short-read data from the same library sample is
recommended
- Characterize alternative splicing with up to 3 Million full-length
transcript reads generated per SMRT Cell 8M
- Each FL transcript read contains single-cell barcode and UMI information
https://www.pacb.com/support/documentation/
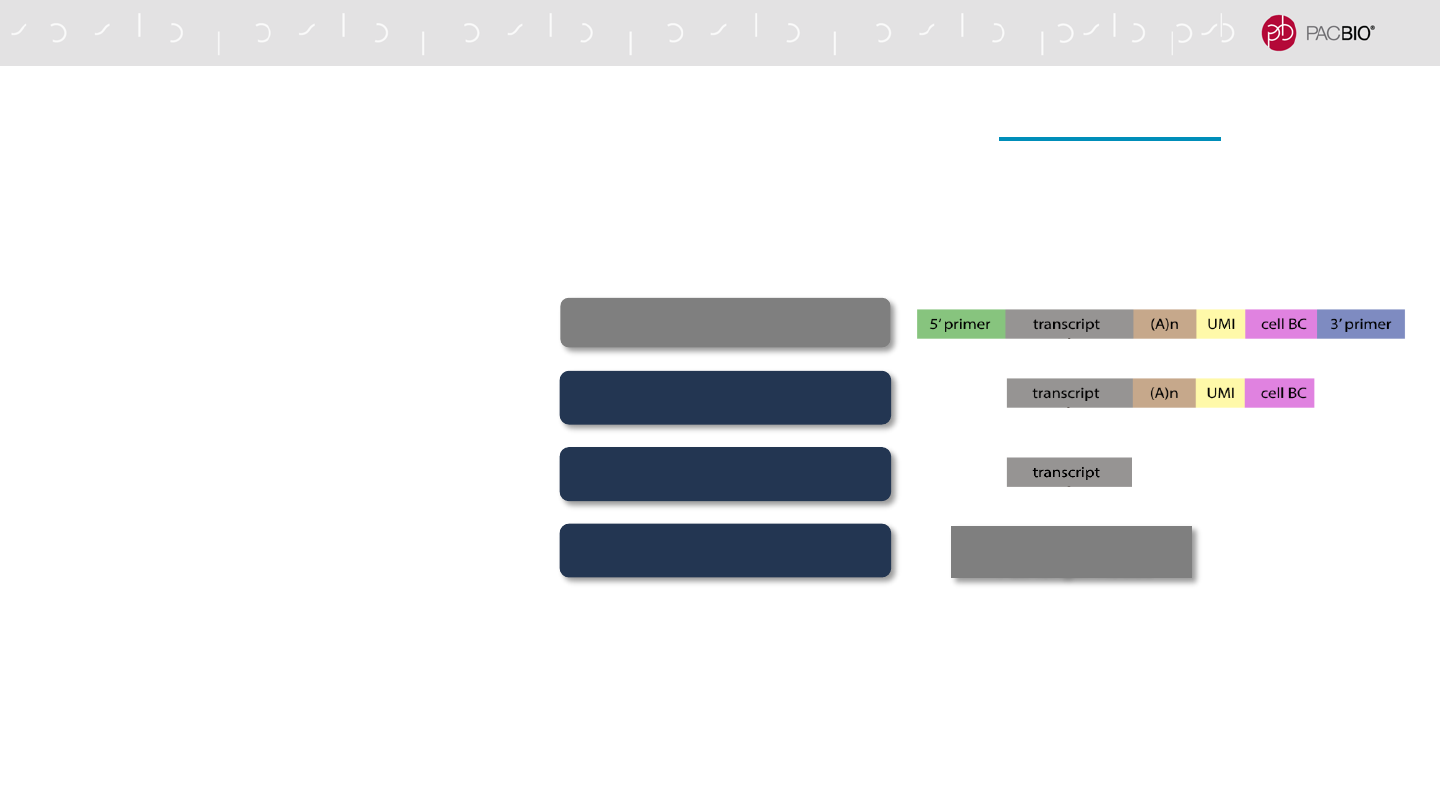
SINGLE-CELL ISO-SEQ ANALYSIS WORKFLOW [BIOCONDA
RELEASE]
1. Remove cDNA 5’ and 3’ primers
2. Extract and trim UMI and cell BC
3. Remove polyA tails and concatemers
4. Cluster by UMI using QV-guided approach
CCS reads ≥QV10
Remove cDNA primers
Extract UMI and BC
Cluster by UMI
Unique Polished
Full-length Reads
Analysis Workflow
78
Supports analysis of Unique Molecular Identifier (UMI) sequence tags in single-cell Iso-Seq
samples

OTHER ANALYSIS APPLICATION UPDATES
Improved Structural Variant Calling Analysis Application
- Improved precision:
▪ Breakend (BND) calls - filtering of short and low-identity alignments
- Added SVLEN annotation for inversion variants
Improvement to Mapping Applications
- Enhanced alignment concordance
▪ Industry-standard BLAST-style alignment identity
79
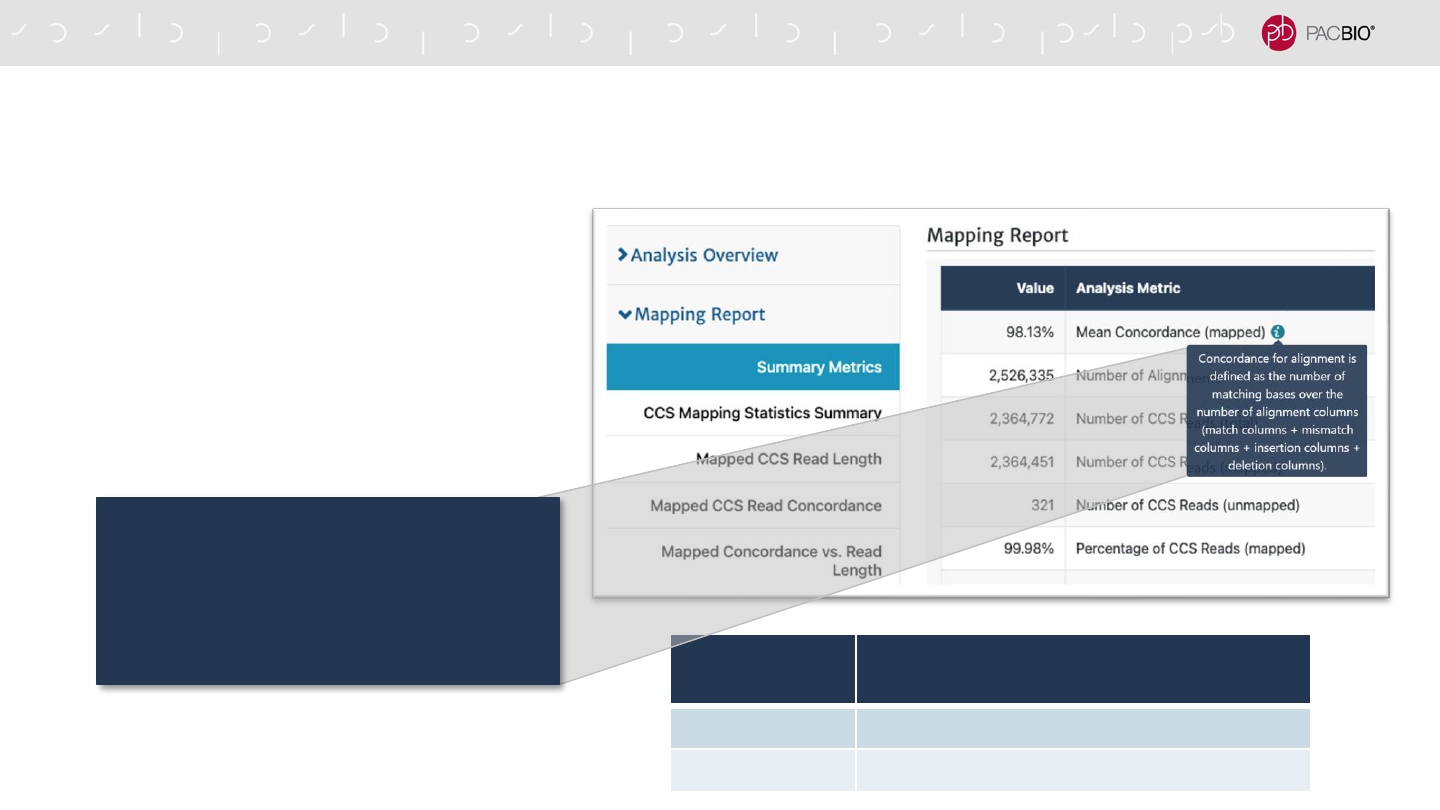
IMPROVED CALCULATION OF ALIGNMENT CONCORDANCE IN
MAPPING APPLICATIONS
- Alignment concordance is now
reported as industry-standard BLAST-
style alignment identity
(matches/alignment columns)
- In earlier versions we used a non-
standard calculation for concordance
DATA TYPE
MEAN MAPPED CONCORDANCE
(OLD VS. NEW CALCULATION)
HiFi Data 0.1 – 0.2% Lower
CLR Data 0.5% Higher
Concordance for alignment is defined as
the number of matching bases over
the number of alignment columns
(match columns + mismatch columns +
insertion columns + deletion columns).
80

