
NICHD Office of Data Science and Sharing (ODSS) Page 1 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
Office of Data Science and Sharing (ODSS)
PRIVACY PRESERVING RECORD LINKAGE (PPRL) FOR
PEDIATRIC COVID-19 STUDIES
FINAL REPORT
SEPTEMBER 2022
Funded by NIH Office of Data Science Strategy
Prepared for NICHD Office of Data Science and Sharing (ODSS) by Booz Allen Hamilton under contract
number GS-35F-386DA
NICHD Office of Data Science and Sharing (ODSS) Page 2 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Table of Contents
1 EXECUTIVE SUMMARY ..................................................................................................................... 4
2 INTRODUCTION ............................................................................................................................... 7
3 INTRODUCTION TO NIH PEDIATRIC COVID STUDIES & THE CASE FOR RECORD LINKAGE ................... 7
4 PROJECT GOAL, OBJECTIVES, AND APPROACH ............................................................................... 13
5 PEDIATRIC COVID STUDIES—PPRL FEASIBILITY ............................................................................... 14
5.1 Studies Selected for the Project .......................................................................................... 14
5.2 Define Questions for PPRL Feasibility .................................................................................. 15
5.3 Summarize Findings ............................................................................................................ 16
6 GOVERNANCE ASSESSMENT & FINDINGS ....................................................................................... 16
6.1 Define Criteria for Governance Assessment ........................................................................ 17
6.2 Select & Research Record Linkage Implementations ........................................................... 19
6.3 Analyze & Summarize Findings ........................................................................................... 21
6.3.1 Types of Data Linked .............................................................................................. 21
6.3.2 Authorization for Linking Data and Sharing Linked Data ......................................... 22
6.3.3 PII Elements Used in PPRL Implementations ........................................................... 24
6.3.4 Two-Party or Three-Party Model for Entity Resolution and Data Linkage ................ 25
6.3.5 Data Linkage Model: Linked Database or Study-Specific Linkage ............................. 26
6.3.6 Controls for Managing Re-Identification Risk with Linked Data ............................... 28
6.3.7 Authorizations and Controls for Accessing the Linked Data..................................... 31
7 TECHNOLOGY ASSESSMENT ........................................................................................................... 32
7.1 Define Criteria for Technology Assessment ......................................................................... 33
7.2 Select & Research Candidate PPRL Technologies ................................................................. 33
7.3 Summarize Findings ............................................................................................................ 34
7.3.1 Hash Generation and Record Linkage ..................................................................... 34
7.3.2 Operating Environment and Licensing Model ......................................................... 35
7.3.3 Usability and Security Features .............................................................................. 35
7.3.4 External System Integration ................................................................................... 35
7.3.5 Data Cleaning/Pre-Processing Features .................................................................. 35
7.3.6 Performance and Scalability ................................................................................... 36
7.3.7 Informational Questions......................................................................................... 36
8 CONSIDERATIONS .......................................................................................................................... 36
8.1 Key Considerations Based on Governance and Technology Assessment .............................. 37
8.1.1 Key consideration 1: Authorization for linking and sharing linked data should
be based on informed consent or approval from the data originator’s
institution and/or their IRB or an equivalent Privacy Board .................................... 37
8.1.2 Key consideration 2: Linkage of certain types of data or data from certain
populations may be subject to additional policies or governance ........................... 42
8.1.3 Key consideration 3: A broad set of PII elements are required to generate high
quality linkage regardless of the tool used, and these PII elements should be
collected early and in a standardized manner ......................................................... 43
NICHD Office of Data Science and Sharing (ODSS) Page 3 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
8.1.4 Key consideration 4: The three-party linkage approach offers researchers the
flexibility to link and use datasets hosted in different data systems ........................ 45
8.1.5 Key consideration 5: The linked database model encompasses a broad scope
of datasets and should be paired with additional controls to protect
participant privacy ................................................................................................. 46
8.1.6 Key consideration 6: Re-identification risk management controls can be
implemented both prior to and after linkage .......................................................... 47
8.1.7 Key consideration 7: All PPRL tools assessed for this Project meet a basic set of
capability requirements, but vary on certain desirable features ............................. 48
8.1.8 Key consideration 8: Certain PPRL tool features better serve robust
implementation approaches and sustainability ...................................................... 50
8.2 Considerations for CARING for Children with COVID ........................................................... 51
8.3 Limitations of this Assessment & Future Directions ............................................................. 54
9 GLOSSARY ...................................................................................................................................... 56
10 ACRONYMS .................................................................................................................................... 62
11 APPENDIX ...................................................................................................................................... 67
11.1 CARING for Children with COVID Studies – Supplemental Information ................................ 67
11.2 Governance Assessment Supplemental Information ........................................................... 74
11.2.1 Governance Summary For Record Linkage Implementations Using PPRL ................ 74
11.2.2 Governance Summary for Record Linkage Implementations Not Using PPRL .......... 97
11.2.3 Example Data Flow Schematic for Record Linkage Implementations ..................... 114
11.2.4 Consent Language for the Record Linkage Implementations Assessed in the
Project ................................................................................................................. 116
11.2.5 Consent Language for the Record Linkage Examples Not Used in the Project........ 121
12 REFERENCES ................................................................................................................................ 124

NICHD Office of Data Science and Sharing (ODSS) Page 4 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
1 EXECUTIVE SUMMARY
The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Office of
Data Science and Sharing (ODSS) undertook a project to assess potential governance and technical
approaches for implementing privacy preserving record linkage (PPRL) across pediatric COVID-19 (COVID
hereafter) studies, with funding from the National Institutes of Health (NIH) Office of Data Science
Strategy (ODSS) and support from Booz Allen Hamilton. The overall goal of the project is to inform an
NIH-wide strategy on the use of PPRL for pediatric COVID studies, based on use cases from the
Collaboration to Assess Risk and Identify LoNG-term outcomes for Children with COVID
, known as
CARING for Children with COVID—an initiative led by NICHD and the National Heart, Lung, and Blood
Institute (NHLBI) in collaboration with the National Institute of Allergy and Infectious Diseases (NIAID).
CARING for Children with COVID is aimed at better understanding SARS-CoV-2 infection in children,
including the multisystem inflammatory syndrome in children (MIS-C), a rare but serious multi-organ
disease.
NIH rapidly mobilized the CARING for Children with COVID initiative by funding new and existing studies
on pharmaceuticals for treatment, cardiovascular complications, immunologic pathway characterization,
and the underlying risk factors that influence the full spectrum of symptomology in children infected
with SARS-CoV-2. To align with the call for rapid data sharing in the January 2021 Executive Order on
Ensuring a Data Driven Response to COVID-19
1
, CARING for Children with COVID leveraged existing and
new NIH data repositories to support data sharing with a broad community of researchers. Study
investigators soon recognized that pediatric COVID patients were likely participating in multiple CARING
for Children with COVID studies and saw value in linking the various data types for each child, yet they
could not link participant data across studies because they were unable to share information about
participant identities with one another. PPRL was identified as the most feasible approach to identifying
which children are participating across CARING for Children with COVID studies.
PPRL is specifically designed to facilitate the linking of records associated with an individual represented
across multiple datasets without exposing any personally identifiable information (PII). PPRL software
uses cryptographic algorithms to generate irreversible, hashed codes (or “tokens”) when PII, such as
name and date of birth (DOB), are entered into the software. The hashed codes can then be compared
across multiple datasets to match records from the same individual. In order to implement PPRL across
multiple data repositories, rules (governance) must be defined including decisions regarding how the
linkage is authorized, which datasets can be linked, which organization is trusted to create the linkage
information, who can access linked datasets and how, and how reidentification risk can be mitigated as
increasingly diverse data types are aggregated on a single research participant. Good governance is
critical to protect research participant privacy and respect participant trust.
The overall goal of this project was to assess and analyze governance and technology approaches in
diverse, existing record linkage implementations, to inform an NIH-wide approach to link data across
pediatric COVID studies, and, more broadly, to inform approaches to linking individual-level datasets
across pediatric research studies. The project achieved this goal through the following activities:
• Summarized information associated with PPRL feasibility for the CARING for Children with
COVID studies
• Analyzed 13 existing record linkage implementations, both PPRL and non-PPRL, funded by NIH,
other federal agencies, and non-government organizations, to fully document end-to-end
governance decisions
NICHD Office of Data Science and Sharing (ODSS) Page 5 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
• Evaluated the capabilities of seven PPRL vendors/organizations, including one NIH-developed
tool, one university-developed tool, and five commercial vendor tools, by extending a recent
technical assessment led by the National Cancer Institute (NCI) and adding facets specific to the
pediatric COVID record linkage use cases
The project analyzed findings across these three activities to develop governance and technology
considerations for a CARING for Children with COVID PPRL implementation, which could serve as a
useful guidepost for the design of any new PPRL implementation. The project concluded that PPRL is a
feasible approach for linking participant data across pediatric COVID studies so long as the involved
parties collaborate prior to implementation to define the governance approaches, technical
requirements, and the data elements required to ensure high-quality linkage.
Prior to implementing PPRL, funders, investigators, researchers, and data repositories should
collaborate to make the following determinations:
• Obtain approval or authorization to link: Studies should consent research participants for the
linkage of their data across studies and data repositories, if feasible, by clearly communicating
the scope of the linkage and how the linked data will be shared. Since CARING for Children with
COVID studies are subject to the NIH Genomic Data Sharing (GDS) Policy, it is appropriate to
seek institutional approval for linkage, with input from an institutional review board (IRB) and/or
equivalent Privacy Board, especially when re-consent is not feasible. It may be possible to link to
data from typically unconsented sources, such as administrative datasets, if explicit consent for
linkage and sharing the linked data is obtained in the context of the research studies (thereby
changing the status of the administrative data to “consented”).
• Identify policies relevant to specific data types or participant populations: Policies or
procedures may apply to certain data types (e.g., genomic data that are subject to the NIH GDS
Policy) or populations (e.g., tribal or international populations).
• Collect and standardize a broad set of PII elements: A broad set of PII elements are required to
generate high-quality linkage, regardless of the PPRL technology used. These PII elements
should be collected at the outset and in a standardized manner. Since certain PII elements are
not typically collected in pediatric research, most, if not all, CARING for Children with COVID
studies would need to collect new PII elements from their study participants.
• Establish which party will link the data: A three-party approach, where the PII is entered into
the PPRL tool by the data originators, the hashed codes are matched by an honest broker or
external server, and approved researchers link the data, offers researchers the flexibility to link
and use datasets that are hosted in different data repositories. By separating the party that
matches the hashed codes from the party that links the data, data use requirements and data
provenance information are retained in all datasets.
• Determine the scope of linkage: All PPRL implementations should make up-front
determinations regarding which datasets would be linked and whether the linkage would be
specific to one study (study-specific) or would encompass multiple datasets from one or
multiple repositories (linked database model), thereby supporting many studies. The linked
database model is the most sustainable and reasonable approach for fostering reproducible
research with CARING for Children with COVID data, as it could encompass multiple current and
future NIH pediatric COVID datasets across multiple repositories, so long as the same PPRL
technology is used.
NICHD Office of Data Science and Sharing (ODSS) Page 6 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
• Use a variety of controls for mitigating re-identifiability risk: For CARING for Children with
COVID, linkage information should be provisioned using access controls (approval from an NIH
data access committee), while the original access tier status of unlinked datasets need not
change. Additional policy controls include using a standard definition of “de-identified” (e.g., the
NIH GDS Policy, which uses the Health Insurance Portability and Accountability Act [HIPAA] Safe
Harbor and the Common Rule) and prohibiting re-identification by users. For certain
implementations, re-identification risk assessments prior to and/or after linkage or applying
modifications to certain data elements could also be considered.
• Select PPRL software that meets basic requirements: To support CARING for Children with
COVID, the selected PPRL tool must accommodate a broad and flexible set of PII, support large
scale implementations, prohibit vendor rights to the data, and appropriately protect PII. This
project determined that nearly all PPRL tools assessed in this report can support these basic
requirements. The PPRL tools diverge on certain desirable features associated with usability,
functionality, and security, which may factor into deciding which software is best for a given
implementation.
• Consider PPRL software sustainability for long-term implementations: Long-term
implementations require that the hashed codes persist over time and may benefit from the use
of NIH-owned software to avoid continual commercial vendor contracts, recurring or use-based
costs, and risk associated with business model modifications.
This assessment represents a snapshot of the landscape of record linkage to support biomedical
research, and additional work is required to assess linkage quality for a given PPRL tool, PII elements,
and the configuration of matching algorithms that would be used for a PPRL implementation, by testing
against a gold standard dataset that is appropriate for the implementation (e.g., pediatric data). Further
investigation is warranted regarding participants’ attitudes towards consent for linkage, as well as actual
PPRL software vendor costs, cross-vendor interoperability capability, and challenges associated with
vendor dependency for long-term implementations.
NICHD Office of Data Science and Sharing (ODSS) Page 7 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
2 INTRODUCTION
The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Office of
Data Science and Sharing (ODSS) undertook a project to assess and analyze approaches for privacy
preserving record linkage (PPRL) to meet the needs of pediatric COVID studies, with funding from the
National Institutes of Health (NIH) Office of Data Science Strategy (ODSS) and support from Booz Allen
Hamilton. The overall goal of the project is to inform an NIH-wide strategy on the use of PPRL for
pediatric COVID studies, which could serve as a useful guidepost for the design of any new PPRL
implementation.
This public NICHD ODSS report provides considerations regarding the use of PPRL for the pediatric
COVID studies based on findings from the following:
• Summary of information associated with PPRL feasibility for pediatric COVID studies that are
part of the Collaboration to Assess Risk and Identify LoNG-term outcomes for Children with
COVID
• Analysis of governance frameworks for existing record linkage efforts implemented across NIH,
other federal agencies, and non-government organizations
• Capability analysis and potential applicability of various PPRL vendors/organizations for pediatric
studies
The intended audience of this public report is any stakeholder considering participating in or
implementing PPRL to address research-based use cases.
3 INTRODUCTION TO NIH PEDIATRIC COVID STUDIES & THE CASE FOR
RECORD LINKAGE
COVID in children
As evidenced by the COVID global pandemic, the novel SARS-CoV-2 virus can cause a broad spectrum of
mild to severe disease, including death. Infection from this virus can also result in a rare but serious
post-infectious hyperinflammatory condition affecting multiple organs called the multisystem
inflammatory syndrome (MIS) in both children (MIS-C) and adults (MIS-A). Some features of MIS-C
overlap with Kawasaki disease (KD), macrophage activation syndrome (MAS), and toxic shock syndrome
(TSS)
2
—diseases that predate COVID.
The Collaboration to Assess Risk and Identify LoNG-term outcomes for Children with COVID, known as
CARING for Childre
n with COVID – an initiative led by NICHD and the National Heart, Lung, and Blood
Institute (NHLBI) in collaboration with the National Institute of Allergy and Infectious Diseases (NIAID) –
was launched to better understand SARS-CoV-2 infection in children and includes the following studies:
• Pharmacokinetics,
Pharmacodynamics, and Safety Profile of Understudied Drugs Administered
to Children per Standard of Care (POP02) focuses on understanding the treatment of children
diagnosed with COVID or MIS-C with medicines that have shown promise in adults with COVID.
• Long-Term Outcomes after th
e MUltisystem Inflammatory Syndrome In Children (MUSIC)
focuses on cardio
vascular complications of MIS-C, but also collects data on all aspects of
childhood and adolescent health in affected participants.
• Pediatric Research
Immune Network on SARS-CoV-2 and MIS-C (PRISM) aims to evaluate the
short- and lon
g-term health outcomes of SARS-CoV-2 infection in children, including MIS-C, and