SMRT Link General Usability Improvements
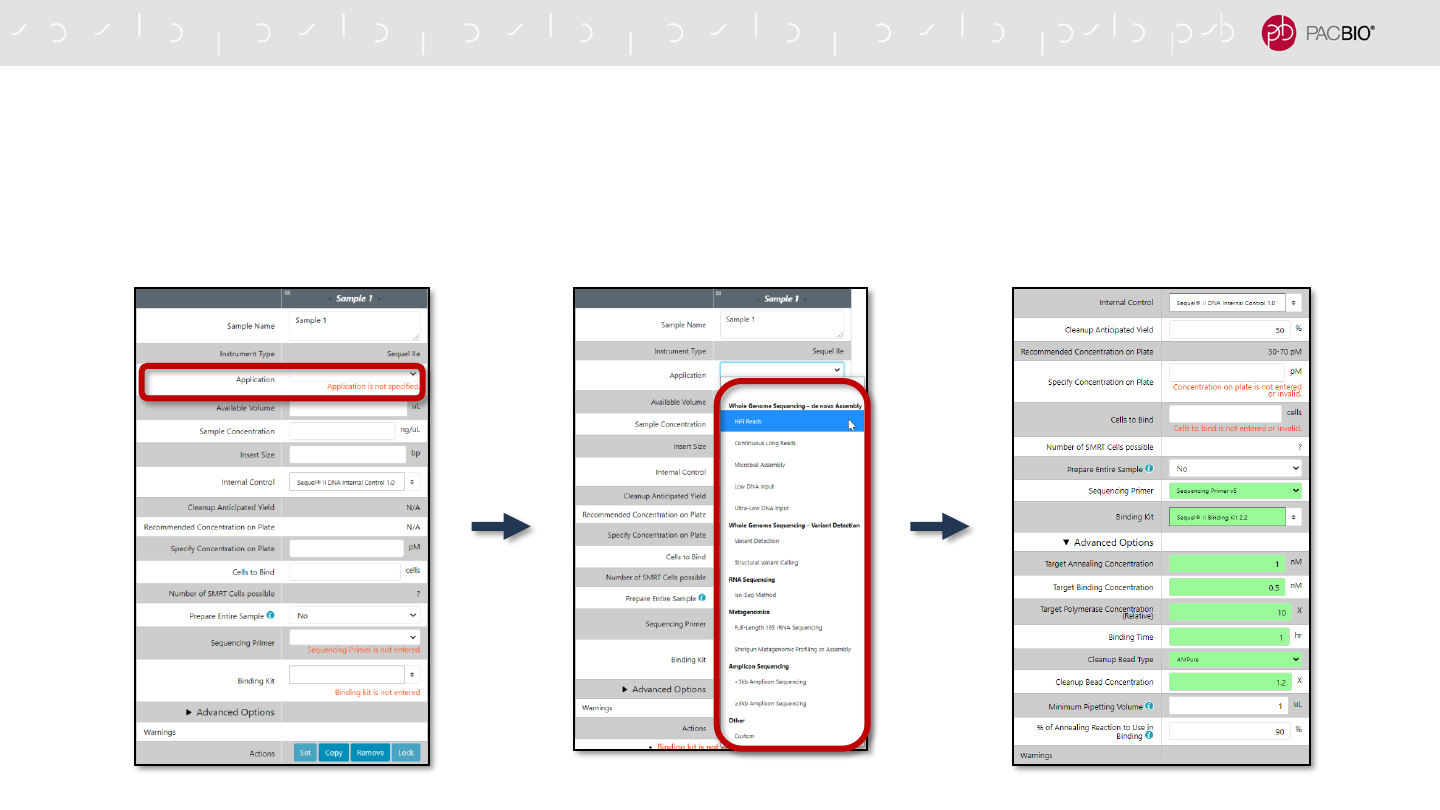
SMRT LINK USER INTERFACE AND USABILITY IMPROVEMENTS
More Streamlined Application-centric Sample Setup and Run Design
- Default protocol and run settings are auto-filled for each selected application type
82

SMRT LINK USER INTERFACE AND USABILITY IMPROVEMENTS
New HiFi Metrics and Data Visualization Reports
- Available in Run QC when On-Instrument CCS is enabled for the Sequel IIe System
83
Displays a histogram distribution of
HiFi Reads (QV ≥20), other CCS
Reads (three or more passes, but QV
<20), and other reads, by read length.
Displays a histogram distribution of
HiFi Reads (QV ≥20) and other CCS
Reads by read quality.
Displays a heat map of CCS Read
lengths and predicted accuracies. The
boundary between HiFi Reads and
other CCS Reads is shown as a
dashed line at QV 20.
HiFi Reads
(≥Q20)
Q20
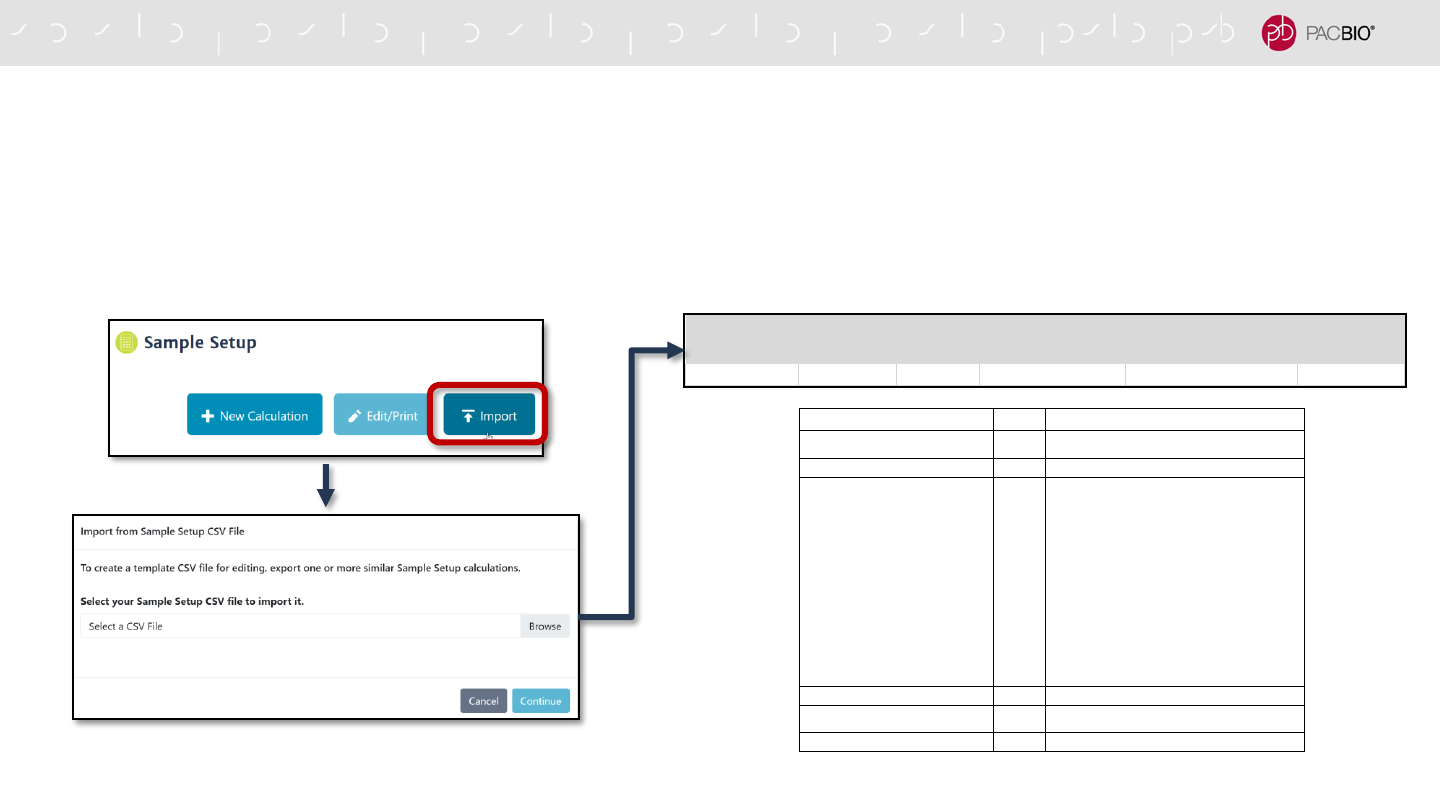
SMRT LINK USER INTERFACE AND USABILITY IMPROVEMENTS
Improved Sample Setup Support for High-Throughput Sequencing
- Sample Setup features enhanced support for high-throughput production environments through new
Sample Setup sheet *.CSV import function
84
Field Name
Required
Description
Sample Name
Yes
Enter alphanumeric characters, spaces, hyphens,
underscores, colons, or periods only.
System Name
Yes
Must be Sequel, Sequel II, or Sequel IIe.
Application
Yes
Enter one of the following values:
• HiFi Reads
• Continuous Long Reads
• Low DNA Input
• Ultra-Low DNA Input
• Microbial Assembly
• Variant Calling
• Structural Variation Calling
• HiFiViral SARS-CoV-2
• Iso-Seq Method
• Full-Length 16S rRNA Sequencing
• Shotgun Metagenomic Profiling or
Assembly
• <3kb Amplicons
• >=3kb Amplicons
• Custom
Available Starting Sample Volume (uL)
Yes
Enter a positive integer. Units are in microliters.
Starting Sample Concentration (ng/uL)
Yes
Enter a positive integer. Units are in nanograms per
microliter.
Insert Size (bp)
Yes
Enter a positive integer. Units are in base pairs.
Sample Name System Name Application
Available Starting
Sample Volume (uL)
Starting Sample
Concentration (ng/uL)
Insert Size (bp)
Sample 1 Demo Sequel IIe HiFi Reads 10 100 18000