NICHD Office of Data Science and Sharing (ODSS) Page 8 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
to characterize the immunologic pathways associated with different disease presentations and
outcomes.
• Eight studies that are part of the Predicting Viral-Associated Inflammatory Disease Severity in
Children with Laboratory Diagnostics and Artificial Intelligence (PreVAIL kIds
), which are part of
the NIH Rapid Accel
eration of Diagnostics Radical (RADx-rad) initiative, focus on developing
cutting-edge approaches for understanding the underlying factors that influence the spectrum
of possible conditions in children infected with SARS-CoV-2.
CARING for Children with
COVID aims to answer the following questions:
• Why are some children more likely than others to get infected with SARS-CoV-2?
• Why do different children show different symptoms of COVID?
• Why do some children who become infected with SARS-CoV-2 have more severe illness, like
MIS-C?
• What are the long-term outcomes for children who have become infected with SARS-CoV-2?
To help address these and other questions and to help develop public health strategies for prevention,
diagnosis, and therapies, NIH is funding additional pediatric COVID studies across the U.S. and other
countries as well. Including but not limited to CARING for Children with COVID, NIH funds approximately
2,966 active projects related to COVID and children, with 449 projects focused on MIS-C
3
. Data collected
from these studies, including electronic health records (EHRs), pathology, laboratory, imaging, and
genomic data, constitute a rich source of information that can be used to develop broad strategies to
address the above questions. Given that MIS-C is a multi-organ disease, addressing these questions will
require a multidisciplinary approach that involves pediatrics, genomics, immunology, cardiology,
hematology, and other disciplines. This interdisciplinary approach towards understanding and treating
MIS-C can best be served if the different types of datasets and records collected for a given patient—for
example, EHRs collected at the various hospital systems, and -omics data generated by the various
COVID studies funded by NIH—can be linked and integrated. Such linkage maximizes NIH’s investments
in research and advances clinical and scientific discoveries not only for MIS-C but also for other related
pediatric conditions.
What is PPRL and how does it work?
Linking two or more records that correspond to the same individual (entity) is called record linkage, a
term introduced in 1946 by Dunn
4
of the United States National Bureau of Statistics: “Each person in the
world creates a Book of Life. This Book starts with birth and ends with death. Record linkage is the name
of the process of assembling the pages of this Book into a volume.” Linking records or datasets
generated from multiple sources and stored in disparate data repositories facilitates creation of
enriched datasets and/or longitudinal datasets for an individual that can then be used to address
multifaceted research questions that a single dataset alone may not be able to answer. Currently two
broad groups of methods exist for record linkage
a
:
• Traditional linkage uses information such as personally identifiable information (PII; for example,
date of birth, Social Security number [SSN], address, which the Health Insurance Portability and
Accountability Act
5
[HIPAA] classified as protected health information [PHI], if present in health
records), or other direct information such as genetics to match records of an individual.
a
Record linking and data linking are used interchangeably in this Final Report.
NICHD Office of Data Science and Sharing (ODSS) Page 9 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
• Privacy preserving record linkage
6
(PPRL) encodes the PII to create one-way hashed codes
(tokens), which are then encrypted and compared so that the resulting matches can be used to
link data or records of an individual.
While both of the above methods use PII as the starting point, the traditional method exposes PII to the
party charged with identifying matches, whereas in PPRL, the PII remains with the data originator and is
not exposed to the party charged with matching the hashed codes/tokens (entity resolver) or linking the
data (record linker).
Regardless of the method used for linking, two types of algorithms
7
are used: deterministic (exact) and
probabilistic (approximate) matching.
• In the deterministic model, all PII elements must match exactly for the record to be considered
to be belonging to the same individual. The model typically uses unique identifiers such as SSN
or medical record number for matching and requires the PII elements in the records to be error-
free, which is a challenge in real-world data.
• In the probabilistic model, the matches are based on the discriminatory power of the PII
elements that are used and the degree of similarity between the elements, resulting in a
likelihood ratio of the entities being a match, non-match, or possible match. This model
tolerates errors and other quality issues often found in real-world data.
These two algorithmic models are routinely and widely used especially when linking administrative,
survey, mortality, health, economic, social, and other types of data collected by various government
agencies such as the Census Bureau
8
, Centers for Disease Control (CDC)/National Center for Health
Statistics (NCHS)
9
, Agency for Healthcare Research and Quality (AHRQ)
10
, and Administration for
Children and Families (ACF)
11
. These agencies typically use the traditional method of linking, which
exposes PII elements to the entity resolver, who typically uses PII elements to create a linked dataset.
However, there are key constraints to utilizing the traditional method for linking patient or research
study participant data, such as EHRs, clinical, non-clinical survey, genomic, image, and viral sequence
data, collected by NIH researchers and stored in a federated data ecosystem:
• The PII elements from each data originator would need to be exposed or shared downstream,
which could present non-compliance with various human subject protection and data privacy
regulations such as the Federal Policy for the Protection of Human Subjects (known as the
Common Rule)
12
, HIPAA, and Privacy Act
13
.
• Appropriate informed consents must be obtained from the study participants if the PII elements
are to be shared beyond the data collector/originator. Participants are generally opposed to the
idea of sharing their PII
14, 15, 16, 17
and usually require assurances that only de-identified data
would be shared externally before agreeing to participate in a study.
• Records/data stored in the NIH data repositories follow a variety of de-identification standards,
such as HIPAA Safe Harbor (de-identified of all 18 HIPAA identifiers), limited dataset (with 16 of
the direct identifiers removed) as per HIPAA guidance
18
, the Common Rule, and/or other
determinations.
PPRL addresses several of these constraints and is specifically designed to facilitate the linking of records
of an individual across multiple data originators without transferring any personal identifiers from the
originating data source system. PPRL always involves at least two parties, the data originator and the
entity resolver. In a “2-party data linkage model,” the entity resolver is also the party that links the data
NICHD Office of Data Science and Sharing (ODSS) Page 10 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
(records) for a given participant (entity) once that participant has been identified in multiple datasets. In
a "3-party data linkage model,” these two functions are performed by different parties. In the 3-party
model, the entity resolver is often referred to as the honest broker as they handle only hashed
codes/tokens (i.e., encoded PII) and are not exposed to any participant-level data.
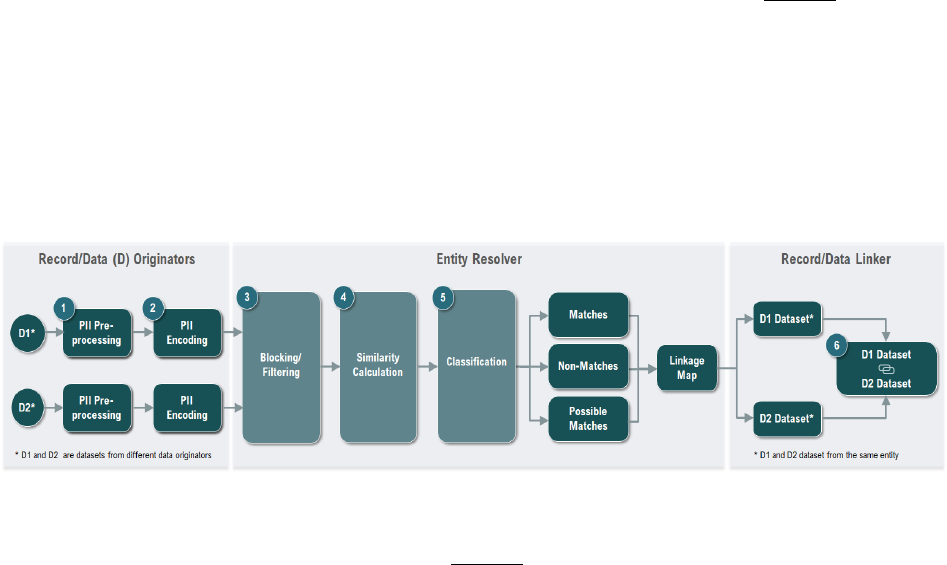
An overview of the PPRL process using the 3-party data linkage model is illustrated in Figure 1
. The data
originato
r uses the PPRL software to process the PII elements and encode them to generate the hashed
codes/tokens—this is the privacy preserving step. The data originator, via the software, then encrypts
the tokens for transfer to the entity resolver, who then performs the matching of the tokens using
deterministic and/or probabilistic algorithms, identifies the tokens that represent same
participant/entity, and documents the matches using the participant IDs provided by the data
originators and generates a linkage map. The matched IDs in the linkage map are then used to create a
linked dataset by the data linker.
Figure 1: PPRL Process across Two Datasets (D1 and D2) in a 3-Party Data Linkage Model
The major steps
in the PPRL process
19
as shown in Figure 1 include:
1. Data Pre-processing (of PII elements): An essential step for generating high-quality linkages, pre-
processing PII elements (sometimes referred to as data in this context) includes cleaning
(handling missing, erroneous, incomplete values) and standardizing to ensure that the PII
elements used for PPRL are transformed into a well-defined and consistent format. Pre-
processing of PII elements (the input in the context of PPRL) is typically performed at the data
originating sites and must be done consistently across sites to achieve the best linkage results.
2. Data Encoding (Hashing): This is the privacy preserving step where the PII elements entered into
the software are cryptographically encoded (hashed) using a one-way hashing algorithm so that
the input of a set string of characters (generated from PII elements) for an individual will
consistently produce a unique and repeatable fixed size output (a deterministic step), known as
hashed codes or tokens. To further preserve privacy and mitigate the possibility of dictionary
attacks aimed at breaking the hashed output and re-identifying the individual, a random known
value known as salt or secret key, is often injected during the encoding process. The salt/secret
key
20
is concatenated with the PII input string prior to hashing and all data originators
participating in the linkage uses the same salt/secret key, which is typically provided by an
external party such as the entity resolver. This ensures that a brute attack to reverse the hash
codes or tokens will not reveal the PII elements or identify the individual. After the data
originators generate the hashed codes/tokens, they are sent to the entity resolver.
3. Blocking: Blocking or filtering is the first step performed by the entity resolver when they receive
the hashed codes/tokens. This process facilitates indexing the records prior to matching by
NICHD Office of Data Science and Sharing (ODSS) Page 11 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
grouping the encoded inputs based on certain PII attributes (e.g., initial of last name and ZIP
Code) to filter out, or reduce, the number of tokens that would need to be compared against
each other. Blocking is critical for scaling up the number of records that can be linked efficiently,
especially across large data sources and can significantly reduce the computational time
required for matching tokens/records
21
.
4. Similarity/Matching Calculation: This step involves comparing tokens either based on
deterministic/exact or probabilistic/approximate matching of hashed codes/tokens.
Deterministic/exact matching results in a similarity score of 1 (match) or 0 (null), whereas
probabilistic/approximate matching is represented by a numerical value ranging from 0 to 1.
Real world data often include errors and inconsistencies, and while data cleaning addresses it to
a certain extent, PPRL algorithms generally use probabilistic matching with specific thresholds
(based on which and the number of tokens that match) set to increase the likelihood of true
positives while minimizing false positives.
5. Classification: The output of the matching process results in classifying two or more entities into
matches, non-matches, and possible matches, based on the similarity scores. This step
represents the actual process of entity resolution. The entity resolver develops a linkage map of
matched participant IDs across data originating sites and may generate a new participant ID or a
globally unique identifier (GUID) based on the matches to facilitate the linking of records across
the data originating sites.
6. Record/Data Linking: The Data Linker uses the linkage map of matching participant IDs across
data originating sites to link the datasets for research use.
Challenges to implementing PPRL in the NIH federated data ecosystem
PPRL has not yet been adopted widely at NIH for linking records across pediatric COVID projects due to
various challenges associated with implementing it within NIH’s federated data sharing ecosystem—
these include:
• Data are distributed across data repositories: The urgent response that was required to address
the COVID pandemic resulted in capturing and storing data for the same participant either in
existing or newly established repositories that are primarily designed for sharing data with the
broader research community. Linking data for the same individual across multiple repositories
was not initially identified as a critical need and would have required more up-front
coordination during the initial phases of the pandemic, when the focus was primarily on data
collection and rapid analysis in line with international
22
and federal
1, 23
calls for rapid data
sharing.
• Data are shared without the required PII elements: In keeping with de-identification standards
and policies (e.g., the NIH Genomic Data Sharing Policy
24
), most, if not all, of the data
repositories within NIH’s data ecosystem hold de-identified data or limited datasets with many
of the PII elements required for PPRL stripped from the data. This makes it difficult, if not
impossible, to link data using information from the repositories alone. Instead, it requires
working with the data originators to collect the required PII elements and assist with PPRL
implementation—such retrofitting for PPRL is not always feasible, and when feasible, it is time
consuming and costly.
• Studies and data repositories follow a diversity of data governance models: Data governance
determines how data can be used at every step of the typical data lifecycle—collection,

NICHD Office of Data Science and Sharing (ODSS) Page 12 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
linking/merging, and sharing—and data governance models vary greatly between NIH studies
and repositories. Data governance models are dictated by regulatory requirements (such as the
Common Rule), the study participant’s informed consent, data submission and sharing policies
and requirements outlined in data submission/sharing agreements, data access tiers (such as
enclave, controlled or open
b
), and ethical considerations that may or may not be addressed by
existing technical or policy frameworks
25
. While such governance controls are critical for data
privacy and security and participant trust, it is not well understood how data governance models
in different data systems can intersect to enable data linkage across a federated data ecosystem
where a variety of technical and non-technical (e.g., policy, regulatory) controls are at play and
effective governance approaches are needed for future data linkages to be defined in advance
of data collection.
Nevertheless, the benefits and impact of using PPRL for health data (where privacy and confidentiality of
PII/PHI are paramount), have led to the development of open source, government-owned, and
commercial PPRL tools in use within a small number of NIH data repositories. One such PPRL tool is the
Global Unique Identifier
26
(GUID) Tool that was developed by the National Institute of Mental Health
(NIMH) to support the NIMH Data Archive. A second GUID tool developed by the Center for Information
Technology (CIT) is being used for a limited number of projects by other NIH Institutes and Centers (ICs),
including National Institute of Neurological Disorders and Stroke (NINDS), National Institute on Aging
(NIA), National Eye Institute (NEI), and National Center for Advancing Translational Sciences (NCATS).
The case for linking pediatric study data
As of August 2022, pediatric COVID cases accounted for approximately 18.4% of the 78,513,599 cases in
the U.S.
27
and among the 8,798 MIS-C cases, 71 children died due to MIS-C complications, as reported
by the CDC
28
. Understanding the full spectrum of symptomology and risk factors underlying COVID in
children is critical for the development or advancement of treatments, which require collecting and
merging data from multiple sources.
Linkage of pediatric studies was identified as a need within the first year of the CARING for Children with
COVID program when pediatric COVID and particularly MIS-C, was relatively rare (especially during the
early phases of the pandemic when schools were closed), and it was strongly suspected that the same
children were being enrolled across multiple studies. Since each study has a different focus
(pharmaceutical data versus immune profiling versus cardiac imaging), the investigators saw value in the
potential opportunity to link the various data types to the appropriate child; however, there was not a
readily available mechanism for facilitating such linkages (i.e., studies were not allowed to share PII with
each other).
Notwithstanding this and other challenges described above regarding linking data within a federated
data ecosystem, an initial assessment conducted by NICHD identified the need to facilitate subject-level
PPRL to support the following use cases across CARING for Children with COVID studies:
• Enable researchers to combine participant-level data collected from multiple studies to merge
multiple data types for each participant and avoid working with inflated sample sizes
• Avoid duplicate data generation (primarily whole genome sequencing)
b
Data access models – Open access: no access restrictions or registration required to access; Registration required: open to all, but users need
to be signed in or registered with the resource to access; Controlled access: application and eligibility requirements need to be met to gain
access (e.g., by a data access committee); Enclave: data cannot leave a specific system boundary (e.g., cannot be downloaded)
NICHD Office of Data Science and Sharing (ODSS) Page 13 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
• Facilitate longitudinal data collection and analysis (e.g., understanding long COVID in children)
Furthermore, participant-level linkages and the subsequent responsible sharing of the linked data is
expected to spawn new research studies and answer new questions long after the data collection has
ended, thus enabling the research community to derive maximal impact from the data to ultimately
benefit children’s health. However, pediatric COVID researchers have identified specific record linking
challenges that would need to be addressed, including:
• Misspellings of names or missing first or last names
• Missing certain commonly required elements such as city/municipality of birth or SSN
• The burden of supporting multiple PPRL tools at single sites
• Considerations for access and security procedures for both the data and the linkage information
in a federated ecosystem where data are shared through multiple repositories (some that
enclaves and others that are more open), and determining if new rules apply for access and use
of linked datasets through these repositories
This project was undertaken to identify technology and governance approaches based on existing record
linkage implementations that could address these challenges and form the basis of an appropriate PPRL
approach for CARING for Children with COVID and other select pediatric COVID projects at NIH.
4 PROJECT GOAL, OBJECTIVES, AND APPROACH
The overarching goal of this PPRL assessment project was to inform an NIH-wide strategy on the use of
PPRL for pediatric COVID studies. To effectively develop a strategy for implementing PPRL for pediatric
COVID studies in NIH’s federated ecosystem, the selection and establishment of two interrelated
components are required:
• Appropriate governance for linking records using PPRL: This includes considerations such as
what authorizations are required for creating linkages and sharing the linked data, whether the
appropriate PII elements required to generate the hash codes or tokens are available at the data
collection sites, who or what system will perform the entity resolution and data linkage, and
which controls should be implemented to mitigate the risk of potential re-identification from
data linkages.
• A tool or technology to implement PPRL: One PPRL tool must be selected to link across pediatric
COVID studies because hashed codes from different tools cannot be matched. The selection of
this tool will largely be determined by the tool’s capabilities, including its flexibility to use
various PII elements for hashing, data pre-processing capabilities, accuracy and other
performance measures of the tool, computational and other resource requirements, ability to
scale to accommodate increasing volumes of records, customization options, compliance with
government security regulations, and flexibility to support the governance needs described
above.
The project goal was achieved through the following three objectives and overall approach:
1. Summarized the current state of pediatric COVID studies selected for the project as related to
PPRL implementation: The Project Team documented critical information such as the PII
elements collected relevant consent language or other agreements potentially relevant to
implementing PPRL, and the interoperability status of the data repositories used to share data
from these studies.
NICHD Office of Data Science and Sharing (ODSS) Page 14 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
2. Develop considerations for key governance components necessary for enabling PPRL for the
selected pediatric COVID studies: The Project Team assessed existing record linkage governance
models and best practices by performing an in-depth environmental scan of existing record
linkage implementations and associated governance frameworks and interviewing relevant
stakeholders to collect additional information, validate accuracy of the information collected,
and fill in gaps.
3. Develop considerations for implementing potential PPRL tools for the selected pediatric COVID
studies. The Project Team assessed available record linkage vendors/organizations against the
needs of the pediatric COVID studies, building off a prior technology analysis performed by
NCI—the Landscape Analysis of Privacy Preserving Patient Record Linkage Software (P3RLS)—
Final Report Version 2 (2020)
29
.
5 PEDIATRIC COVID STUDIES—PPRL FEASIBILITY
5.1 Studies Selected for the Project
NICHD ODSS identified a total of 11 NIH-funded pediatric COVID studies that are part of the CARING for
Children with COVID
initiative to assess the feasibility of implementing PPRL for linking data within and
across these studies. An overview of the studies is shown in
Table 1.
These studies aim to address a broad set of clinical questions related to diagnosing and treating COVID
and MIS-C in children, including diagnostic methods for predicting disease severity, cardiological and
immunological response profiles, and potential drug efficacy and safety. These multifaceted studies are
expected to generate diverse types of data, including demographic, clinical, EHR, laboratory, genetic,
imaging, other biomarkers, social, economic, and other survey data. These data present a rich source of
information on an individual participant/patient and appropriate linkage of data across studies and data
repositories using PPRL would enable the broader research community to derive answers beyond the
initial set of questions posed by each of the respective primary studies.
Table 1: Pediatric COVID Studies Selected for the Project
POP02 MUSIC PRISM PreVAIL kIds
Full Study
Name
Pharmacokinetics,
Pharmacodynamics,
and Safety Profile of
Understudied Drugs
Administered to
Children Per Standard
of Care (POPS or
POP02)
Long-Term Outcomes
after the MUltisystem
Inflammatory
Syndrome In Children
(MUSIC)
COVID: Pediatric Research
Immune Network on SARS-
CoV-2 and MIS-C (PRISM)
Predicting Viral-Associated
Inflammatory Disease
Severity in Children with
Laboratory Diagnostics and
Artificial Intelligence
(PreVAIL kIds)
Study
Description
The study investigators
are interested in
learning more about
how drugs given to
children by their health
care provider act in the
bodies of children and
young adults in hopes to
find the most safe and
The COVID MUSIC
Study, funded by NIH
and the National
Heart, Lung, and
Blood Institute, is an
observational study
that aims to
understand
cardiovascular
The primary objectives of
this study are to determine:
o The proportion of children
with Severe Acute
Respiratory Syndrome
Coronavirus 2 (SARS-
CoV-2) related death,
rehospitalization, or major
complications after
The PreVAIL kIds initiative
funded eight studies that
will evaluate genes,
immune system proteins,
and other biomarkers, and
examine how the virus
interacts with the body and
how the immune system
responds to it. These
-

NICHD Office of Data Science and Sharing (ODSS) Page 15 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
POP02 MUSIC PRISM PreVAIL kIds
effective dose for
children. The primary
objective of this study is
to evaluate the
pharmacokinetics of
understudied drugs
currently being
administered to children
per standard of care
(SOC) as prescribed by
their treating provider
30
.
outcomes after MIS-C,
as well as other
outcomes in children
and adolescents
31
.
infection with SARS-CoV-
2 and/or Multisystem
Inflammatory Syndrome
in Children (MIS-C)
o Immunologic
mechanisms and immune
signatures associated
with disease spectrum
and subsequent clinical
course during the year of
follow-up
32
.
studies will rely on artificial
intelligence and machine
learning to interpret the
data they acquire, to
understand risk factors
underlying the severity of
COVID and MIS-C
33
.
Years 2020-2024 2020-2025 2020-2023 2020-2024
Funding
Agency
NICHD NHLBI NIAID NIH
Program/
Network
Pediatric Trials Network Pediatric Heart
Network
Pediatric Research Immune
Network
RADx-rad Program
Study Sites U.S.
42 study sites
U.S., Canada
32 study sites
U.S.,
20 study sites
U.S, U.K., Canada,
Colombia
58+ study sites
Data
Coordinating
Center
Name
The Emmes Company,
LLC
HealthCore Vanderbilt University Medical
Center
University of California San
Diego (UC San Diego) and
the University of Texas
Health Science Center at
Houston (UTHealth)
5.2 Define Questions for PPRL Feasibility
In consultation with NICHD ODSS, the Project Team defined a set of questions for documenting PPRL
feasibility for the CARING for Children with COVID studies, as shown in Table 2, given that these studies
are underway and could have certain constraints around data linking (e.g., based on consents, IRB
approved protocols, or other existing governance or logistical constraints).
Table 2: Analysis Questions for PPRL Readiness of CARING for Children with COVID Studies Selected for the
Project
PPRL Readiness Analysis Questions
1 Is there a broad set of PII elements collected by the study?
2 Does the consent obtained from study participants address the following?
o Linking data across study sites within the study and with other pediatric COVID studies
o Sharing the linked data
If not, are other agreements in place for approval to link and share linked data?
3 Are there any tribal or international data collected in the study? If so, are the agreements required for linking and
sharing data in place?
4 What technical and non-technical controls do the data repositories used for sharing study data have in place to
provide access to the linked data?
5 Can each study’s repository interoperate with other study repositories?
-
The project reviewed both public (study websites, funding announcements, clinicaltrials.gov, repository
websites) and internal study documentation (study protocols, data dictionaries, informed consent

NICHD Office of Data Science and Sharing (ODSS) Page 16 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
forms, and other study materials) as well as conducted stakeholder interviews with the Project Officers
(POs) and data coordinating centers (DCCs) for each of the studies and their repositories.
5.3 Summarize Findings
The project summarized the current state of CARING for Children with COVID studies as it relates to
feasibility for PPRL implementation using the questions listed in Table 2. Overall, the pediatric COVID
studies all collecte
d the PII elements first name, last name, date of birth, and sex while they rarely
collected email, address, and SSN. Most studies also collected ZIP Codes.
The Project Team also documented language from the informed consent forms (ICFs) relevant to linking
data and sharing linked data within a study and/or across multiple studies (see details of the language in
the consent forms in Appendix Table 1). Since many of these studies are multi-site stud
ies, the consent
forms have language for linkage across study sites within a study. However, only two of the 11 studies
include specific language regarding linkage of data across a broader network of studies that would
encompass all of the CARING for Children with COVID studies. The study consents mainly focused on the
broad sharing of de-identified data through NIH designated data repositories. Additionally, Institutional
Certifications
34,35
used by these studies (which certify that data submissions to the repositories are
appropriate) do not explicitly address record linkage and there are no additional agreements explicitly
addressing PPRL that have been established for any of the CARING for Children with COVID studies.
Some studies that incorporate data from international sites specifically address sharing these data with
organizations in the US in a manner consistent with the rest of the study data, while others use the same
study documents to communicate consistent data sharing expectations regardless of whether the data
are collected from non-tribal US, tribal, and international populations.
The data repositories for POP02, MUSIC, PRISM and PreVAIL kIds studies – the Kids First Data Resource,
BioData Catalyst, ImmPort, and the RADx Data Hub – are planned to be interoperable so that CARING for
Children with COVID data stored in one repository will be findable and accessible from any of the other
repositories. The data access permissions for these repositories range from registered tier to controlled
access. Genomic data, which has not yet been generated, will be shared through an NIH designated
controlled data repository such as the Kids First Data Resource or BioData Catalyst. To access genomic
data in these NIH controlled repositories, researchers must submit a Data Access Request and document
their eligibility requirements in the
NIH Database of Genotypes and Phenotypes Authorized Access
System
where requests are reviewed and approved by NIH Data Access Committees.
6 GOVERNANCE ASSESSMENT & FINDINGS
Governance as defined in this Report comprises of the policies, processes, and controls that address
ethics, privacy protections, compliance, risk management, or other requirements for a given record
linkage or PPRL implementation. PPRL governance is multifaceted—it involves the who (the people and
organizations), the what (the policies, processes, and controls), the when (at what stage in the data
lifecycle), and the how for implementation of processes and controls. Based on the recommendation
from NICHD ODSS, the scope of the Governance Assessment included both PPRL and record linkage
implementations not only in systems merging biomedical and healthcare data but also in other NICHD-
prioritized systems where PPRL or record linkage has been employed successfully. The rationale behind
expanding the scope to non-PPRL implementations and non-health data was to learn from the
experience of those who have been performing record linking for an extended period of time (for
NICHD Office of Data Science and Sharing (ODSS) Page 17 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
example, Census), to gather best practices, and to understand how they addressed various technical and
non-technical challenges.
Gaining a detailed understanding of the critical governance elements necessary to implement PPRL,
specifically within the NIH federated data ecosystem and with a pediatric focus, was fundamental to
developing PPRL approaches for the selected pediatric COVID studies. The Project Team’s overall
approach for the Governance Assessment is described in Section 4
. Briefly, team members defined
assessment crite
ria, researched a variety of publicly available web pages and documentation,
interviewed key stakeholders of 13 record linking and PPRL implementations, and, finally, analyzed and
summarized the information to inform the development of considerations for PPRL in pediatric COVID
studies. The sections below describe the various steps in more detail.
6.1 Define Criteria for Governance Assessment
The criteria examined the people, policies, processes, and controls along the data life-cycle continuum—
from data collection through linking to data access—for record linkage/PPRL implementations that are
currently operational and successful, paying particular attention to how linkage was operationalized in
pediatric use cases. The criteria and the rationale for selecting the criteria are shown in Table 3
. The key
criteria incl
uded authorizations and controls in place for linking, sharing, and accessing the linked data,
as well as the processes and methodologies in place to maintain participant privacy, data confidentiality,
security, and other data use requirements.
Table 3: Crite
ria for Record Linkage/Governance Assessment
Criteria Rationale for Selection
1 Sources and types of data used for linkage
o Where do the data originate and what types of data
are linked?
o Understand the breadth and scope of data linked,
specifically if pediatric data were linked
o Identify any limitations or constraints for data linking based
on the type or source of data—for example, genomic, tribal
data, international data, etc.
2 Authorizations for linking
o
Has the individual/participant given permission to link
their records/data?
o If not, who authorizes the linkage?
o Addresses whether the participant was aware of and agreed
to the linkage, and/or whether some other entity approved
the linkage
3 Record linking methodology
o What method was used for linking the records?
o If PPRL methodology, what tool or software was
used?
o Determine if different methods used for matching records
might dictate or limit how governance is implemented
o Identify PPRL tools that are used in the NIH ecosystem and
with healthcare and biomedical data
4 PII elements used for the linking
o Which PII elements were used, if any, for linking
the records?
o Examines the breadth of PII elements, if used, for record
linking, and identifies, if possible, the most common
combinations used for matching
Note: Although PII elements used for linking may be a
technical component of PPRL implementation, this criterion is
included in the governance assessment because policies and
regulations could impact which PII elements are collected
and/or used.
NICHD Office of Data Science and Sharing (ODSS) Page 18 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Criteria Rationale for Selection
5 Entity resolving party/organization/system
o Who or which organization/system does the
matching of the records?
o Identifies the methodology and controls in place to access
PII, hashed tokens based on PII, and/or other participant
information to resolve entities
o Examines whether there is physical separation between
data generators who have access to PII and entity resolving
processors
6 Data linking party/organization/system
o Who or which organization/system does the actual
matching of the records/data?
o Identifies methodology and controls in place to link
participant records/data
o Examines whether there is physical separation among data
generators who have access to PII, entity resolving
processors, and record linking parties
7 Data linkage model
What datasets are within the scope of the linkage?
o Linked database model, where the linkage
information created and/or provided encompasses
all datasets in a given database
o Study-specific model, where linkage information is
created and/or provided for the purposes of a
specific study
o Identifies the scope of datasets linked—i.e., datasets
associated with a database or datasets associated with a
specific study
8 Authorizations for sharing the linked data
o Has the individual/participant given permission to
share their linked data?
o If not, who authorizes the sharing of linked
records?
o Addresses whether the participant agreed to the sharing of
linked data, and/or whether some other party approved the
sharing of linked data
9 Re-identification Risk Management
o Is deductive disclosure review of the linked data
performed? If so, what is/are the process/criteria?
o If not, what other re-identification risk
management controls are in place?
o Examines whether and which controls are in place for
sharing the linked dataset, which may have a higher risk
profile compared to an individual dataset alone
10 Authorizations for accessing the linked data
o Who authorizes whether the linked records can be
accessed by researchers?
o Identify the controls in place for users to access the linked
data and allowable use of that data
11 Data access model
How are the data provisioned to the users? Data
access models identified include:
o Open access: no access restrictions or
registration required to access
o Registration required: open to all, but users need
to be signed in or registered with the resource to
access
o Controlled access: application and eligibility
requirements need to be met to gain access (e.g.,
by a data access committee)
o Enclave: data cannot leave a specific system
boundary (i.e., data cannot be downloaded locally)
o Identify specific conditions under which users access the
linked data
NICHD Office of Data Science and Sharing (ODSS) Page 19 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
6.2 Select & Research Record Linkage Implementations
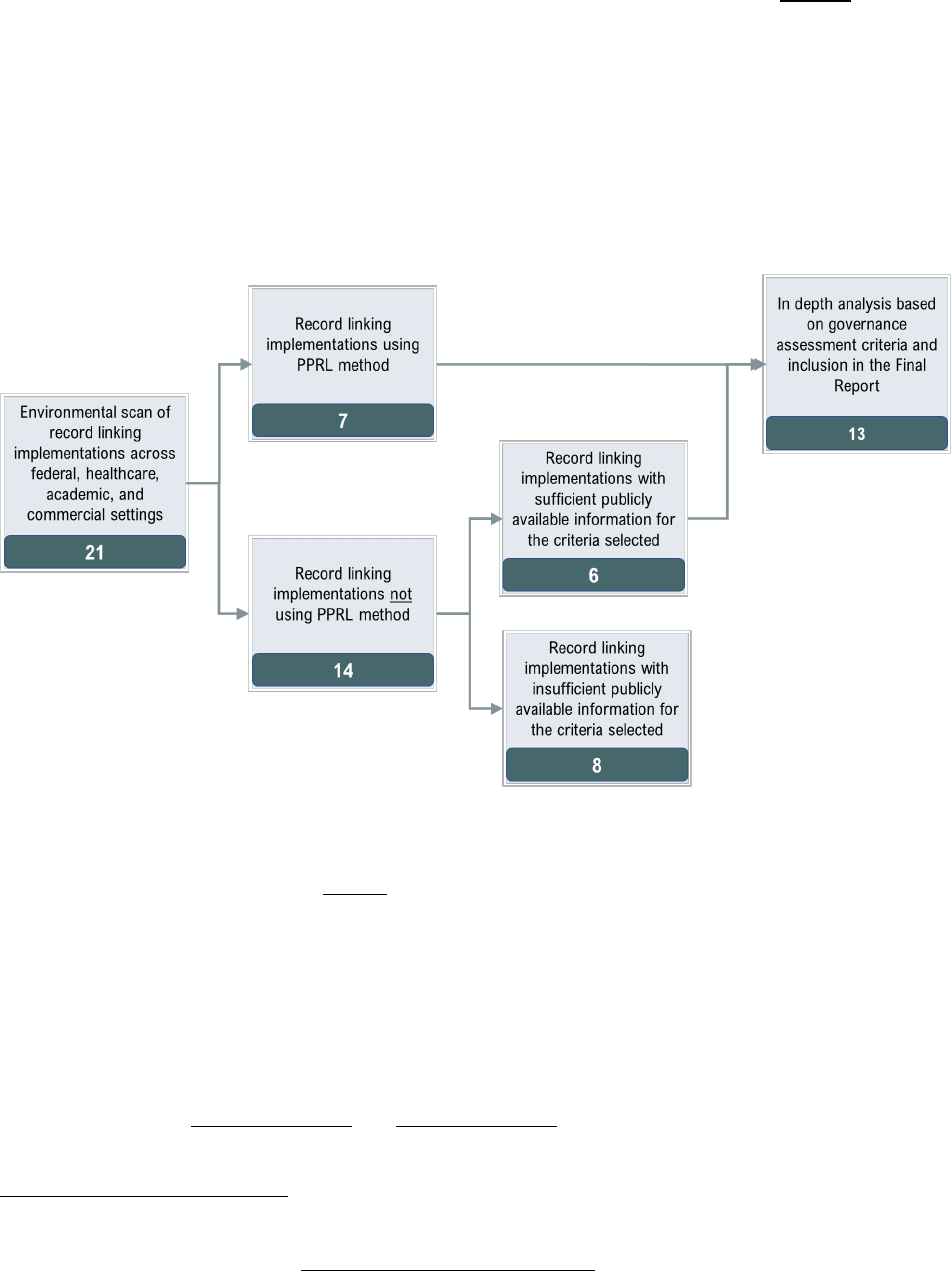
The overall process for selecting candidate record linkage implementations is shown in Figure 2. Briefly,
the Project Team conducted an environmental scan of various record linkage examples across federal,
academic, and commercial sectors using Google search and PubMed search to gather an initial list and
obtain an understanding of the breadth of record linkage implementations. The Project Team then
conducted more comprehensive research of the resulting 21 implementations to gather available
information from public sources, including web pages, documentation, research articles, publications,
and white papers, and from NICHD ODSS Team based on their knowledge and experience of record
linking. These 21 were further categorized into those using or not using PPRL tools/software for record
linking.
c
Figure 2: Process for selecting record linkage implementations for the assessment
A set of 13 PPRL and non-PPRL implementations were selected as the final set of candidates for in-depth
analysis against the criteria listed in Table 3. The non-PPRL examples (often referred to as “clear text”
d
linkages, since direct PII are used) were also included to ensure that the project gains from the
experiences of others with record linking in general and to collect broader best practices and lessons
learned. The 13 included all seven of the record linking candidates using PPRL and six of the 13 non-PPRL
implementations (the remaining eight were not selected due to lack of sufficient publicly available
information). To facilitate a full understanding of the 13 record linkage implementations, the Project
Team also prepared overview graphics to illustrate the flow of data from collection through linking to
sharing to identify the various points of governance—two examples of data flow overview in the context
of linking is shown in
Appendix Figure 1 and Appendix Figure 2. After researching the 13 record linkage
implementations, the Project Team identified gaps in the information needed to perform a thorough
c
Major key words used for G
oogle search included: record linking, data linking, data combining, data merging, entity resolution, identity
resolution, identity management, PPRL, governance, and technology.
d
Clear text: information that is not encrypted
(https://csrc.nist.gov/glossary/term/clear_text)