SMRT LINK USER INTERFACE AND USABILITY IMPROVEMENTS
85
We recommend notifying PacBio of your successful SMRT Link v10.1 installation and sending ongoing
SMRT Link analysis usage information to PacBio in order to expedite case troubleshooting and to help us
continually improve our products

SMRT Link Fixed & Known Issues

SMRT LINK V10.1 FIXED ISSUES HIGHLIGHTS
See the latest SMRT Link Release Notes for an updated list of fixed issues
87
- SV calling: Joint SV calling on demultiplexed Data Sets from the same cell/collection – demultiplexed Data Sets are now
analyzed separately, and the Bio Sample name is used.
- Sample Setup: Columns in the edit/print view can now be drag-and-dropped.
- Copying to the clipboard now works as expected.
- Exporting large analysis directories now works correctly and does not fail.
- Absolute file paths are now included in the subreadset.xml file.
- Login for local SMRT Link WSO2 users is now enabled.
- The outputs analysis directory now includes symbolic links to the BAM files.
- BAM files consolidation for microbial assemblies now works correctly.
- Using the hyphen character "-" in barcode and Bio Sample names no longer causes the Demultiplex Barcodes application
to fail.
- HGAP4 analysis no longer fails if the Genome Size is set to more than 2.0 GB.

- Run Design: When opening a saved Run Design, you will sometimes be asked to save changes when no changes were
made.
- Run Design: The Import from Sample Setup feature does not distinguish between Sample Setup designs created for Sequel II
and for Sequel IIe, instead showing both.
- A cached URL containing the string /welcome at the end of the SMRT Link URL (Example: https://URL/sl/welcome) in the
browser’s history causes an error when accessing SMRT Link.
- Motif detection is designed for microbial genomes and has not been tested on non-microbial genomes; it may run out of
memory on large genomes.
- When copying an analysis using the Demultiplex Barcodes application (using Copy from an Analysis Results page or Copy
From on the New Analysis page), the input of sample names for each barcode is not preserved from the copied analysis. In
the second step of the New Analysis wizard, users must either re-enter the sample names using the Interactive Barcode
Selector and Sample Name Editor, or re- upload a Barcoded Sample File. The Start button is not enabled until users do so.
- When creating a user, ensure that the new user profile has the Username attribute populated with the account/login name.
This is required for the user search in the configuration and project pages to find local users. (See SMRT Link Software
Installation (v10.1) for details.)
- When using Bio Sample Names with a PacBio analysis application, you can enter names that include spaces. Please avoid
using spaces in Bio Sample Names as spaces may lead to third-party compatibility issues.
SMRT LINK V10.1 KNOWN ISSUES HIGHLIGHTS
See the latest SMRT Link Release Notes for an updated list of known issues
88