NICHD Office of Data Science and Sharing (ODSS) Page 20 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
assessment of the governance based on the criteria listed in Table 3 and reviewed them with NICHD
ODSS to identify relevant stakeholders for interviews. The purpose of the interviews was to collect
additional information, validate accuracy of the information collected, and fill in gaps. Ten of the 13
record linkage implementations were selected for the interviews (
Table 4). All of the PPRL and non-PPRL
implementations at NIH were selected for interviews.
Table 4: Record Linkage Implementations Included in Governance Assessment
Record Linkage Implementations Pediatric
Focus
PPRL Stakeholders
Interviewed?
1 NIH Center for Information Technology (CIT)/
The Biomedical Research Informatics Computing
System (BRICS) Instances
e
Secondary Data
Repository
No Yes Yes
2 National Institute of Mental Health (NIMH) Data
Archive Repository (NDA)
Secondary Data
Repository
No Yes Yes
3 National Center for Advancing Translational
Sciences (NCATS)/National COVID Cohort
Collaborative (N3C) – EHR Data Linkage
Clinical Data
Infrastructure
No Yes Yes
4 National Center for Advancing Translational
Sciences (NCATS)/National COVID Cohort
Collaborative Class 0 (N3C) – EHR Data Linkage
with Data from an external enclave
Clinical Data
Infrastructure
No Yes
5 National Center for Advancing Translational
Sciences (NCATS)/National COVID Cohort
Collaborative Class 2 (N3C) – EHR Data Linkage
with external datasets ingested into N3C
Clinical Data
Infrastructure
No Yes
6 PEDSnet Clinical Data
Infrastructure
Yes Yes Yes
7 Centers for Disease Control and Prevention
(CDC)/The Childhood Obesity Data Initiative
(CODI)
Study Yes Yes No
8 National Institute of Health National Center for
Biotechnology Information (NIH NCBI)/Database
of genotypes and phenotypes (dbGaP)
Secondary Data
Repository
No No Yes
9 National Institute of Health (NIH) All of Us (AoU) Study No No Yes
10
The National Patient-Centered Clinical Research
Network & Down Syndrome Connect (PCORnet-
DS Connect)/DS-DETERMINED Study
Study No No Yes
11 Georgetown University/Federal Statistical
Research Data Center (FSRDC) – Census
Administrative Data
Infrastructure
No No Yes
12 National Center for Health Statistics (NCHS) with
National Death Index (NDI)
Study No No No
13 The Administration for Children and Families
(ACF) – The Child Maltreatment Incidence (CMI)
Data Linkages project: Alaska Department of
Health and Social Services/Oregon Health
Sciences University (ADHHS/OHSU)
Study Yes No No
System Type
e
Includes implementations at the following institutes/programs (instances): NINDS/Parkinson’s Disease Biomarker Program, NIA, NEI, NCATS/
Global Rare Diseases Data Repository (
GRDR), NINR/Common Data Repository for Nursing Science (cdRNS), The Federal Interagency Traumatic
Brain Injury Research (
FITBIR).
NICHD Office of Data Science and Sharing (ODSS) Page 21 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
6.3 Analyze & Summarize Findings
The initial governance analysis of the 13 record linkage implementations was based on publicly available
information, and the subsequent deep dive of these for the project included distilling and documenting
information collected from public sources and supplemental interviews to address each of the criteria
shown in Table 3
. Below are some high-level findings from the analysis:
• Only three of the 13 implementations selected for this project were focused on pediatric record
linking (Table 4).
• Review of publicly available literature and documentation for the original 21 record linkage
implementations, including the 13 that were chosen for this project, showed that publicly
available information relating to governance is limited and not consistently documented or
shared.
• Stakeholder interviews were a critical component of performing a comprehensive Governance
Assessment—through these interviews, the Project Team identified discrepancies, gaps, and
outdated information in the public documentation for the record linkage implementations.
A summary of the analysis for these 13 record linkage implementations categorized by PPRL vs non-PPRL
examples are presented in Appendix Table 2 and Appendix Table 3
. These tables summarize the
assessment using the 11 criteria for each of the record linkage implementations, with additional
descriptive details below the overview table. The Project Team identified the following key findings
based on detailed governance analysis of these 13 record linkage implementations.
6.3.1
Types of Data Linked
Table 5 shows the types of data collected/linked in the 13 record linkage implementations—it is
important to note that in all of these cases, while a specific type of data is collected, one cannot
automatically infer that these data are all linked to each other. Further, the data types listed in the table
may not be exhaustive. Nevertheless, the findings from the analysis show that a majority of the PPRL
implementations collect and link EHR (or EHR-derived), clinical, and other types of data typically
collected during clinical research, such as demographics, genetic, and imaging data. On the other hand,
the majority of the non-PPRL record linkage implementations collect and link primarily administrative
and survey data. A variety of other types of data are also collected and linked in these
implementations—these range from data from mobile devices to disease registry data, vital statistics,
geocoded data, longitudinal household records, economic data, and workforce data. Further details are
available in
Appendix Table 2 and Appendix Table 3.
Table 5: Types of Data Collected/Linked in the Record Linkage Implementations
[Note: the list below may not be completely exhaustive]
Implementation EHR Clinical Demographic Genetic Imaging Mortality
data
Administrative
data
Survey
data
Other
PPRL Implementations
1 NIH BRICS
Instances
X X X X X
2 NIMH NDA
X X X X X X
f
3 N3C EHR Linkage
X X X
f
Neurosignal recordings data
-

NICHD Office of Data Science and Sharing (ODSS) Page 22 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Implementation EHR Clinical Demographic Genetic Imaging Mortality
data
Administrative
data
Survey
data
Other
4 N3C Class 0
Linkage
X X X X
5 N3C Class 2
Linkage
X X X
X X
g
6 PEDSnet
X X X X X
h
7 CDC/CODI
X X X X
i
Non-PPRL Implementations
8 dbGaP
X X
j
X
9 All of Us
X X X
X X
k
X
l
10 DS-DETERMINED
X X
X
m
11 Georgetown
FSRDC – Census
X X
n
X
o
12 NCHS/NDI
X X
p
13 ACF/CMI –
ADHHS/OHSU
X X X
q
X
r
6.3.2
Authorization for Linking Data and Sharing Linked Data
Authorizations are an important and foundational element for appropriate data linking and sharing.
Analysis of the 13 record linkage implementations showed the following mechanisms for authorization
to link and share linked data; these authorizations were not mutually exclusive as some
implementations used multiple mechanisms (Table 6
):
• Explicit consent from the participant for linking and sharing linked records: Analysis of the 13
record linkage implementations showed that explicit consent for participant-level record linkage
is verified in a minority (4/13) of implementations—All of Us, DS-DETERMINED, NCHS/NDI, and
some, but not all, PEDSnet studies. When consent is obtained in PEDSnet, the consent language
is broad and addresses sharing linked data. For some studies, PEDSnet also obtains consent for
linkages using clear text. Available examples of consent language for some of these record
linkage implementations that were available from various sources are in Appendix Table 4
.
• Waiver of consent from the data originator’s IRB: All three implementations of N3C and most of
the PEDSnet studies operate under a waiver of consent.
• Determination by an IRB or an equivalent Privacy Board: Three implementations—CODI,
Census/FSRDC, and ACF/CMI-ADHHS/OHSU—relied on the IRB of the institution or organization
submitting participant data to provide authorization for linkage. CODI requires IRB approval for
g
Viral variant summary data
h
Data from health plans, disease specific registries, vital statistics, and geocoded data
i
Community invention data including longitudinal household records
j
Also, genome wide association (GWAS) data, Short Read Archive (SRA) data, and expression data
k
Surveys topics include sociodemographic, overall health, lifestyle, and health care access and utilization
l
Data from mobile devices
m
Survey topics include the Initial Health Questionnaire (IHQ) from the DS-Connect Registry and the Self-Determination survey from the Self-
Determination Inventory System Data Dashboard
n
Survey topics include the American Community Survey (ACS), Population Survey, and the Survey of Income and Program Participation (SIPP)
o
Also, health data, economic data, U.S. labor/workforce data, science and engineering and technology workforce data
p
The linked dataset comprised of the following populated-based health surveys: National Health Interview Survey (NHIS): 1985-2014,
Continuous National Health and Nutrition Examination Survey (NHANES): 1999-2014, NHANES III (1988-1994), NHANES II (1976-1980), NHANES
I Epidemiologic Follow-up Study (NHEFS), Second Longitudinal Study of Aging (LSOA II), Supplement on Aging (SOA), National Home and Hospice
Care Survey (NHHCS): 2007, National Nursing Home Survey (NNHS): 1985, 1995, 1997, 2004.
q
Oregon Pregnancy Risk Assessment Monitoring System (PRAMS) survey data
r
Child protective services record data

NICHD Office of Data Science and Sharing (ODSS) Page 23 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
linking longitudinal patient records for research using PPRL. Record linking at Census/FSRDC is
limited to statistical purposes only; while Census performs the linkage, the researcher
requesting the linked data must certify and show proof (via the data sharing agreement with the
data owner) that they have permission to link data from a specific agency or multiple agencies
with Census data and with each other. Also, since the linkage is based on PII that must be shared
with the FSRDC, an IRB from the organization contributing the data may need to make a
determination regarding whether the intended use of the data is appropriate. Several NIH
programs that use BRICS also require that an IRB and/or Privacy Board has verified that the
submission and associated use of the GUID tool and data sharing is consistent with informed
consent, that the data are de-identified according to the respective repository standards, risk to
the study population has been considered, and the data were collected in a manner consistent
with NIH/DOD regulations; however, the IRB is not asked to sign the submission request.
• Data submitter authorization for linking and sharing the linked data: Six of the record linkage
implementations rely on data submitters to authorize linkage and sharing. For NDA and BRICS
instances, the data submitters and their institutions must agree to linkage via the use of the
GUID as part of the data submission requirements of each of the respective repositories, but
whether the informed consent specifically addresses the use of the GUID is not confirmed by the
repositories. dbGaP requires data submitters to submit an Institutional Certification and a
subject consent file that describes various data use limitations based on consent, but it is up to
the submitter to determine whether their subject IDs should be linked with existing dbGaP
datasets. N3C, PEDSnet, and CODI allow data submitters/owners to participate or permit
linkages for certain data sources (e.g., mortality data or viral variant data for N3C) or studies
(PEDSnet and CODI sites can engage on a study-by-study basis).
• Federal authorization: Both Census/FSRDCs and NCHS/NDI operate under federal laws that
authorize them to collect and link data with PII. The Census Bureau is authorized to collect and
link data for statistical purposes based on Titles 13 and 26 of the United States Code and the
Office of Management and Budget (OMB) Guidance M-14-06 (Guidance for Providing and Using
Administrative Data for Statistical Purposes). In the case of NCHS/NDI linkage, specific federal
laws authorize the National Health Interview Survey (NHIS) to ask for PII for linkage purposes.
These include Section 308(d) of the Public Health Service Act (42 United States Code 242m(d)),
the Confidential Information Protection and Statistical Efficiency Act (Title V of Public Law 107-
347), and the Privacy Act of 1974 (5 U.S.C. § 552a). As noted above under consent, the NCHS
survey data is deemed eligible to link based on whether a survey participant gives consent for
data linkage in the survey and whether adequate PII is present for linkage.
Regardless of the mechanism for authorizing sharing, most of the data sharing approaches allow the
withdrawal of data for a participant—in such instances, no future linking and sharing of the data for that
participant will occur, but the data that have been already linked and shared will not be withdrawn.
Further details on the authorizations for linking and sharing linked data are available in Appendix Table 2
and Appendix Table 3.

NICHD Office of Data Science and Sharing (ODSS) Page 24 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Table 6: Authorizations for Linking and Sharing for the 13 Record Linkage Implementations
Record Linkage Implementations Authorization for
Linking Data
(C/A: Consent/Assent, W: Waiver of
Consent, I: IRB, S: Data Submitter
Agreement, F: Federal Authorization)
Authorization for
Sharing Linked Data
(C/A: Consent/Assent, W: Waiver of
Consent, I: IRB, S: Data Submitter
Agreement, F: Federal Authorization)
PPRL Implementations
1 NIH BRICS Instances S C or S
2 NIMH NDA S C or S
3 N3C EHR Linkage W W
4 N3C Class 0 Linkage W + S W + S
5 N3C Class 2 Linkage W + S
W + S
6 PEDSnet [W or C] + S [W or C] + S
7 CDC/CODI I + S I + S
Non-PPRL Implementations
9 dbGaP S C
s
or S
10 All of Us C C
11 DS-DETERMINED C C
12 Georgetown FSRDC – Census F [+ I
t
]
F [+ I
17
]
13 NCHS/ NDI F + C F + C
14 ACF/CMI – ADHHS/OHSU I I
6.3.3
PII Elements Used in PPRL Implementations
The Project Team analyzed the PII elements used in the seven PPRL implementations and examined
whether the pediatric PPRL implementations used a unique set of PII elements. The 25 PII elements
utilized across these implementations are shown in Table 7
and can be summarized as follows:
• First name, last name, and date of birth are used in all seven implementations.
• All seven implementations use some form of geographical location information; five use ZIP
Code, two use city of birth, and one uses household street address.
• Gender or sex is used in all seven implementations; three use gender, three use sex, and one
uses “physical sex at birth.”
• Cell/phone number is used in four of the seven implementations.
• SSN and email are used in all three N3C implementations, but not in other implementations.
• Combinations of five or more PII elements are used in all of the record linkage implementations.
Comparison of the PII elements used by the various PPRL tools showed the following:
• Two government-owned GUID tool implementations at NIH (BRICS instances and NIMH Data
Archive) and an open source tool, Anonlink (used by CODI), use similar PII elements, including
first name, last name, date of birth, and sex. Anonlink also uses ZIP Code and household street
address and BRICS/NDA each use city of birth.
• The Datavant tool used by four of the seven record linkage implementations incorporated
mostly the same PII elements: first name, last name, date of birth, gender/sex, SSN, email, cell
phone number, and ZIP Codes.
s
dbGaP obtains Study Consent files from submitters which denotes the consent groups.
t
Linkages for statistical purposes only

NICHD Office of Data Science and Sharing (ODSS) Page 25 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
The two pediatric-focused record linkage implementations, PEDSnet and CODI, did not use identical PII
elements.
Table 7: PII elements used in PPRL based Record Linkage Implementations
PII Elements NIH BRICS
Instances
NIMH NDA N3C
(3 Implementations)
PEDSnet CODI Total
PPRL Tool Used BRICS
GUID
NDA GUID Datavant Datavant Anonlink
1 First name
X X X X
X
7
2 First initial of first name
(X)
u
(1)
3 Middle name
X X
2
4 Last name
X X X X X
7
5 Date of birth
X
v
X X X X
7
6 Day of birth
(X)
1
7 Month of birth
(X)
(1)
8 Year of birth
(X)
(1)
9 City of birth
X X
2
10 Country of birth
X
1
11 Place of birth
(X) (X)
(2)
12 ZIP3
X
1
13 ZIP5
X
3
14 ZIP9
X
3
15 ZIP Code
X
1
16 Household street
Address
X
1
17 Gender
X
3
18 Sex
X X
X
3
19 Physical sex at birth
X
1
20 SSN
X
3
21 Email
X
3
22 Cell phone number
X X
w
4
23 Phone number
(X)
(1)
24 Government or national
issued ID
X
x
1
25 Country issuing
government issued or
national ID
X
x
1
6.3.4
Two-Party or Three-Party Model for Entity Resolution and Data Linkage
The 13 record linkage implementations were analyzed for the separation of parties who perform entity
resolution and who perform the data linking. In a two-party model, the entity resolution and the data
linking are done by the same organization whereas in a three-party model, these two activities are
u
(X): Derived PII elements
v
Day, month, and year of birth used for BRICS instances were categorized as ‘date of birth’ in this table.
w
Phone number was considered same as cell phone number for Anonlink in this table.
x
PII field is not required for PPRL linkage.
NICHD Office of Data Science and Sharing (ODSS) Page 26 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
performed by separate organizations (the first party is the PII holder). In three-party situations, often a
trusted neutral party serves as the honest broker
y
who performs entity resolution.
Analysis of the 13 record linkage implementations show that six of them used the three-party model and
the remaining seven used the two-party model, as shown in Table 8
. The three-party model was used by
a majority of the PPRL implementations (five of seven), whereas most of the non-PPRL implementations
used two-party. Details of the data linkage approaches for all 13 record linkage implementations are in
Appendix Table 2 and Appendix Table 3.
Table 8: Two-party or Three-party and Data Linkage Models used in the Record Linkage Implementations
Record Linkage
Implementations
Entity Resolution & Data Linkage
Two-Party (2)
or
Three-Party (3)
Entity Resolver Data Linker Data Linkage
Model
z
(D: Linked database,
S: Study-specific)
PPRL Implementations
1 NIH BRICS Instances 3 GUID server Researchers D
2 NIMH NDA 3 GUID server
Researchers D
3 N3C EHR Linkage 3 Regenstrief Researchers D
4 N3C Class 0 Linkage 3 Regenstrief Researchers D
5 N3C Class 2 Linkage 3 Regenstrief Researchers D
6 PEDSnet 2 PEDSnet Data
Coordinating Center
(DCC)
PEDSnet DCC S
7 CDC/CODI 2 CODI DCC
CODI DCC S
Non-PPRL Implementations
8 dbGaP 3 dbGaP Data
Curation Team
Researchers D
9 All of Us 2 Raw Data
Repository
Raw Data
Repository
D
10 DS-DETERMINED 2 Study Team Study Team
S
11 Georgetown FSRDC -
Census
2 Census
Census S
12 NCHS/NDI 2 NCHS Data
Linkage Program
NCHS Data
Linkage Program
D
13 ACF/CMI – ADHHS/OHSU 2 Integrated Client
Services
Integrated Client
Services
D
-
6.3.5
Data Linkage Model: Linked Database or Study-Specific Linkage
The data linkage model refers to the scope of the data that is linked and provisioned to users in the
various record linkage implementations. For this project, two models were identified:
• Linked database model, where the linkage information that is created and/or provided
encompasses all datasets in a given database
y
A party that holds de-identified tokens (“hashes”) and operates a service that matches tokens generated across disparate datasets to
formulate a single Match ID for a specific use case.
z
Linked database model: the linkage information is created and/or provided encompasses all datasets in a given database
Study-specific linkage model: linkage information is created and/or provided for the purposes of a specific study

NICHD Office of Data Science and Sharing (ODSS) Page 27 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
• Study-specific model, where linkage information is created and/or provided for the purposes of
a specific study
Table 8
shows that a majority (nine) of the 13 record linkage implementations operate as a linked
databa
se model by default and the remaining four are study-specific linkages. Details of the data linkage
approaches for all 13 record linkage implementations are in
Appendix Table 2 and Ap
pendix Table 3 and
a summary of the data linkage
approaches for key record linkage implementations is included below.
BRICS and NDA use globa
l unique identifier/s (GUID) servers/systems to perform the matching and
entity resolution, where the PPRL tool automatically checks the database for existing GUID and returns
an existing one (if a match is identified) or a new one (if no match is detected). Approved researchers
receive access to the GUIDs for all of the data they are approved to access (no additional requests are
needed); however, it is up to the researcher to use the GUID to link data in their analyses. Some
instances
aa
of BRICS use a common GUID server, which means that users with approval to access data
from multiple instances could use the GUIDs to link data across multiple instances. All three N3C PPRL
implementations —internal EHR to EHR linkage and external Class 0 and Class 2 linkages —follow the
linked database model where entity resolution occurs as the data are received by N3C and tokens are
sent to the linkage honest broker from the data partners and the external sources. Only externally linked
Class 2 data are currently available to users whereas the internal EHR data linkage and external Class 0
data linkages are in pilot phase. In all three cases, entity resolution is performed by a linkage honest
broker (LHB), Regenstrief, based on matching the hashed tokens generated by N3C EHR data partners
and external enclaves/data sources (for Class 0 and 2). The LHB then creates a linkage map with a new
MATCH_ID mapping to the original de-identified subject IDs (Pseudo_IDs) provided by N3C data partners
across all datasets that participate in PPRL. Once fully launched, the linkage map will be made available
to researchers who have approval to access the HIPAA limited dataset (level 3) in the N3C enclave to use
in their analysis. Entity resolution is performed across all participating EHR, Class 0, and Class 2 datasets,
but linkage maps between EHR data and Class 0 or Class 2 will only be created for EHR data partners
who have specifically approved linkage with a Class 0 or Class 2 dataset, and these linkage maps will only
be shared with approved users of the specific Class 0 or Class 2 dataset.
dbGaP follows the linked database model by checking all incoming subject IDs provided by the submitter
in the subject consent file against existing studies in the database using a custom string matching
analysis. If the subject ID of the incoming data matches with the subject ID of existing datasets, dbGaP
curators notify the incoming submitter to verify the origin of the matched subjects, and if there is a true
match, dbGaP asks the submitter to add the existing study’s IDs (in dbGaP) to the subject consent file
and resubmit. When there is a match, dbGaP links the incoming data with the existing dbGaP Subject ID,
and when there is no match, creates a new dbGaP Subject ID, which is openly available to all data users.
Researchers can use the dbGaP Subject ID to find participants who are represented in multiple studies
and link the data during analysis after approval to access the individual datasets from the respective
data access committees. dbGaP also performs entity resolution within a study through the use of the
Genetic Relationship and Fingerprinting (GRAF) tool, to assess inconsistencies between molecular data
sample IDs and phenotype sample IDs, unintended data duplications, incorrect pedigree information,
and subject relationships within a given data submission. DbGaP does not use GRAF to identify cross-
study linkage, but GRAF could be leveraged for this purpose if approved for a specific implementation.
aa
An instance is a collection of services for a managed data repository software platform; each instance is a
distinct data repository in BRICS.
NICHD Office of Data Science and Sharing (ODSS) Page 28 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
In the case of All of Us, all datasets collected from participants for the All of Us Research Program are
assigned a participant identifier (PID) and are automatically linked using the PID when deposited into the
raw data repository (RDR). The PID is an internal linking ID and is converted to a research ID for data
users. All of Us is currently exploring linkage with external datasets, where the data linkage will be
performed by All of Us across all All of Us participants and provisioned in the All of Us enclave.
PEDSnet, CODI, and FSRDC/Census use the study-specific linkage model where users must submit a
research proposal detailing the proposed use of the linked data, and only after approval of the proposal
does the study team or organization provide the linked data to the researchers. In PEDSnet and CODI
implementations, their DCC performs the entity resolution using hashed codes and generates a new
unique linking ID that is then used by the DCC to link across datasets for each study. In both these
implementations, the linking ID is replaced with a study-specific ID for provisioning to the users. Census
uses a study-specific linkage model within the FSRDC for linking Census data with data from other
external sources for provisioning to approved users. The external data can be from other agencies
(Agency for Healthcare Research and Quality, National Center for Health Statistics, Bureau of Economic
Analysis, Bureau of Labor Statistics, etc.) or other sources that users bring into the FSRDC. Once the
linkage is approved by both Census and the external source/s, Census links the data by generating a
Personal Identification Key (PIK) for all data imported into Census or via a PIK bridge (provided by
external agencies), after which the linked data is provisioned to the user.
6.3.6
Controls for Managing Re-Identification Risk with Linked Data
While the benefits of using linked data for addressing more complex and/or new research questions
may be greater compared to the individual datasets, linking datasets at the individual record level raises
the potential re-identification risk even if the data that are being linked are considered fully de-
identified (e.g., stripped of all 18 HIPAA identifiers
18
). Therefore, managing re-identification risks is
especially important when sharing linked data. While many controls typically used for sharing research
data are also applicable when sharing linked data, there might be additional controls that are
appropriate based on the nature of the datasets that are linked, such as whether they are pediatric data,
the de-identification status of the data, the types of data (e.g., linkage with administrative data or social
media data), the granularity of the data, and the sensitivity of the data (drug use, criminal record, etc.).
Multiple controls—applied both prior to linking and after linking—can be used to manage the risk of re-
identification. Two categories of re-identification risk management controls were examined in the 13
record linkage examples based on the information available publicly or gathered via stakeholder
interviews.
De-identification status of the data: De-identification of the linked data serves as a control mechanism
before sharing the linked data. Methods typically used to de-identify datasets include masking,
perturbing, suppressing variables, and collapsing small cell sizes; the method chosen will depend on
whether the data can continue to be meaningfully used after it has been de-identified. While some
implementations use other de-identification standards, the HIPAA Privacy Rule specifies the following
standards for de-identifying PHI data and identifies two levels of de-identification:
• De-identified dataset: refers to Expert Determination or Safe Harbor (removal of all 18
identifiers enumerated at section 45 C.F.R. 164.514(b)(2) (the HIPAA Privacy Rule))
• Limited dataset: refers to PHI that excludes 16 of the direct identifiers but may include
geographic information (city, state, ZIP Code), elements of dates, and other values that are not
direct identifiers

NICHD Office of Data Science and Sharing (ODSS) Page 29 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
The NIH Genomic Data Sharing Policy follows the definition for de-identified data in the HHS Regulations
for Protection of Human Subjects (also known as the Common Rule) and the HIPAA Safe Harbor method.
An examination of the de-identification status of the linked data that are shared in the 13 record linkage
implementations shows that a majority (9 of 13) share exclusively de-identified datasets whereas all
three N3C implementations share linked data only as limited datasets, and one (CODI) shares a mix of
limited and de-identified linked datasets (Table 9
). Additional details for all 13 record linkage
implementations are in
Appendix Table 2 and Appendix Table 3.
Disclosure review and other re-identification risk management controls: Disclosure "relates to
inappropriate attribution of information to a data subject, whether an individual or an organization”
36
,
and a disclosure review is designed to prevent potential disclosures. Disclosure reviews and other re-
identification risk management protocols and procedures serve as controls that can be implemented
before or after data linkage, prior to sharing linked data. Disclosure review usually comprises some
combination of an expert review of variable combinations and statistical analysis of potential re-
identifiability. Other controls include requiring a letter of determination (LOD) from the data user’s IRB
and including specific terms in the data use agreement/certification established with the user’s
institution that explicitly prohibit re-identification of study participants and limit the period of data use.
The Project Team assessed disclosure reviews and other re-identification risk management controls in
place for sharing linked data in 12 of the 13 record linkage implementations (Table 9
) and drew the
following conclusions (Note: DS-DETERMINED is not sharing the linked data beyond the study team yet):
• Six of the 12 record linkage implementations—N3C Class 0 and Class 2 linkages, PEDSnet, All of
Us, Census and NCHS/NDI—have some form of re-identification assessment process in place. In
some of these implementations, special committees perform a formal re-identification risk
analysis—these committees include the Tools and Resources Review Committee for N3C Class 0
and Class 2, the PEDSnet Steering Committee for PEDSnet, the Committee on Access, Privacy,
and Security (CAPS) for All of Us internal datasets, and a Disclosure Review Board for Census.
These assessments may result in the exclusion of sharing certain datasets or data elements or
modifying specific data elements to share less detailed information.
• PEDSnet Policy also requires masking cell counts <11 in reports and manuscripts. However,
smaller cell counts can be reported if five or more institutions contributed to the dataset and
these institutions agree to allow the small cell sizes to be revealed.
• Of the 12 implementations, an LOD from the user’s IRB is required for all three N3C
implementations. There are specific datasets in dbGaP, NDA, and BRICS that require IRB
approval as a component of the data access request, but this is based on consent-based data
use limitations and not related to record linkage, nor is it standard practice across all datasets
that reside in these repositories. Finally, NDA and BRICS data access guidance advises that data
submitters who still have access to PII for the datasets they shared, may need to update their
IRB approved protocol because access to additional data obtained through use of the GUID
increases the amount of data they are accessing for their participants.
• All 12 record linkage implementations require the user and their institution to execute a Data
Use Agreement/Certification, which explicitly prohibits re-identification of study participants.
Additional details for these 12 record linkage implementations are in Appendix Table 2 and
Appendix
Table 3.