For Research Use Only. Not for use in diagnostic procedures. © Copyright 2021 by Pacific Biosciences of California, Inc. All rights reserved.
Sequel II and IIe Systems Applications Support
Resources
Subhead should be no longer than 1 line
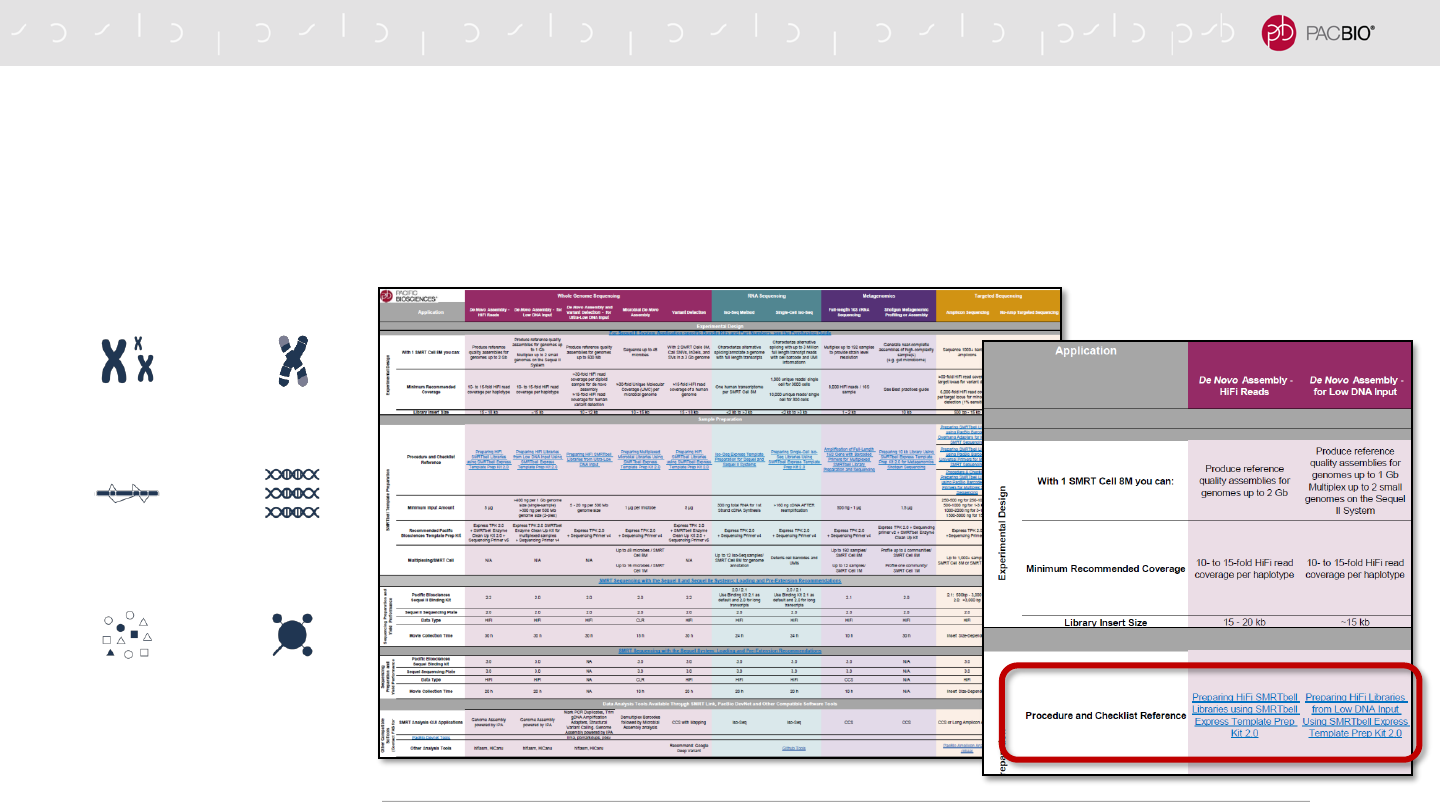
OVERVIEW – SEQUEL SYSTEMS APPLICATION OPTIONS AND
SEQUENCING RECOMMENDATIONS
This document provides high-level application workflow guidance and links to protocols for
preparing samples for sequencing on the Sequel Systems and analysis.
https://www.pacb.com/wp-content/uploads/Overview-Sequel-Systems-Application-Options-and-Sequencing-Recommendations.pdf
Whole Genome
Sequencing
Variant Detection
RNA Sequencing Targeted
Sequencing
Complex
Populations
Epigenetics
90
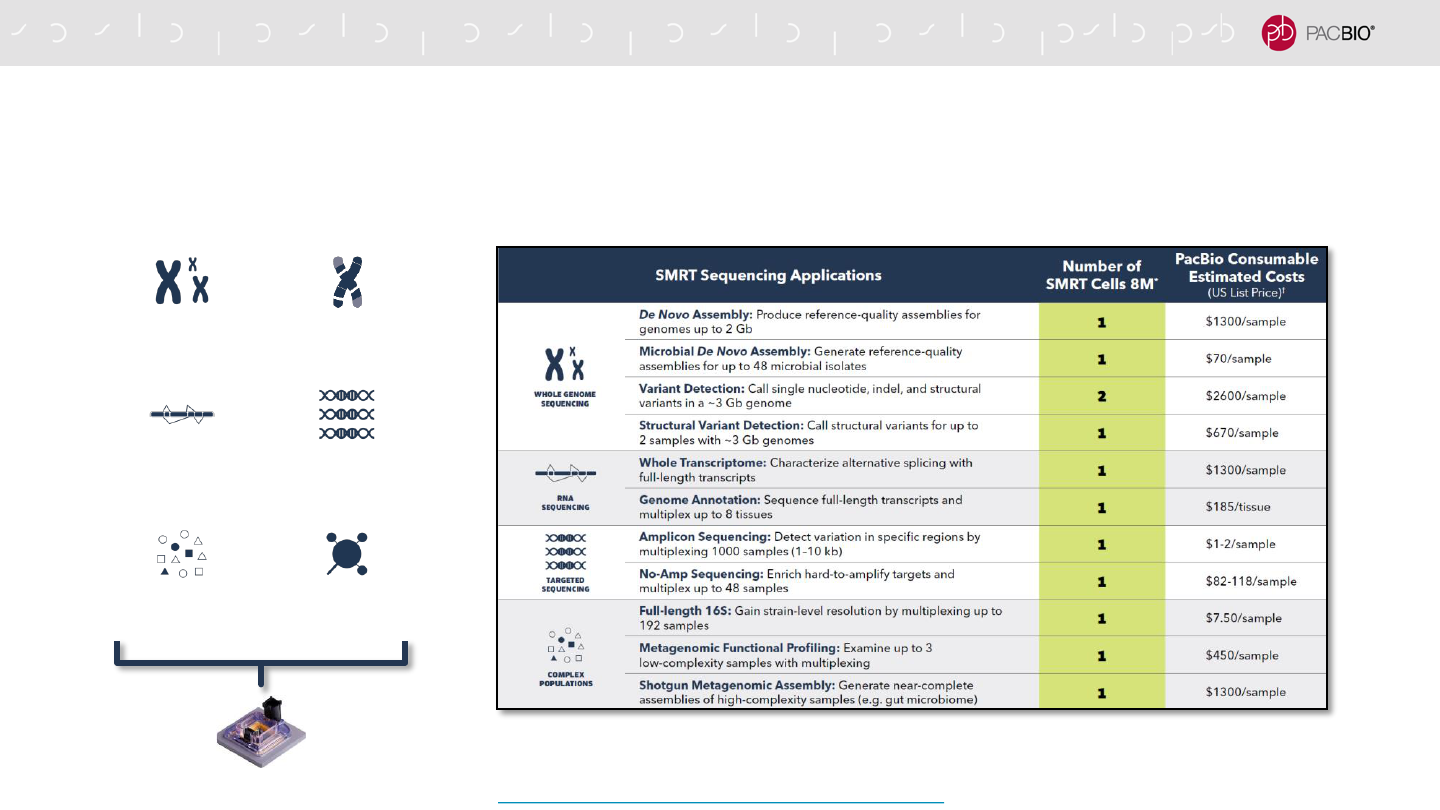
WHAT CAN YOU DO WITH ONE SMRT CELL 8M?
With PacBio Single Molecule, Real-Time Sequencing on the Sequel II and IIe Systems you can
characterize whole genomes and transcriptomes with just one SMRT Cell 8M.
*Study design, sample type, and level of multiplexing may affect the number of SMRT Cells 8M required.
†
All prices
are listed in USD and cost may vary by region. Pricing includes library and sequencing reagents run on your Sequel
II System and does not include instrument amortization or other reagents.
Application Brochure: What Can You Do with One SMRT Cell?
91
SMRT Cell 8M
Whole Genome
Sequencing
Variant Detection
RNA Sequencing Targeted
Sequencing
Complex
Populations
Epigenetics