NICHD Office of Data Science and Sharing (ODSS) Page 30 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Table 9: Disclosure and Re-identification Risk Management of Linked Data
Record Linkage
Implementations
De-identification Status
of the Linked Data
bb
(Limited Dataset,
De-identified, Synthetic)
Disclosure Review/
Other
Re-Identification Risk
Management Controls
IRB Letter of
Determination
cc
Data Use
Agreement/
Certification
dd
PPRL Implementations
1 NIH BRICS
Instances
De-identified data No No Yes
2 NIMH NDA De-identified data No No Yes
3 N3C EHR Linkage Limited dataset
(retains dates and ZIP
Codes)
No Yes Yes
4 N3C Class 0
Linkage
Limited dataset
(retains dates and ZIP
Codes)
Re-identification risk
assessed by the Tools
and Resources Review
Committee
Yes Yes
5 N3C Class 2
Linkage
Limited dataset
(retains dates and ZIP
Codes)
Re-identification risk
assessed by the Tools
and Resources Review
Committee
Yes Yes
6 PEDSnet De-identified Risk review is conducted
for each proposed study,
but no separate deductive
disclosure review of the
linked data is performed
No Yes
7 CDC/CODI De-identified or Limited
dataset
No No Yes
Non-PPRL Implementations
8 dbGaP De-identified (HIPAA
Safe Harbor + Common
Rule per NIH Genomic
Data Sharing Policy)
No No Yes
9 All of Us De-identified Risk of re-identification is
routinely assessed by the
All of Us Committee on
Access Privacy and
Security (CAPS)
No Yes
10 DS-DETERMINED De-identified Linked data sharing
process yet to be defined
Linked data not
yet shared
outside of study
team
Linked data not
yet shared
outside of
study team
bb
Limited dataset refers to PHI that excludes 16 categories of direct identifiers; De-identified data refers to removal of all 18 identifiers
enumerated at section 45 C.F.R. 164.514(b)(2) (the HIPAA Privacy Rule); Synthetic data refers to data that are computationally derived from the
limited dataset and that resemble patient information statistically but are not actual patient data.
cc
Determination that an activity IS NOT Human Subject Research: the project does not meet either the definition of “research” as defined in 45
CFR 46.102(l) or 21 CFR 56.102(c); or the definition of “human subject” at 45 CFR 46.102(e)(1) or 21 CFR 56.102(e) – or –
Determination that an activity IS Human Subject Research: proposed activity is human subject research because it meets the DHHS definition of
research [45 CFR 46.102(l).
dd
Data Use Agreement or Certification established with the user’s institution explicitly prohibiting re-identification of participants

NICHD Office of Data Science and Sharing (ODSS) Page 31 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Record Linkage
Implementations
De-identification Status
of the Linked Data
bb
(Limited Dataset,
De-identified, Synthetic)
Disclosure Review/
Other
Re-Identification Risk
Management Controls
IRB Letter of
Determination
cc
Data Use
Agreement/
Certification
dd
11 Georgetown
FSRDC – Census
De-identified Approval from the
Disclosure Review Board
(DRB) prior to
disseminating statistical
products or publications
derived from the analysis
No Yes
12 NCHS/NDI De-identified NCHS Research Data
Center (RDC) reviews
research proposals from
users requesting access
to restricted use linked
mortality files (LMFs) to
identify potential
disclosure risks
No Yes
13 ACF/CMI –
ADHHS/OHSU
De-identified No No Yes
6.3.7
Authorizations and Controls for Accessing the Linked Data
As mentioned earlier, linked data might present a higher risk profile than the individual datasets. It is
critical therefore to ensure that such data are provisioned to users in a manner by which participant
confidentiality and data privacy are protected and maintained. Two criteria were used to examine the
mechanisms established by the 13 record linkage implementations to provision the linked data to users:
• Authorization for data access: Analysis of the 12 record linkage implementations that are
currently sharing linked data showed that a majority (10 out of 12) relied on an internal group—a
Data Access Committee, Steering Committee, Governance Bo
ard, or a Resource Access Board
(Table 10) (Note: DS-DETERMINED is not sharing the li
nked data beyond the study team yet).
These committees were responsible for reviewing the proposed research plan from data users
or requestors to ensure it is consistent with
data use limitations if any, imposed by the consent or
by t
he submitter, or other requirements. For accessing linked data
within N3C, which is
considered level 3 (limited dataset), users must attest to the N3C Code of Conduct, IT
security
trai
ni
ng,
a
nd h
uman subjects protection
training. In t
he
c
ase o
f Census/FSRDCs, users requesting
access t
o the linked data via FSRDCs are required to obtain Special S
worn
Statu
s
37
,
c
omplete
background
c
heck
a
nd
t
raining,
a
nd sign an agreement prohibiting attempts to re-identify the
participants and
requiring using the data only for statistical purposes. A Census Bureau data
access team confirms that all researchers have completed required trainings and then provides
access within a secure computing environment. Use of data provisioned by Census through the
FSRDCs are governed by laws and policies
from agencies that supplied data. For example, Titles
13 and 26 of the U.S. Code protect the privacy and confidentiality of the participants and their
data. Violation of these laws are federal crimes are punishable by serious penalties including
prison time and fines.
• Data access
m
odels: The four types of access options examined
in the record linkage
implementations included open, registration required, controlled, and e
nclave (Table 10). All 12
of the implementations that are currently sharing individual-level linked data provision those
data either in a control
led manner or through an enclave. For linkage of EHR data with external

NICHD Office of Data Science and Sharing (ODSS) Page 32 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
datasets that are categorized as Class 0, N3C provisions the linked data in an ephemeral
workbench, which is a temporary extension of the N3C enclave. All of Us provisions data in the
secure curated data repository (CDR) via three tiers of access—an open tier where summary
statistics and aggregate information are provided, and registered and controlled tiers, where
data with differing levels of granularity are provided. Re-identification risk analysis is performed
before provisioning the All of Us data through the registered and controlled tiers. Although the
controlled tier (with genomic and more granular demographics, EHR, and survey data) tolerates
a higher level of risk than the registered tier, both the registered and controlled tiers within All
of Us also function as enclaves (i.e., data cannot be downloaded or analyzed in other systems).
Table 10: Authorizations and Controls for Accessing the Linked Data
Record Linkage
Implementations
Authorization for Data Access Data Access Model
ee
(O: Open, R: Registration required,
C: Controlled, E: Enclave)
PPRL Implementations
1 NIH BRICS Instances Respective Program’s Data Access
Committee (DAC)
C
2 NIMH NDA NDA DAC C
3 N3C EHR Linkage N3C DAC E
4 N3C Class 0 Linkage N3C DAC E
ff
5 N3C Class 2 Linkage N3C DAC E
6 PEDSnet PEDSnet Steering Committee E
7 CDC/CODI CHORDS Governance Committee C
Non-PPRL Implementations
8 dbGaP Respective Dataset’s DAC C
gg
9 All of Us All of Us Resource Access Board O, R/E, C/E
hh
,
10 DS-DETERMINED N/A
20
N/A
ii
11 Georgetown FSRDC – Census Census Bureau’s Data Access Team E
12 NCHS/NDI NCHS Research Data Center (RDC) E
13 ACF/CMI – ADHHS/OHSU Project specific approving authority
including an advocate from one state
agency (Oregon Health Authority)
C
7 TECHNOLOGY ASSESSMENT
In support of the overall project goal to inform an NIH-wide strategy on the use of PPRL for pediatric
COVID studies, a third objective was to develop considerations for implementing potential PPRL tools in
support of the selected use cases identified by the pediatric COVID studies. To meet this objective, the
Project Team assessed the landscape of broadly used PPRL vendors/organizations, with a focus on those
used within the NIH data ecosystem. To further understand the existing tools, the Project Team
expanded upon a prior technology analysis conducted by NCI—the Landscape Analysis of Privacy
ee
Data access models–Open access: no access restrictions or registration required to access; Registration required: open to all, but users need
to be signed in or registered with the resource to access; Controlled access: application and eligibility requirements need to be met to gain
access (e.g., by a data access committee); Enclave: data cannot leave a specific system boundary (e.g., cannot be downloaded).
ff
Ephemeral Work Bench (A temporary extension of the N3C Enclave)
gg
Controlled Data but Public dbGAP IDs
hh
Both the registered and controlled tiers of access are within All of Us’s enclave environment.
ii
Linked data sharing process yet to be defined

NICHD Office of Data Science and Sharing (ODSS) Page 33 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Preserving Patient Record Linkage Software (P3RLS)
jj
—Final Report Version 2 (2020)
29
. The high-level
approach for performing the analysis is further discussed in the sections below.
7.1 Define Criteria for Technology Assessment
The Project Team developed a PPRL technology survey questionnaire based off the NCI Landscape
Analysis to address the pediatric data linkage use cases presented in Section 3. The survey comprised 57
questions organized into seven capability categories. The categories and descriptions can be found in
Table 11. A subset of these questions was designated as essential as they address pediatric-specific
needs and are related to the ability to implement PPRL governance models.
Table 11: Technology Capability Categories for Survey Questions
Capability Category Category Description
1 Hash Generation and Record
Linkage
Describes the overall process required in the generation of hashes from one
or more combinations of input data fields.
2 Operating Environment and
Licensing Model
Describes deployment considerations such as the ability to deploy to the
cloud, availability on different operating systems, and licensing parameters
such as what software or features the license is for, how long it is valid for,
how many users can use the software, the computers on which the software
can be used.
3 Usability & Security Features Describes the difficulty or ease for users to become proficient in using the
software effectively, quality of the user interface, robustness for error
handling, accessibility, installation and maintenance. In addition, this
category captures requirements for protection of information such as
confidentiality, integrity, non-repudiation, and accountability.
4 External System Integration Describes the ability of the software to interface with other systems (such as
the ability to ingest data).
5 Data Cleaning/Pre-Processing
Features
Describes the ability of transforming data into consistent formats and
performing semantic cleanups (e.g., phonetic spellings of names and
substitution of nicknames) to enable more consistent generation of
identifiers and improving matching performance.
6 Performance and Scalability Describes whether processing times and resources fall within specified
constraints and whether the software scales to the required data sizes.
7 Use Cases, Applications, and Future
Capabilities (Informational
Questions)
Describes current use cases and applications that the vendor has
supported/is currently supporting as well as any future capabilities that are
being planned.
7.2 Select & Research Candidate PPRL Technologies
The Project Team defined the scope for the Technology Assessment to include only vendors/
organizations with products that are widely used, able to support large use cases (greater than 1 million
records), and/or currently used within NIH data systems. Based on these criteria, a total of seven
vendors/organizations were included in the Technology Assessment (Table 12
). An eighth product,
Anonlink, which is an open source tool used in the federal space for PPRL linkage (for example in the
CDC/CODI governance example) was also contacted for the survey but was not included in this
assessment due to lack of response. The tools identified are or have been used for record linkage
implementations within health systems, academic medical centers, research institutions, commercial
data aggregators, biopharma stakeholders, population health analytic platforms, medical device
vendors, registries, government entities, national clinical laboratories, Electronic Medical Record (EMR)
jj
This report uses the term privacy preserving record linkage (PPRL) to refer to the concept of P3RL in the NCI report.
NICHD Office of Data Science and Sharing (ODSS) Page 34 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
vendors, retailers, Prescription Benefit Managers (PBMs), health information technology companies,
pharmaceutical manufacturers, health insurers, health services providers, and healthcare suppliers.
Table 12: PPRL Vendors and Tools
Number
PPRL
Vendor/Tool
Prior/Current Use at NIH/Other Federal Agencies
1
HealthVerity
Used by NIH, NCI, CDC, and FDA
2
Datavant
Used by N3C and for NIH funded studies in PEDSnet
3
Senzing
Currently being assessed for use at NIH
4
Crossix
Unknown
5
IQVIA
Identified during the N3C stakeholder interview as another vendor that NIH
is using for certain projects
6
BRICS GUID
Used by many NIH institutes/program data repositories, including NINDS,
NIA, NCATS, NEI, NINR, and DOD/FITBIR
7
NHash
A hashed subject ID generator that is being used by the RADx-rad PreVAIL
Kids Initiative to generate subject IDs for data submission to the RADx-rad
Data Hub
The Project Team sent the capability questions to the seven vendors/organizations. The vendors who
had participated in the NCI Landscape Analysis were given the opportunity to update any original
answers to the NCI request. All vendors/organizations completed the questionnaires by self-assessing
their capabilities in the seven question categories.
The Project Team received the survey responses and reviewed all responses and explanations provided
by the vendors/organizations to summarize their key capabilities to address the pediatric PPRL use
cases.
7.3 Summarize Findings
Results of the complete and detailed analysis of survey responses performed by the Project Team are
not shared in this public report as they are intended to inform internal NIH decision making. General
observations and findings are documented in the sections below. In general, most PPRL tools responded
favorably to a majority of the essential capabilities criteria. The tools that fulfilled the majority of these
criteria differentiated themselves by offering data cleaning and pre-processing capabilities and by their
superior security certifications in their array of features. Tools that did not address the essential
capabilities criteria primarily were hard-coded with inflexible hash generation and little additional
functionality.
7.3.1
Hash Generation and Record Linkage
Hash generation capabilities, such as the ability for the user to specify which variables are used in the
hashes and the ability to link on multiple hashes, are crucial for accurate record linkage without sharing
PII. All vendors/organizations generally responded favorably to the evaluation questions in the hash
generation and record linkage category. All tools support two-party and three-party protected linkage.
To support three-party protected linkage, each tool handles the transmission of additional information
to an honest broker (sometimes also referred to as a trusted broker) differently, including sending a
metadata file that provides a data quality profile of the tokenizing site, sharing a list of the top
candidates of matches, allowing a third party to generate confidence scores to map matches, or sending
metadata about the match. The majority of PPRL tools allow for linking based on multiple concatenated
input variable hashes that are generated from different arrangements of concatenated input PII
NICHD Office of Data Science and Sharing (ODSS) Page 35 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
elements, with no limit on the number of these variable combinations. However, some tools require
certain PII elements for generating a hash, such as first name, last name, DOB, location elements, and/or
gender.
Each tool also has its own unique tunable characteristics for identifying matches between hashes. Some
allow for all matching parameters to be tuned through different matching designs or plugins and
configurations, whereas others only permit the adjustment of certain parameters, such as risk ratios,
error rates, and/or toggling between close match checking.
7.3.2
Operating Environment and Licensing Model
All PPRL tools assessed are implementable in a variety of operating environments, including all major
platforms (Windows, MacOS and Linux). Although the vendors/organizations assessed utilize various
licensing models, all tools demonstrated vendor-rights policies that align to NICHD ODSS requirements
(i.e., vendors do not have rights to the data ingested). This includes PII, metadata, derivative software
data, and hashes along with the associated metadata.
7.3.3
Usability and Security Features
Several vendors/organizations responded favorably in the Usability and Security Features category, as
their tools met various security certification thresholds (e.g., SOC 2, NIST) while providing a low
technology barrier for non-technical users. With regard to usability, several vendors/organizations have
implemented numerous features to improve usability such as Graphical User Interfaces (GUIs) while
others are configurable from the start. These tools can be configured to automate a significant portion
of the PPRL operations and include a default configuration to be used “out of the box.”
In terms of security, several PPRL tools have or are in the process of obtaining a U.S. government
security certification like the Federal Information Security Modernization Act (FISMA) or the Federal Risk
and Authorization Management Program (FedRAMP), although not all tools that are being used by the
government have FISMA or FedRAMP status. Additionally, all the PPRL vendors/organizations ensure
that unencrypted PII elements are not exposed. The most common hash generation algorithm was SHA-
256 followed by SHA-512. A couple of tools incorporate AES-128 encryption, a two-way encryption
algorithm which can both encrypt and decrypt, within its hash generation pipeline.
7.3.4
External System Integration
Only a limited number of tools offer the capability to easily customize and flex input and output formats.
Among the assessed vendors/organizations, the CSV file format was the most popular usable format for
providing PII elements followed by JSON file format. Other supported formats include flat/gzipped
delimited/positional files, input files encoded in UTF-8 delimited by pipe, comma, tab, or semicolon, and
delimited or fixed width text files.
7.3.5
Data Cleaning/Pre-Processing Features
Data cleaning and pre-processing improve the quality of the PII data elements used to generate hashes.
Since real world data are messy, data that are cleaned and standardized prior to hashing returns more
consistent hashes and in return, higher quality record linkages
38
.
A couple of the tools included in the technology assessment are currently only intended for hashing and
linking and therefore did not have data cleaning/pre-processing features. Of the vendors/organizations
NICHD Office of Data Science and Sharing (ODSS) Page 36 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
with capabilities beyond hashing and linking, additional data cleaning/pre-processing features include
handling cases of data abnormalities, phonetic encoding of names (Soundex), international naming,
metadata cleaning, substitution name expansion, noise detection, and collision scores. Certain tools also
allow for the specification of data cleaning by field. In addition to data cleaning, probabilistic matching,
aggregation techniques, bloom filters, and frequency tables and other statistical analysis features have
been implemented by a couple of tools to ensure the quality of the data linkages.
7.3.6
Performance and Scalability
All tools have little to no restrictions for handling large quantities of data for PPRL. Significant advances
in software development in the last decade have enabled scalable and high-performance software for
PPRL applications. Several of the assessed vendors/organizations can handle an uncapped maximum
number of records (up to tens of billions), with the limiting factor instead being hardware performance.
7.3.7
Informational Questions
A few tools currently support or have previously supported initiatives with pediatric data. Additionally,
one vendor/organization reported that they are developing the ability for their tools to interoperate
with other PPRL vendors/organizations’ tools. However, technical documentation was not provided for
review. Any interoperability approaches between different vendors/organizations’ PPRL tools warrant
careful analyses, given the complexity of the technical challenge.
8 CONSIDERATIONS
The NIH CARING for Children with COVID initiative was established to better understand SARS-CoV-2
infection in children who display a broad spectrum of the symptomology, with some infected children
exhibiting a serious multi-organ disease called MIS-C. CARING for Children with COVID study
investigators identified the need to link data from these studies early in the pandemic when the cases
were relatively rare due to school closures and it was suspected that the same children were being
enrolled across multiple studies. The investigators recognized the potential value and impact of linking
different types of data collected for an individual child across these studies, given the multi-organ nature
of the disease and each study having a different focus (pharmaceutical data versus immune profiling
versus cardiac imaging). PPRL was identified as the most feasible approach to address this need, given
that the studies are unable to share PII with one another or with the data repositories used for sharing
the de-identified study data and while genetic matching was another possibility, study investigators
wanted to reduce the number of times the same child underwent genetic sequencing.
The overall goal of this project was to assess and analyze governance and technology approaches in
diverse, extant record linkage implementations to inform an NIH-wide approach to using PPRL to link
data across pediatric COVID studies and, more broadly, to inform approaches for linking individual-level
datasets across pediatric research studies. The project achieved this goal by:
•
Summarizing the
status of the CARING for Children with COVID studies for PPRL feasibility
•
Analyzing 13 recor
d linkage implementations—both PPRL and non-PPRL—funded by NIH, other
federal agencies, and non-government organizations, to fully document end-to-end governance
decisions, including authorization for linking and sharing, data linkage models, re-identification
risk management, and access controls