APPLICATION CONSUMABLE BUNDLES & PURCHASING GUIDE
https://www.pacb.com/wp-content/uploads/Application-Consumable-Bundle-Purchasing-Guide.pdf
Purchasing Guide brochure enables users to easily order required consumables needed to run
a specific type of application on the Sequel II and IIe Systems.
-Customers can use a single part number to order a
consumables bundle containing PacBio-branded reagents
needed for SMRTbell library construction, primer annealing &
polymerase binding
-Exclusions:
- Core PacBio-branded SMRT Sequencing consumables (SMRT
Cells, Sequencing Kits & SMRT Oil), plastics and other 3rd-party
reagents are not included in the application bundles
- For Barcoded Adapter bundles that support >16-plex, PacBio
recommends customers purchase barcoded adapters directly from
a third-party oligo synthesis company.
92
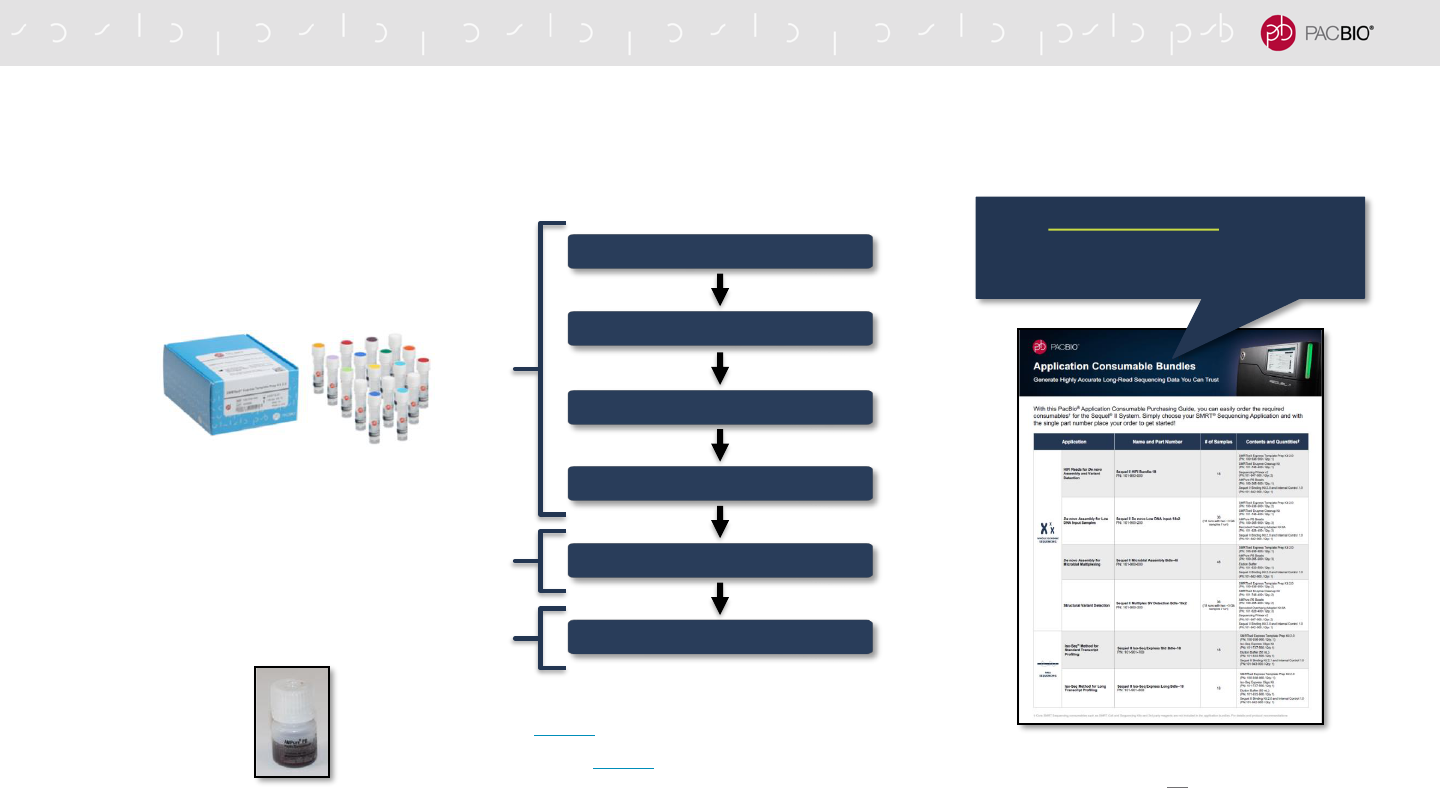
CORE PACBIO REAGENTS & CONSUMABLES REQUIRED
FOR SMRTBELL EXPRESS LIBRARY CONSTRUCTION
93
DNA End Repair / A-Tailing
Adapter Ligation
Purify SMRTbell Templates**
Remove Single-Strand Overhangs
DNA Damage Repair
Nuclease Treatment*
SMRTbell Express Template Prep Kit 2.0
SMRTbell Enzyme Cleanup Kit 2.0
* A Nuclease Treatment step is included in some
protocols to remove non-intact SMRTbell templates
AMPure PB Beads
** Some protocols may specify the use of ProNex Beads
(instead of AMPure PB Beads) for SMRTbell purification
PacBio Purchasing Guide brochure enables
users to easily order required consumables needed
to prepare a SMRTbell library to run a specific type
of application on the Sequel II/IIe System.***
*** Core PacBio-branded SMRT Sequencing consumables
(SMRT Cells, Sequencing Kits & SMRT Oil), plastics and
other 3rd-party reagents are not included in the application
bundles
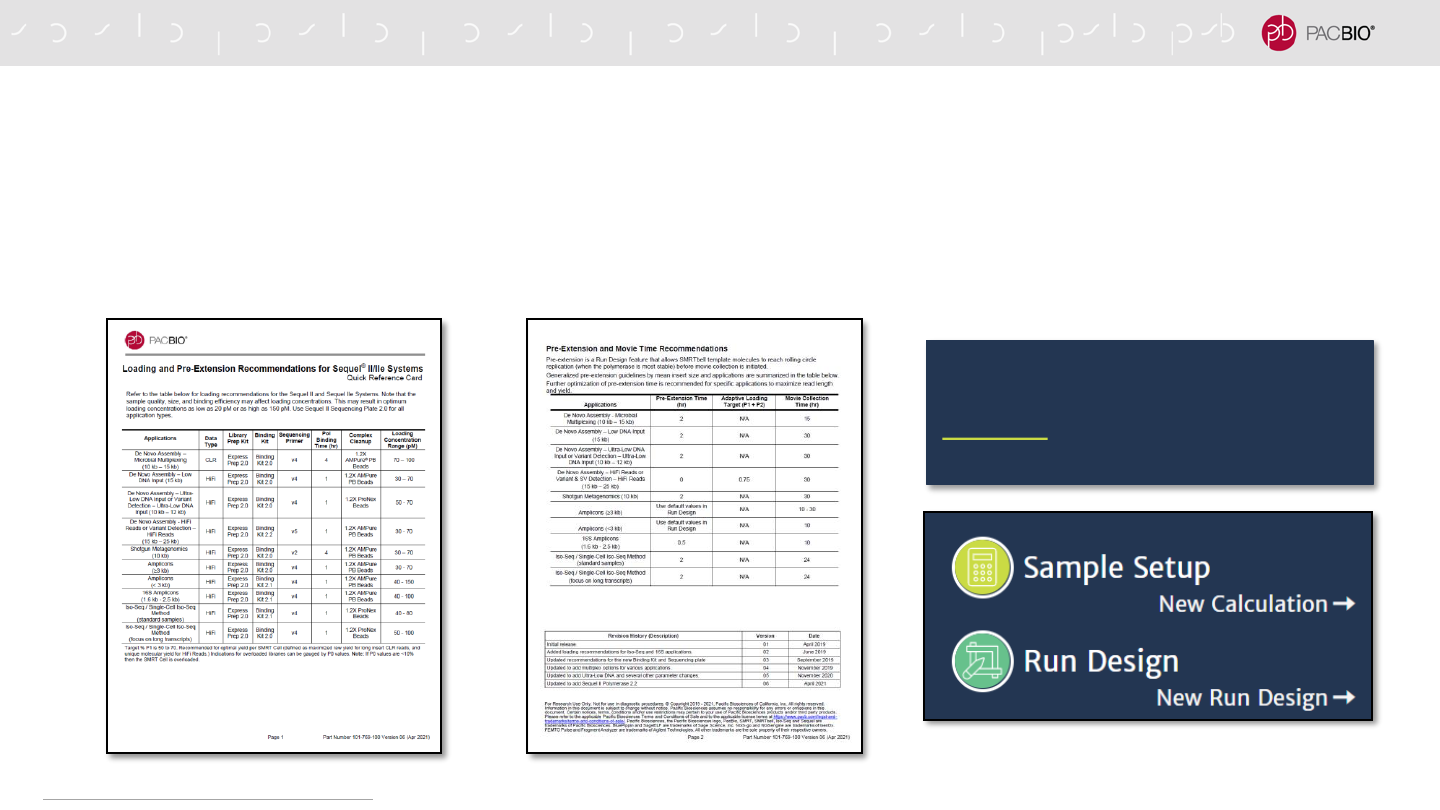
SEQUEL IIe SYSTEM QUICK REFERENCE CARD – DIFFUSION
LOADING AND PRE-EXTENSION RECOMMENDATIONS
Follow SMRT Link Sample Setup & Run Design instructions using the recommendations provided in the
Quick Reference Card – Loading and Pre-Extension Time Recommendations for the Sequel II/IIe System
unless specified otherwise in the relevant Procedure & Checklist
94
In SMRT Link v10.1, most Sample Setup
and Run Design parameter fields are
auto-filled with the recommended settings for
each application type.
https://www.pacb.com/support/documentation/

TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES
- Sequel II and Sequel IIe Systems Operations Guide (PN 101-774-700)
- Sequel II/IIe System v10.1 Release Notes (PN 102-041-700)
- Sequel IIe System: Location of HiFi Reads Files (PN 102-110-200)
- Quick Reference Card – Loading and Pre-Extension Recommendations for the Sequel II/IIe Systems (PN 101-769-100)
- Pacific Biosciences Glossary of Terms (PN 000-710-267)
Sequel IIe System Documentation
https://www.pacb.com/support/documentation/
- SMRT Link v10.1 Software Download Site: https://www.pacb.com/support/software-downloads/
- SMRT Link v10.1 Software Installation Instructions (PN 102-036-900)
- SMRT Link v10.1 Release Notes (PN 102-040-000)
- SMRT Link v10.1 User Guide (PN 102-037-000)
- SMRT Link Cloud Reference Guide (v10.1) (PN 102-043-900)
- SMRT Link Web Services API Use Cases (v10.1) (PN 102-040-300)
SMRT Link Documentation
95

TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES (CONT.)
- Overview – Sequel Systems Application Options and Sequencing Recommendations (PN 101-851-300)
- Procedure & Checklist – Using AMPure PB Beads for Size-Selection (PN 101-854-900)
- Whole Genome Sequencing Applications
- De Novo Assembly – HiFi Reads
- Procedure & Checklist – Preparing HiFi SMRTbell Libraries using SMRTbell Express Template Prep Kit 2.0 (PN 101-853-100)
- De Novo Assembly – Low DNA Input
- Procedure & Checklist - Preparing SMRTbell Libraries Using Express Template Prep Kit 2.0 With Low DNA Input (PN 101-730-400)
- De Novo Assembly – Ultra-Low DNA Input
- Procedure & Checklist – Preparing HiFi SMRTbell Libraries from Ultra-Low DNA Input (PN 101-987-800)
- Microbial De Novo Assembly
- Procedure & Checklist – Preparing Multiplexed Microbial Libraries Using SMRTbell Express Template Prep Kit 2.0 (PN 101-696-100)
- Variant Detection
- Procedure & Checklist – Preparing HiFi SMRTbell Libraries using SMRTbell Express Template Prep Kit 2.0 (PN 101-853-100)
Sample Library Preparation Documentation
https://www.pacb.com/support/documentation/
96

TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES (CONT.)
- RNA Sequencing Applications
- Iso-Seq Method
- Procedure & Checklist – Iso-Seq Express Template Preparation for Sequel and Sequel II Systems (PN 101-763-800)
- Procedure & Checklist – Preparing Single-Cell Iso-Seq Libraries Using SMRTbell Express Template Prep Kit 2.0 (PN 101-892-000)
- Metagenomics Applications
- Full-length 16S Sequencing
- Procedure & Checklist – Amplification of Full-Length 16S Gene with Barcoded Primers for Multiplexed SMRTbell Library Preparation and Sequencing (PN 101-599-700)
- Metagenomics Shotgun Sequencing
- Procedure & Checklist – Preparing 10 kb Library Using SMRTbell Express Template Prep Kit 2.0 for Metagenomics Shotgun Sequencing (PN 101-800-800)
- Targeted Sequencing Applications
- Amplicon Sequencing
- Procedure & Checklist – Preparing SMRTbell Libraries using PacBio Barcoded Overhang Adapters for Multiplexing Amplicons (PN 101-791-700)
- Procedure & Checklist – Preparing SMRTbell Libraries using PacBio Barcoded Universal Primers for Multiplex SMRT Sequencing (PN 101-791-800)
- Procedure & Checklist – Preparing SMRTbell Libraries using PacBio Barcoded M13 Primers for Multiplex SMRT Sequencing (PN 101-921-300)
- No-Amp Targeted Sequencing
- Procedure & Checklist – No-Amp Targeted Sequencing Utilizing the CRISPR-Cas9 System (PN 101-801-500)
Sample Library Preparation Documentation (Cont.)
https://www.pacb.com/support/documentation/
97

TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES (CONT.)
- Whole Genome Sequencing Applications
- Application Brief: Whole genome sequencing for de novo assembly – Best Practices (PN BP102-121219)
- Application Brief: Variant detection using whole genome sequencing with HiFi reads – Best Practices (PN BP106-092419)
- Application Brief: Microbial whole genome sequencing – Best Practices (PN BP101-013020)
- RNA Sequencing Applications
- Application Brief: Long-read RNA sequencing – Best Practices (PN BP103-062619)
- Application Brief: Single-cell RNA sequencing with HiFi reads - Best Practices (PN BP109-102020)
- Metagenomics Applications
- Application Brief: Metagenomic sequencing with HiFi reads – Best Practices (PN BP108-030220)
- Targeted Sequencing Applications
- Application Brief: Targeted sequencing for amplicons – Best Practices (PN BP105-071919)
- Application Brief: No-Amp targeted sequencing – Best Practices (PN BP107-092319)
Applications Best Practices Guides
https://www.pacb.com/support/documentation/
98

TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES (CONT.)
- Whole Genome Sequencing Applications
- Technical Overview: HiFi Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-855-400)
- Technical Overview: Low DNA Input Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-781-000)
- Technical Overview: Ultra-Low DNA Input Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (101-998-000)
- Technical Overview: Multiplexed Microbial Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-742-600)
- RNA Sequencing Applications
- Technical Overview: Iso-Seq Express Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-814-400)
- Technical Overview: Single-Cell Iso-Seq Library Preparation Using SMRTbell Express TPK 2.0 (PN 101-925-400)
- Metagenomics Applications
- Technical Overview: Metagenomics Shotgun Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-894-900)
- Technical Overview: Full-Length 16S Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-916-900)
- Targeted Sequencing Applications
- Technical Overview: Multiplexed Amplicon Library Preparation Using SMRTbell Express Template Prep Kit 2.0 (PN 101-814-300)
- Technical Overview: No-Amp Targeted Sequencing Library Preparation and Data Analysis Technical Overview (PN 101-840-800)
- PacBio HiFiViral Workflow Overview: Multiplexed Amplicon Library Preparation for Full-Viral Genome Sequencing of SARS-CoV-2 (PN 102-084-800)
Applications Technical Training Documentation
https://www.pacb.com/support/documentation/
99

TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES (CONT.)
Data Analysis Documentation
https://www.pacb.com/support/documentation/
- Analysis Procedure – Multiplexed Microbial Assembly with SMRT Link v8.0 and SMRTbell Express Template Prep Kit 2.0
(PN 101-855-300)
- Analysis Procedure – No-Amp Data Preparation and Repeat Analysis (PN 101-801-400)
- Brief Primer and Lexicon for PacBio SMRT Sequencing Webpage (v10.0)
- PacBio Bioinformatics File Formats Documentation Webpage (v10.0)
- SMRT Analysis Barcoding Overview (v9.0) (PN 101-923-200)
- SMRT Tools Reference Guide (v10.1) (PN102-037-300)
- Guide – Step-by-Step Run Performance Evaluation (For Sequel II and Sequel IIe Systems) (PN 101-993-600)
Sequencing Performance Troubleshooting Documentation
100
TECHNICAL DOCUMENTATION & SOFTWARE DOWNLOAD
RESOURCES (CONT.)
Technical Notes
https://www.pacb.com/support/documentation/
- Technical Note: Preparing samples for PacBio whole genome sequencing for de novo assembly – Collection and storage
(PN TN100-040518)
- Technical Note: Preparing DNA for PacBio HiFi sequencing – Extraction and quality control (PN TN101-081420)
101
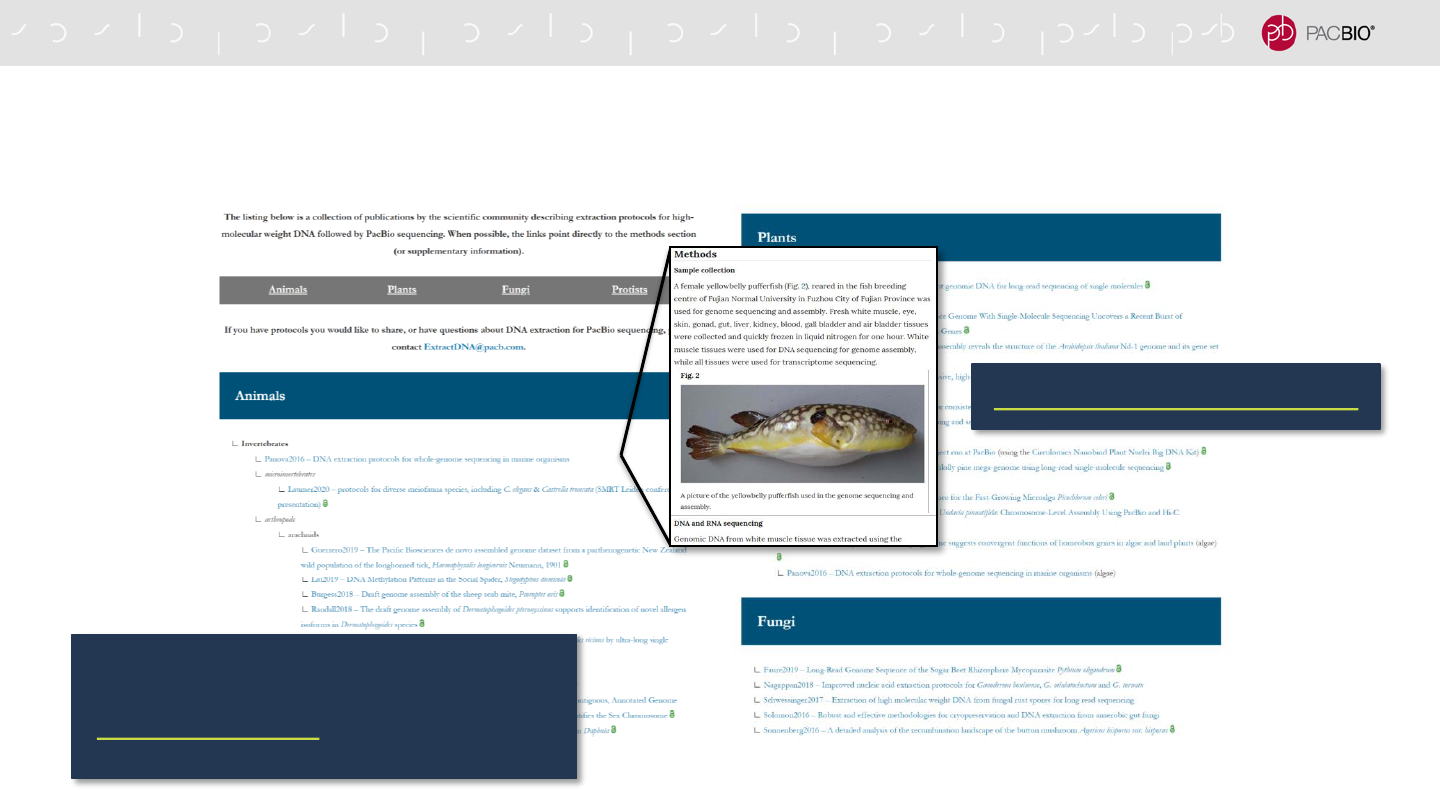
DNA SAMPLE PREPARATION ONLINE RESOURCE
Literature resource for sample collection and DNA extraction protocol references
www.ExtractDNAforPacBio.com
102
PacBio does not assume responsibilities/guarantees for
these external publications/protocols, but we are happy to
help as best as we can to guide / connect. Please contact
ExtractDNA@pacb.com for more discussions around
your particular species & sequencing project!

SEQUEL II AND IIe SYSTEM BEST PRACTICES OVERVIEW GUIDES
Variant Detection Using Whole Genome Sequencing with HiFi Reads
Whole Genome Sequencing for De novo Assembly
RNA Sequencing / Single-Cell RNA Sequencing
16S / Metagenomics Shotgun Sequencing of Complex Populations
https://www.pacb.com/smrt-science/smrt-resources/pacbio-literature/
No-Amp Targeted Sequencing
103

10
4
BEST PRACTICES: WHOLE GENOME SEQUENCING FOR DE NOVO
ASSEMBLY
SMRTbell Template Preparation
- Start with unamplified genomic DNA input (≥5 µg for a ~3-Gb sample genome size) from any sample type (blood, tissue, cell lines)
- Using SMRTbell Express Template Prep Kit 2.0, prepare libraries for HiFi sequencing of up to 16 samples at a time with manual prep, or 96 samples
using an automation-friendly workflow
- Enrich for ~15 kb - 20 kb inserts with size selection.
Sequence on the Sequel II or IIe Systems
- With highly accurate long reads (HiFi reads) from the Sequel II or IIe Systems, you can assemble up to a 2 Gb genome in a single SMRT Cell 8M for
~$1,300* or scale up for larger genomes
- Run up to 200 samples (2 Gb) per year, per Sequel II or IIe System
- Sequence to desired coverage depth based on the complexity of the genome sample:
- Recommend aiming for 10- to 15-fold HiFi read coverage per haplotype for phased de novo assembly
Data Analysis Solutions with the PacBio Analytical Portfolio
- Use SMRT Link Genome Assembly (powered by IPA), or open-source tools including HiCanu or hifiasm to assemble and phase the genome
- Example datasets are available at pacb.com/dataset
* Read lengths, reads/data per SMRT Cell 8M and other sequencing performance results vary based on sample quality/type and insert size.
†
Prices, listed in USD, are approximate and may vary by region. Pricing includes library and sequencing reagents run on a Sequel II or IIe System and does not include instrument
amortization or other reagents.

10
5
SMRTbell Template Preparation
- Start with unamplified genomic DNA input (≥5 µg for a ~3-Gb sample genome size) from any sample type (blood, tissue, cell lines)
- Using SMRTbell Express Template Prep Kit 2.0, prepare libraries for HiFi sequencing of up to 16 samples at a time with manual prep, or 96 samples
using an automation-friendly workflow
- Enrich for ~15 kb - 18 kb inserts with size selection. Inserts larger than this range may reduce read and variant calling accuracy
Sequence on the Sequel II or IIe Systems
- With highly accurate long reads (HiFi reads) from the Sequel II or IIe Systems you can comprehensively detect variants in 100s to 1000s of genomes
in a year
- Sequence to desired coverage based on study needs:*
- Aim for ≥15-fold HiFi read coverage of a Human genome for variant detection applications
- Recommend 2 SMRT Cells 8M to achieve ≥15-fold coverage of a human genome for comprehensive variant detection for $2600
†
Data Analysis Solutions with the PacBio Analytical Portfolio
- Detect all variant types – including SNVs, indels, SVs, and CNVs – with the highest precision and recall using SMRT Link Structural Variant Calling
analysis application (powered by pbsv) and Google DeepVariant (PacBio model)
- Use joint calling in pbsv and DeepVariant for multiple samples
- Expand variant calling into previously inaccessible regions of the genome, including repetitive regions and medically relevant genes that are difficult to
map
- Phase small variants into phase blocks using WhatsHap and Confirm variant calls visually with IGV and GenomeRibbon
BEST PRACTICES: VARIANT DETECTION USING WHOLE GENOME
SEQUENCING WITH HIFI READS
* Read lengths, reads/data per SMRT Cell 8M and other sequencing performance results vary based on sample quality/type and insert size.
†
Prices, listed in USD, are approximate and may vary by region. Pricing includes library and sequencing reagents run on a Sequel II or IIe System and does not include instrument
amortization or other reagents.

BEST PRACTICES: RNA SEQUENCING (ISO-SEQ ANALYSIS)
SMRTbell Template Preparation
- Prepare full-length cDNA from 300 ng of total RNA using the NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module kit
- Use the SMRTbell Express Template Prep Kit 2.0 to prepare libraries in one day
- Multiplex up to 12 samples with barcoding
Sequence on the Sequel II or IIe Systems
- Maximize output and turn-around-time with adjustable sequencing parameters
- Sequel IIe System: 24 hour movies with 2 hours pre-extension is recommended
- Use the Sequel IIe System to generate up to 4 million* full-length, non-concatemer (FLNC) reads per SMRT Cell 8M
- Scale throughput based on project needs – With a single SMRT Cell 8M you can:
- Characterize a whole transcriptome
- Multiplex multiple tissues for genome annotation
Data Analysis Solutions with the PacBio Analytical Portfolio
- Generate highly accurate long reads (HiFi reads), with single-molecule resolution using circular consensus sequencing (CCS) mode
- Use the Iso-Seq analysis in SMRT Link to output high-quality, full-length transcript FASTA sequences, with no assembly required, to characterize
transcripts and splice variants
- Run Iso-Seq analysis with or without a reference genome, and annotate the genome using community tools such as SQANTI2, TAMA, and
LoReAn
* Read lengths, reads/data per SMRT Cell 8M and other sequencing performance results vary based on sample quality/type and insert size.
106