NICHD Office of Data Science and Sharing (ODSS) Page 37 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
• Evaluating the capabilities of seven PPRL vendors/organizations, including one NIH-developed
tool, one university-developed tool, and five commercial vendor tools, by extending a recent
Technology Assessment led by NCI
29
and adding facets specific to the pediatric COVID record
linkage use cases
The Project Team considered the current state and context of the CARING for Children with COVID
studies when identifying and evaluating existing record linkage implementation approaches, including
the following:
• The CARING for Children with COVID studies selected for the project—POP02, MUSIC, PRISM,
and PreVAIL kIds—are already underway, and participant enrollment has closed for some
studies.
• The studies are depositing data to multiple NIH data repositories that are or will be
interoperable (data will be findable and accessible across multiple repositories) and use multiple
access tiers to share data.
• None of the studies currently uses a PPRL tool to link data within the study or across studies.
The Project Team determined that PPRL is a feasible approach for linking participant data across
pediatric COVID studies so long as the involved parties collaborate prior to implementation to define the
governance approaches, technical requirements, and the data elements required to ensure high-quality
linkage. The Project Team developed eight key considerations for governance and technology
implementations derived from the findings presented in three sections of this report: Pediatric COVID
Studies – PPRL Feasibility (Section 5), Governance Assessment & Findings (Section 6
), and Technology
Assessment (
Section 7). Based on these key considerations, the Project Team identified potential
approaches for PPRL implementation across the CARING for Children with COVID studies. Most of these
key considerations are generalizable to other record linkage implementation efforts. These
considerations and approaches are primarily targeted to NIH and HHS agency staff, who are considering
implementation of PPRL to address research-based use cases. They are also applicable to stakeholder
communities that might participate in or implement PPRL, including investigators conducting pediatric
research and their institutions, and data repositories and data centers. Other audiences for these
considerations may include PPRL software vendors and IRBs, privacy boards, or equivalent bodies.
8.1 Key Considerations Based on Governance and Technology Assessment
8.1.1
Key consideration 1: Authorization for linking and sharing linked data should be based
on informed consent or approval from the data originator’s institution and/or their IRB
or an equivalent Privacy Board
Authorization for record linkage and sharing linked data is a foundational element of a record linkage
governance model. The 13 record linkage implementations revealed a wide range of authorization
mechanisms that are sometimes employed in combination, including:
• Informed consent from participants (and assent in the case of children, where applicable)
• Waiver of consent
• Data originator/submitter authorization
• Approval/determination from an IRB or an equivalent Privacy Board
• Federal authority

NICHD Office of Data Science and Sharing (ODSS) Page 38 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
8.1.1.1
Informed Consent from Participants
Federally funded human subjects research in general must abide by the informed consent requirements
of the Common Rule
39
. However, record or data linkage activities involving de-identified data that do
not qualify as “human subjects research” are not subject to the requirements
40
of the Common Rule. A
majority of the record linkage implementations analyzed in this project share data that are considered
de-identified. Nevertheless, it is ideal to obtain explicit consent for record linkage from study
participants to foster transparency of data use and to honor participant trust
25,41,42, 43
. Where feasible,
such consent should address the scope of linkage—that is, which datasets will be linked—and how the
linked data will be shared without overly restricting the scope in a way that could pose challenges for
answering future unanticipated valuable scientific questions.
In the record linkage implementations analyzed in this project for which data were collected under the
auspices of consent, consent language often does not explicitly address record linkage. Further, while
data sharing often is addressed in consent, there is usually no separate language for sharing of linked
data. In examples where the consent does address record linkage, the language is sometimes very
specific to a particular study or linkage. For example, the DS-DETERMINED study consent describes
linking to associated EHR data and DS-Connect surveys, and in the NCHS/NDI implementation, the NCHS
survey data are deemed eligible to link with NDI based on whether a survey participant gives consent for
data linkage in the survey. Consent language for some implementations, such as the recommended
language from NDA, anchors the scope of linkage to the use of a specific PPRL tool (i.e., the NDA GUID
tool) and covers sharing of linked data across all studies hosted within one data repository (NDA). While
All of Us participants consent for linking to data from “other sources,” the linked data are shared
specifically through the All of Us Research Workbench, which includes two tiers that both function as
enclaves and do not allow data exchange with external data systems.
The CARING for Children with COVID study datasets span multiple repositories, and as additional
pediatric COVID studies generate and share data, the desired scope of linkage could grow beyond the
datasets and repositories known today. The two CARING for Children with COVID studies that do
address record linkage in their consents (MUSIC and one of the PreVAIL kIds studies) do so in a broad
manner anchoring the scope of linkage to “multiple projects and databases” or “other research studies”
(Appendix Table 1
).
Pediatric COVID research highlights the value of this broad manner of consent when it is difficult to
anticipate the scope of studies and data repositories to be incorporated into a record linkage
implementation. Additionally, rather than naming a specific data repository for future sharing, consent
language could describe baseline expectations for sharing data in repositories designated or controlled
by NIH from which those data might be shared for future use, as has been done in several of the consent
examples shared in this report.
There are also distinct considerations for linking consented data with data collected without consent,
such as data from health care systems or public health surveillance and other administrative sources.
The Project Team documented an approach to authorizing this type of linkage by specifically obtaining
consent for linkage to unconsented/administrative data sources. The All of Us consent process explicitly
describes planned linkage with “other sources” and lists specific administrative data sources as
examples (e.g., pharmacy records, health insurance records, or cancer registries) and an additional
form
44
is required to obtain and link with EHR records (Appendix Table 4). Another example not fully

NICHD Office of Data Science and Sharing (ODSS) Page 39 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
assessed in this report is the Health and Retirement Survey, for which participants consent specifically
for linkage with CMS records (Appendix Table 5).
For pediatric studies that require consent, in addition to obtaining consent from a legal guardian, the
minor should be given age-appropriate information and included in the permission/assent process to
the greatest extent possible
45
. Reconsent is required when the minor reaches the age of majority if the
protocol is still active and the study team continues to hold the personal information (e.g., PII) of the
study participants. This multi-level consent adds an additional layer of consideration to obtaining
consent for immediate linkages, and more so when the scope includes potential linkages in the future.
However, a record linkage implementation with pediatric data would not require reconsent to continue
linking to new data that are submitted to a repository if all data in the repository(ies) are de-identified
and the longitudinal linkages are derived from PPRL-based hashed codes rather than PII itself. While the
Project Team did not analyze it as part of this project due to limited information available publicly, the
National Survey of Child and Adolescent Well-Being (NSCAW) administered by the Administration for
Children and Families (ACF) provides language that addresses the various consent/assent scenarios for
linking and sharing linked data, including reconsenting as an adult
46
. Consents for both All of Us and
NSCAW also address the option for participants to withdraw their consent/assent and prevent future
data sharing, with the caveat that data that have already been linked and shared with users will not be
withdrawn (Appendix Table 5
).
Based on the analysis of existing implementations, the Project Team developed example consent (for
the legal guardian, the language refers to “your child”) and assent language (for the child, the language
refers to “you”) to address broadly scoped linkage and sharing of linked data that could be used for
studies like CARING for Children with COVID. Note that the scope of this example language is limited to
other studies that the child may participate in, meaning this language may not be appropriate for linkage
with data that are collected for other purposes (e.g., unconsented administrative data sources), which
may warrant additional data source-specific language. Additionally, example language should be
considered in the context of other key elements and requirements of consent
47,48
.
If [you/your child] join this study, we will gather data about [you/your child]. What we learn
in this study will be put in a secure NIH-designated storage location, called a data repository,
where these data would be shared for future research. Information about [you/your child]
will be “de-identified,” which means it will not include anything that identifies [you/your
child]. [NIH] will approve researchers from all over the world to access information from the
repository. Researchers will agree not to attempt to identify [you/your child]. It is possible
that if [you/your child] participate[s] in more than one study, researchers may be able to
combine de-identified data from multiple studies to ease the burden on researchers and
participants alike. The purpose of sharing this information is to make more research possible
that may improve children’s and everyone’s health. This sharing of information will be done
without obtaining additional permission from [you/your child].
If [you/your child] no longer want [your/your child’s] de-identified data to be shared with
researchers and combined with other data about [you/your child], you can request
[your/your child’s] data to be withdrawn from the data repository and destroyed. Please
note that any data that has already been shared with researchers cannot be withdrawn.
NICHD Office of Data Science and Sharing (ODSS) Page 40 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
If [you/your child] turns 18 years old while taking part in this study, [you/your child] will be
asked to review and sign an informed consent form as an adult if [you/your child] wants to
continue to be in the study.
Across all the record linkage implementations, the Project Team observed that the governance
frameworks for linking and sharing linked data are often more complex when consent for data collection
is lacking such that additional controls are needed. Examples include restricting access to a specific
enclave (N3C) or a physically restricted area (Census and administrative datasets accessed at the FSRDC),
or creating the linked dataset only for a specific research project and then destroying the linkages and
dataset after use (PEDSnet).
8.1.1.2
Data Originator/Submitter and/or IRB or an Equivalent Privacy Board Determination
For data that are collected under the auspices of consent for research, but for which the consent may
not necessarily include explicit language addressing record linkage, other mechanisms have been
employed to authorize the record linkage. These mechanisms include authorization from the data
originator/submitter and/or their institution, which may include approval or determination from an IRB
or an equivalent Privacy Board. In many cases, the authorization to share linked data is addressed by the
general requirements for data sharing for a given implementation; therefore, this discussion focuses on
the linkage-specific authorizations.
BRICS and NDA require record linkage (via the use of the GUID) for all data submissions. This
requirement is clearly described in data submission agreements and associated policies, although
exceptions are reviewed case-by-case
49
. The NDA Data Submission Agreement requires an Authorized
Organizational Representative to sign on behalf of the data originator/submitter’s research institution,
confirming that the data submission is consistent with informed consent. By agreeing to the terms of the
submission agreements, the data submitter and their institution authorize the GUID-based record
linkage. Several NIH programs that use BRICS also expect that an IRB and/or an equivalent Privacy Board
has verified that the data submission is consistent with informed consent. This expectation means that
the submitter’s IRB and/or an equivalent Privacy Board weighs in on the decision to link data as part of
the submission. An IRB and/or an equivalent Privacy Board could, in theory, then require updates to the
informed consent or administer a waiver of consent for the linkage; however, such behind-the-scenes
procedures are typically not tracked by these NIH data repositories.
While dbGaP does not implement PPRL, it does implement some cross-dataset linkage based on
submitted subject IDs that are detected to likely represent the same participant in an existing study. In
such cases, it is up to the submitter to decide whether the linkage should be incorporated into the
dbGaP data repository. While under the NIH GDS Policy
50
, the Institutional Certification
34
used for dbGaP
expects that an IRB and/or an equivalent Privacy Board has ensured that the data submission is
consistent with the informed consent, cross-study record linkage is not an element of this agreement. In
fact, the topic of data linkage was raised in a recent Request for Information (RFI) on Proposed Updates
and Long-Term Considerations for the NIH GDS Policy
51
, specifically, whether data linkage should be
addressed when obtaining consent for sharing and future use of data under the GDS Policy, as well as in
IRB consideration of risks associated with submission of data to NIH genomic data repositories.
For datasets that are collected under a waiver of consent and use PPRL, the waiver of consent is often
combined with approval from the data originator/submitter (e.g., N3C data partner, PEDSnet site, CODI
data partner) and/or some other governing body (e.g., PEDSnet Data and Steering Committees, CODI’s