BEST PRACTICES: SINGLE-CELL RNA SEQUENCING (SINGLE-CELL
ISO-SEQ ANALYSIS)
SMRTbell Template Preparation
- Enrich for single-cell cDNA using a single-cell sorting platform that generates full-length cDNA*
- Start library preparation with at least 160 ng of input cDNA (post-single-cell platform PCR reaction) for 1-2 SMRT Cells 8M
- Use the SMRTbell Express Template Prep Kit 2.0 to prepare libraries in one day
Sequence on the Sequel II or IIe Systems
- Use HiFi sequencing on the Sequel II or IIe Systems to generate 3 million full-length transcript reads from one SMRT Cell 8M to obtain ~1,000
unique molecules for 3,000 single cells**
- Use a 24-hour movie collection time with a 2-hour pre-extension time
- For human samples, run up to 240 SMRT Cell 8M/year at a cost of ~$1,300/SMRT Cell 8M, excluding single-cell
- enrichment cost
†
Data Analysis Solutions with the PacBio Analytical Portfolio
- Analyze HiFi reads which allow accurate single-cell barcode and UMI identification
- Use the single-cell Iso-Seq analysis tools on GitHub to output high-quality, full-length transcript FASTA sequences per UMI, with no assembly
required, to characterize transcript variants for each cell
* Number of usable reads, containing the UMI and cell barcode, vary by single-cell platform. Any platform that generates full-length cDNA is compatible with the single-cell RNA sequencing
workflow.
** Read lengths, reads/data per SMRT Cell type and other sequencing performance results vary based on single-cell platform, sample quality/type and insert size.
† Prices, listed in USD, are approximate and may vary by region. Pricing includes library and sequencing reagents run on a Sequel II or IIe System and does not include instrument
amortization or other reagents.
107

BEST PRACTICES: NO-AMP TARGETED SEQUENCING
SMRTbell Template Preparation
- Start with high-quality genomic DNA (~1-20 μg / SMRT Cell)
- Prepare SMRTbell libraries in 2-days with stream-lined protocol
- Block 5’ & 3’ ends to prevent off-target ligation
- Use custom-design guide RNAs to enrich for target regions of interest
- Multiplex up to 48 samples SMRT Cell 8M using barcoded adapters to analyze 5 or more targeted regions per sample
Sequence on the Sequel II or IIe Systems
- Sequence multiplexed targets and/or samples on the Sequel II or IIe Systems using the latest chemistry*
- Run up to 48 samples per SMRT Cell 8M at ~$82-118/sample on the Sequel IIe System
†
Data Analysis Solutions with the PacBio Analytical Portfolio
- Use command line tools to perform de-multiplexing and circular consensus sequencing (CCS) analysis to generate highly accurate long reads
(HiFi reads)
- Output data in FASTQ format for results summary reporting on repeat counts and on-target rates
- Visualize results with IGV and command-line scripts for easy review of repeat count of both alleles, mosaic characterization, identification of
interruption sequences and CRISPR / Cas9 off-targets
* Read lengths, reads/data per SMRT Cell 8M and other sequencing performance results vary based on sample quality/type and insert size.
†
Prices, listed in USD, are approximate and may vary by region. Pricing includes library and sequencing reagents run on a Sequel II or IIe System and does not include instrument
amortization or other reagents.
108

BEST PRACTICES: MICROBIAL WHOLE GENOME SEQUENCING
SMRTbell Template Preparation
- Start with 1.0 µg of high-quality input gDNA per microbial sample and construct a 10 – 15 kb library using SMRTbell Express TPK 2.0
- Reduce costs by multiplexing samples to assemble most bacterial genomes into 5 contigs or fewer, exclusive of plasmids
- Simplify equimolar pooling with Microbial Multiplexing Calculator
- Adjust multiplexing depth to balance cost per genome with genome completeness
- Note: Closure of class III complexity genomes with large repeat regions may require 20–30 kb library preparations & size-selection and may not be
suitable for multiplexing
Sequence on the Sequel II or IIe Systems
- Maximize output and turn-around-time with adjustable sequencing parameters*
- Multiplex up to 48 isolates per SMRT Cell 8M for $70/sample
†
with a 15 hour collection time
- Recommend generating ≥30-fold unique molecular coverage (UMC) per microbial genome
Data Analysis Solutions with the PacBio Analytical Portfolio
- Use SMRT Link for fully automated demultiplexing, assembly, circularization, and polishing of both chromosomes and plasmids to produce gold
standard references
- Achieve high-quality consensus accuracies >99.999%
- Detect and annotate active m6A and m4C Restriction-Modification system motifs with the ‘Base Modification and Motif Analysis’ application in SMRT
Analysis [m4C (≥25-fold coverage per strand) / m6A (≥25-fold coverage per strand)]
* Read lengths, reads/data per SMRT Cell 8M and other sequencing performance results vary based on sample quality/type and insert size.
†
Prices, listed in USD, are approximate and may vary by region. Pricing includes library and sequencing reagents run on a Sequel II or IIe System and does not include instrument
amortization or other reagents.
109

BEST PRACTICES: 16S AMPLICON SEQUENCING
SMRTbell Template Preparation
- Perform library construction with SMRTbell Express TPK 2.0
- PacBio full-length 16S protocol available with recommended barcoded 16S primer sequences; OR use third-party Shoreline Biome kits for DNA
extraction and PCR amplification
- Multiplex up to 192 16S samples per SMRT Cell 8M
Sequence on the Sequel II or IIe Systems
- Produce HiFi reads with circular consensus sequencing (CCS)
- Generate up to 2 Million 16S HiFi (Q30) reads per Sequel SMRT Cell 8M* using a 10-hour movie collection time
- Recommend generating ≥8000 HiFi reads per multiplexed 16S sample for analysis
Data Analysis Solutions with the PacBio Analytical Portfolio
- De-multiplex barcodes within SMRT Link GUI or on the command line
- Output data in standard file formats, (BAM and FASTA/Q) for seamless integration with downstream analysis tools
- Analyze 16S HiFi data using third-party analysis tools like Shoreline Biome SB Analyzer or DADA2
* Read lengths, number of reads, data per SMRT Cell 8M, and other sequencing performance results vary based on sample quality/type and insert size, among other factors
110

BEST PRACTICES: METAGENOMICS SHOTGUN SEQUENCING
SMRTbell Template Preparation
- Start with recommended amount of input gDNA per sample (1.5 μg) and construct a 10 kb library with SMRTbell Express TPK 2.0
- The size distribution of the starting genomic DNA is critical for shearing and PacBio recommends working with samples where the majority of the
input gDNA is greater than 15 kb whenever possible.
Sequence on the Sequel II or IIe Systems
- Produce HiFi (≥Q20) reads with circular consensus sequencing (CCS)
- Generate up to 2.4 Million 16S HiFi (Q20) reads per Sequel SMRT Cell 8M* using a 30-hour movie collection time
- Target coverage recommendations:
- 5-fold coverage (~3000 HiFi reads) of least abundant species for profiling intact genes and operons
- 20-fold coverage (~12,000 HiFi reads) for near-complete genome assemblies
Data Analysis Solutions with the PacBio Analytical Portfolio
- De-multiplex barcodes within SMRT Link GUI or on the command line
- Analyze metagenomic shotgun HiFi reads using third-party analysis tools
- Perform taxonomic classification and functional gene profiling using QIIME and MEGAN
- Perform gene prediction and discovery using FragGeneScan and Prodigal
- Perform metagenomic shotgun assembly directly with HiFi reads using Canu
- Bin contigs and plasmids originating from the same strain by leveraging epigenetic signatures
* Read lengths, number of reads, data per SMRT Cell 8M, and other sequencing performance results vary based on sample quality/type and insert size, among other factors
111

For Research Use Only. Not for use in diagnostic procedures. © Copyright 2021 by Pacific Biosciences of California, Inc. All rights reserved. Pacific Biosciences, the Pacific Biosciences logo,
PacBio, SMRT, SMRTbell, Iso-Seq, and Sequel are trademarks of Pacific Biosciences. Pacific Biosciences does not sell a kit for carrying out the overall No-Amp Targeted Sequencing method.
Use of these No-Amp methods may require rights to third-party owned intellectual property. FEMTO Pulse and Fragment Analyzer are trademarks of Agilent Technologies Inc.
All other trademarks are the sole property of their respective owners.
www.pacb.com