NICHD Office of Data Science and Sharing (ODSS) Page 41 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
CHORDS Research Council) to authorize the linkage on behalf of the participants. In the CODI
implementation, for example, the linkage also requires approval from the data originator’s IRB or an
equivalent Privacy Board or a designation as non-human subject research.
While institutional oversight is a critical component of making decisions regarding sharing data
52
and
data linkage (as described above), the Secretary’s Advisory Committee on Human Research Protections
(SACHRP) response
40
to the above-mentioned RFI on the NIH GDS Policy suggests that IRB or an
equivalent Privacy Board approval may not be an appropriate path when making determinations about
de-identified (i.e., non-human subjects) data submissions that fall outside the scope of an IRB’s
jurisdiction. However, for both consented and unconsented scenarios, this report uncovered several
potential reasons why it might be appropriate for an IRB or an equivalent Privacy Board to engage in the
decision to link data:
• While a given data submission (and downstream secondary use of the data) may not qualify as
"human subjects research," data linkage could result in the data originator obtaining access to
additional information about participants for whom identities are known, if the data originators
still hold PII for their study. Therefore, when the IRB protocol for a given study is still active, it is
appropriate for the IRB or an equivalent Privacy Board representing the data originator/
submitter to weigh in on the decision to link because the breadth and depth of the data
collected under the protocol could be expanded through linkage to additional data accessible in
the repository. The NDA Data Use Certification (DUC) describes this situation for researchers
accessing data from NDA who may have also submitted data to the NDA (Appendix Table 2
).
• IRB approval is often appropriate for non-PPRL record linkage approaches where the direct PII
are sent to other locations to identify matches (e.g., FSDRC/Census). In addition to explicit
consent regarding record linkage that occurs within the All of Us system boundary, the consent
informs participants that if PII is shared with an outside entity to link records from external
sources, All of Us will file an amendment with the All of Us IRB.
• For PPRL implementations where the PPRL software is installed locally on the data originator’s
server, PII does not leave the boundary of the data originator’s organization. Studies that are
not able to use or install PPRL software locally at the data source may need local IRB approval to
send the PII to another location such as a DCC to generate the information needed for PPRL.
• Data that are subject to the NIH GDS Policy require an IRB and/or an equivalent Privacy Board to
ensure that the data sharing is consistent with the informed consent and to determine whether
the use of the data are limited to certain purposes (i.e., data use limitations). Therefore, it may
also be appropriate for the IRB to weigh in on the decision to link de-identified genomic data
with other datasets.
• Finally, even for implementations that follow the HIPAA Safe Harbor method
18
of de-
identification, linkage of individual datasets comprising multiple data types may alter the status,
whereby the “identity of the human subjects cannot readily be ascertained” as defined in the
Common Rule, is no longer applicable
53,54,55
. This report did not uncover any scenarios where a
record linkage implementation evidently crossed this threshold based on the richness and/or
diversity of data types that were linked, but the implementations already have multiple re-
identifiability risk mitigation processes in place (see Section 8.1.6
). Providing an IRB or
an
equivalent Privacy Board the opportunity to weigh in on the decision to link for a given
implementation affords the opportunity for them to assess potential re-identification risk. In
scenarios where the planned scope of the linkage is known, this review may provide the IRB or
NICHD Office of Data Science and Sharing (ODSS) Page 42 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
equivalent Privacy Board an opportunity to determine the appropriateness of linkages with
specific datasets (e.g., similar to when an N3C data partner can opt out of linking with specific
external datasets such as mortality data); however, potential re-identification risks can be
difficult to predict and determine when the scope of data linkage is constantly expanding
through the addition of new datasets.
8.1.1.3
Federal Authority
Federal authorizations based on U.S. Code laws and regulations may permit the unconsented collection
and linkage of individual-level datasets containing PII for statistical use, public health surveillance, or
public health emergency response. Of the 13 record linkage implementations assessed, two non-PPRL
implementations—Census/FSRDC and NCHS/NDI—use PII matching for data linkage under federal
authority for statistical use. However, any external data sources that are ingested to be linked to these
federally authorized datasets are expected to be authorized for linking and sharing based on informed
consent and/or IRB approval. The federal authorization mechanism and PII-based record linkage
approach is not applicable to the CARING for Children with COVID studies.
8.1.2
Key consideration 2: Linkage of certain types of data or data from certain populations
may be subject to additional policies or governance
The analysis of the 13 record linkage implementations did not uncover many specific considerations for
addressing unique linkage requirements for certain data types or modalities (e.g., EHR, survey, imaging,
genomic), as for the most part, the rules that govern a given implementation or repository apply to all
data modalities represented in that implementation. However, the NIH GDS Policy
50
and the recent RFI
51
contain two conditions for sharing NIH-funded genomic data that relate to data linkage:
• “For studies initiated after the effective date of the GDS Policy, NIH expects investigators to
obtain participants’ consent for their genomic and phenotypic data to be used for future
research purposes and to be shared broadly.”
• “(D)ata should be de-identified to meet the definition for de-identified data in the HHS
Regulations for Protection of Human Subjects
12
and be stripped of the 18 identifiers listed in the
HIPAA Privacy Rule
5
.”
While the current GDS policy does not explicitly address record linkage, sharing genomic data that are
linked with a HIPAA limited dataset would result in a violation of the GDS definition of “de-identified."
This topic warrants further consideration pending the outcomes of the RFI
51
.
The Project Team sought to understand how various implementations address record linkage for data
from participants from U.S. tribal and international settings. The Project Team’s findings indicate that for
the 13 record linkage implementations assessed, either: 1) the scope of the implementation was limited
to data from the United States and non-tribal populations, or 2) the implementation did not have
unique requirements for these populations (i.e., they would be subject to the same data submission and
data access/use agreements and policies as all other datasets represented in the implementation). Some
tribal or international data requirements may not be consistent with the linkage and data sharing
policies of a given implementation
56
. Such cases may require additional agreements and governance
approaches or may preclude data from inclusion in any type of data linkage or data sharing activities.
NICHD Office of Data Science and Sharing (ODSS) Page 43 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
8.1.3
Key consideration 3: A broad set of PII elements are required to generate high quality
linkage regardless of the tool used, and these PII elements should be collected early
and in a standardized manner
To successfully implement PPRL, certain PII elements must be collected by the data originator/submitter
based on both software requirements and linkage quality considerations. IRB approval is generally
required to collect the PII needed for generating record linkage in the research setting. This requirement
for IRB review may constrain the PII elements available for linking if the study is already underway and
the protocol has already been approved (i.e., submitting protocol modifications to an IRB to collect
additional PII elements could place study timelines at risk).
While some of the PPRL tools assessed in this report are extremely flexible in terms of the PII elements
they can use for record linkage (i.e., they do not require specific PII elements to use their products for
PPRL), the Project Team observed that most tools typically rely on first name, last name, date of birth,
and sex or gender. Most tools also use some sort of “location” information, either in the form of
city/municipality of birth or household address or ZIP Code. In the seven PPRL implementations the
Project Team reviewed for governance approaches, first name, last name, date of birth, and gender or
sex were common to all seven. Four out of seven PPRL implementations the Project Team reviewed also
relied on cell or phone numbers, including one of the pediatric-focused studies (CODI), although the
initial NICHD-led pilot assessment identified phone number as a potential challenge for linking data from
children. The CARING for Children with COVID studies all collected first name, last name, date of birth,
and sex that are used by most tools.
8.1.3.1
PII Element Standardization
For any PPRL implementation, standardization of all PII elements, including clear definitions, should
improve linkage quality
57,58
. For example, gender or sex are collected in all 11 of the CARING for Children
with COVID studies and gender or sex is used in all seven PPRL implementations the Project Team
assessed. However, the definitions of gender versus sex were not assessed in this Report and there is
evidence that these elements are used interchangeably in studies such as RADx (“sex” is a common data
element for RADx-rad and “gender” is used in its NHash tool). In general, sex and gender are often
conflated, and a majority of current studies/systems use “Birth Sex,” “Administrative Gender,” or just
“Sex.” The HL7 Gender Harmony Project
59
recommends expanding the elements to include: gender
identity, recorded sex or gender ("used to more accurately identify sex values or gender values that are
specified in a particular source or documents such as identity cards or insurance cards"), sex for clinical
use (defined as "a summary sex classification element based on one or more clinical observations such
as an organ survey, hormone levels, and chromosomal analysis"), name to use, and pronouns. Given
that the CARING for Children with COVID studies are already collecting sex or gender, using both sex and
gender as distinct PII elements might be preferable when implementing PPRL for these studies.
Of the 11 CARING for Children with COVID studies included in this project, only two collect
city/municipality of birth (required for BRICS and NDA GUIDs). An alternative to city/municipality of birth
is to use either ZIP Code or address. Eight CARING for Children with COVID studies collect and four of the
seven PPRL implementations use some iteration of ZIP Code and one study used household street
address. The Project Team assumes, but could not confirm, that these refer to the participants’ current
home ZIP Code and address.

NICHD Office of Data Science and Sharing (ODSS) Page 44 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
For the use case “facilitate longitudinal data collection and analysis,” using current home location
information could be challenging given that people move an average twice before turning 18 and 11.7
times across their lifetime
60
. As records of home location information are collected at various time
points, this information could be unreliable for longitudinal data linkage. If the ZIP Code collected refers
to the location of study enrollment rather than home address, it would not enable linking data from
multiple studies and sources. ZIP Codes that refer to the location of birth may be more stable, but this
element is not significantly different from city/municipality of birth (used in NDA and BRICS), which may
in fact be easier for a participant or their parent/guardian to remember and easily provide when
enrolling in a study.
Pediatric data may not contain some of the PII elements typically available for adult linkage, such as
phone number and email address. Phone number was noted as a challenge with the CARING for
Children with COVID studies. For the youngest research participants, hospital record naming
conventions such as “Baby Girl Jones” are yet another limitation to PPRL. Researchers have proposed
collecting additional PII elements to facilitate pediatric record linkages, such as mother’s maiden
name
61
.
8.1.3.2
PII Element Preprocessing
Preprocessing of PII elements prior to using PPRL software is a common practice to account for data
entry errors or misspellings in study datasets. Some PII elements useful for PPRL are truncated or
derived versions of PII elements already collected, such as first three characters of first name, Soundex
of first and/or last name, and a less specific ZIP Code (e.g., ZIP2 or ZIP3). Of the seven PPRL
implementations the Project Team reviewed, only PEDSnet and the three N3C implementations
reported using Soundex to account for possible name misspellings. Although not documented in this
assessment, PEDSnet anecdotally expressed that Soundex introduces and raises the noise levels leading
to increased false positives. Some of these results may be due to the fact that PEDSnet used populations
that included families (i.e., multiple people with similar last names) for PPRL. Similarly, N3C indicated
that email address and SSN led to high match failure rates.
8.1.3.3
PII Element Combinations for Tokenization
The Project Team compared the PII elements that are collected and those that can be derived in the
CARING for Children with COVID studies to the PII elements required for existing NDA, BRICS, N3C and
PEDSnet PPRL implementations assessed in the Governance Assessment (Section 6
). The Project Team
fo
und that none o
f these PPRL implementations could be leveraged for CARING for Children with COVID
as many of these studies lack location information like ZIP Code and municipality of birth/country.
Furthermore, tools that currently require use of email, phone number, and/or SSN, would not work for
CARING for Children with COVID as these elements are generally not collected by the studies. In general,
analysis of the seven PPRL implementations showed that combinations of five or more PII elements are
used in all of them. While the existing PII elements collected in the CARING for Children with COVID
studies could enable linkage in PPRL tools that allow for flexible PII element inputs, the currently
available PII may not be robust enough to support high-quality linkage. The CARING for Children with
COVID studies will likely have to revise their IRB protocols to obtain additional PII elements, possibly
including city/municipality of birth or some sort of location at birth information, or possibly other
elements (mother’s name) to yield high-quality record linkage, regardless of the PPRL tool adopted.
Prior to any PPRL impl
ementation, the selected PII elements, data standardization and preprocessing
approaches, and matching algorithms should undergo rigorous statistical assessments using relevant
NICHD Office of Data Science and Sharing (ODSS) Page 45 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
gold standard datasets to identify the configuration that delivers an acceptable threshold for false
positive and false negative matches. In the case of pediatric studies, an adult-focused dataset would not
serve as a sufficient gold standard.
8.1.4
Key consideration 4: The three-party linkage approach offers researchers the flexibility
to link and use datasets hosted in different data systems
For each record linkage implementation, the Governance Assessment documented which parties have
access to the PII needed to generate hashed tokens in PPRL, which parties use the hashed codes/tokens
(that typically do not qualify as “PII” but are often treated as sensitive) to identify matches (entity
resolution), and which parties use the resulting linkage information (either in the form of GUIDs or
linkage maps) to create a linked and de-duplicated dataset (data linking). In a two-party model, the
entity resolution and the data linking are performed by the same organization whereas in a three-party
model, these two activities are performed by separate organizations (the first party is always the PII
holder). PPRL implementations that use a three-party model utilize either an honest broker or a
separate GUID server to perform entity resolution (i.e., match hashed codes/tokens) and generate
GUIDs or linkage maps for repositories and researchers. The GUID server or honest broker does not
share the matched hash codes/tokens with the repositories or the researchers using the data.
The Project Team observed that the record linkage implementations using a three-party linkage model
(e.g., dbGaP, NDA, BRICS, N3C) typically do so by providing researchers access to individual datasets
alongside either GUIDs or linkage maps. This approach means that linking of data (i.e., merging and
deduplicating) is not performed centrally, but the information needed to “link” is made available to
researchers who are then responsible for linking and de-duplicating the data as part of their analysis
approach. Each individual dataset is treated separately, which enables the user to:
• Make decisions about potentially conflicting data from multiple sources (e.g., different
diagnoses from different data originators)
• Track new data that are added for a given participant over time (and determine which studies
have long-term follow-up information)
• Attribute the data to the original studies
• Follow requirements of any specific data use limitations that may be associated with a given
dataset (e.g., a dbGaP dataset that has a disease specific data use limitation for the study of
COVID is more constrained than a dataset that is approved for General Research Use, even if the
same participant is represented in both)
In two-party linkage implementations the entity resolver is also the data linker. For several of the two-
party linkage implementations the Project Team reviewed, the entity resolver and data linker creates a
merged and deduplicated dataset without providing researchers any provenance information for the
original records nor any decisions made regarding deduplication (e.g., PEDSnet, CODI, All of Us,
Census/FSDRC). In these cases, a common data access governance approach is applied to the merged
data product. For example, in CODI, all data contributors agree that the merged dataset is the property
of the DCC—the dataset provided is stripped of information linking specific participants to the data
sources/original sites and it can be used by approved data requestors (both within and outside of CODI).
This two-party approach could be difficult to accommodate in a federated ecosystem designed to share
many study datasets in a manner that is consistent with consent-based data use limitations, where such
limitations should travel with the data such that two separate datasets have different approved uses
NICHD Office of Data Science and Sharing (ODSS) Page 46 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
even if the same participants are represented in both
52
. Consent-based data use limitations, such as
those described in the NIH GDS Policy
24
can require very specific research use for a given dataset and
should be conveyed to end users to reduce risk for data management incidents.
8.1.5
Key consideration 5: The linked database model encompasses a broad scope of
datasets and should be paired with additional controls to protect participant privacy
The majority of the PPRL implementations the Project Team reviewed utilize a linked database model,
meaning the scope of linkages created and made available to researchers is defined by the datasets
associated with a given database. The linked database model can also span one or multiple data
repositories (e.g., multiple BRICS instances that use the same GUID server, N3C linking with Class 0
datasets like MIDRC) or encompass only a subset of datasets within a given repository. For example, N3C
EHR data partners choose to participate in PPRL and can opt out of linkage with specific external
datasets; however, the hashed codes/tokens generated are designed to link all participating N3C
datasets.
Study-specific linkages are typically created as needed for a specific research study or scientific question.
In PEDSnet and CODI, for example, each data contributor chooses whether to participate in a specific
study and associated linkage. Study-specific linkage provides data originators the opportunity to choose
on a case-by-case basis whether to contribute their data to a given research study. However, such study-
specific linkage presents scaling challenges given the time to review/approve each request and to re-run
linking processes. Further, this approach introduces reproducibility challenges, as linkage information is
only recreated in response to each study request and is not maintained over time.
The linked database model approach of maintaining a persistent database of GUIDs or linkage maps
fosters reproducibility and continuous tracking of longitudinal data without having to recreate the
linkage information for every use. Further, the linkage information in a linked database model can be
shared using a variety of access controls. For example, while most dbGaP data are controlled-access,
dbGaP shares cross-study dbGaP subject IDs in an open manner, whereas BRICS and NDA provide GUIDs
as part of an approved data access request. N3C requires an additional approval (Letter of
Determination from the requester’s IRB) to access PPRL-generated linkage maps across participating
EHR datasets. While the scope of the N3C linkage encompasses external (Class 2/Class 0) datasets,
access to those datasets and associated linkage information must be specifically requested and
combined analysis with data from other repositories (e.g., MIDRC/Class 0) must be performed in an
ephemeral workbench.
PPRL implementations could take similar modular approaches by generating linkage information that
spans a broad scope of datasets while requiring that the researchers who would like to obtain the GUIDs
or linkage maps specifically apply for and/or request this information. Controlling access to GUIDs or
linkage maps could be a component of a data access request for each dataset; however, rather than
generating a unique set of linkage information on-demand for every request, the linkage information
that is provided would be the same for all approved requesters. Regardless of the method chosen, the
scope of the linkage should be clear from the beginning of a project so it can be communicated to study
participants (via consent) and/or to data originators/submitters, and so a plan for making the linkage
information available to users can be strategized with the parties sharing the data (i.e., data
repositories).

NICHD Office of Data Science and Sharing (ODSS) Page 47 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
8.1.6
Key consideration 6: Re-identification risk management controls can be implemented
both prior to and after linkage
While all record linkage implementations the Project Team reviewed shared data that are considered
de-identified, six of the 12 implementations that are currently sharing linked data (DS-DETERMINED is
currently not sharing the linked data) also used some form of re-identification risk assessment (where
the datasets that are to be linked are assessed for potential re-identification when combined) or other
risk management controls at various stages in the linking and sharing process. Some of these risk re-
identification assessments/controls take place prior to data linkage or data submission to a repository,
which may result in excluding the dataset or modifying the data to reduce the potential for re-
identifiability (e.g., N3C Tools and Resources Committee classification of linkage with external datasets).
Other assessments are performed after the data have been linked but prior to sharing (e.g., All of Us,
FSDRC/Census) and may result in modifications to the linked dataset prior to research use or data
export. In the case of N3C, for example, an internal Tools and Resources Review Committee reviews the
linkage of the EHR data from within N3C to external datasets (Class 0 and Class 2) for any potential re-
identification risks, and data may be modified for certain datasets (e.g., using zip3 instead of zip5 for
tribal reservations). All of Us takes multiple steps to fully strip all data of PII prior to sharing, and
additionally performs a number of transformations on data that are shared in their registered tier versus
the controlled tier (e.g., share more detailed demographic information and genomic data in the
controlled tier). In contrast to other data repositories, however, All of Us’s registered tier functions more
like an enclave and currently the requirements for accessing data from All of Us are not different
between the registered and controlled tier. The FSDRCs only allows users to access read-only de-
identified versions of approved files and further require the application of “Disclosure Avoidance
kk
”
techniques prior to removing analytical results and statistical products from the FSDRC workspaces and
approval from the Disclosure Review Board prior to publishing or disseminating findings.
While the NICHD-funded Data Sharing for Demographic Research (DSDR)
62
repository was not included
in this assessment, DSDR reviews all data for disclosure risk from both direct identification and
inferential re-identification. DSDR also checks data for sensitivity and special populations such as
children. When possible, DSDR remediates the data sufficiently (e.g., by masking direct identifiers,
suppressing certain variables, collapsing categories, and perturbing responses) to allow public access via
download from the DSDR website. For data that still have inferential disclosure risk (e.g., geographic
identifiers and detailed personal histories) or sensitive information (e.g., drug abuse and sexual activity),
DSDR provides controlled access via its restricted tiers. DSDR considers data linkage projects as
restricted. For access to restricted data, researchers must submit a research plan, IRB or Ethics Panel
review and pledges to protect the confidentiality of the data. Restricted access requires the execution of
a data use agreement (DUA) with the researcher’s organization to cover any mishandling of the data.
While this approach is extremely robust, it may not be scalable or feasible to implement a full disclosure
review for all linkage scenarios or requests to access or download linked data across a federated
ecosystem.
kk
At the U.S. Census Bureau, disclosure avoidance is defined as a process used to protect the confidentiality of
respondents' personal information.

NICHD Office of Data Science and Sharing (ODSS) Page 48 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
In addition to these specific re-identification risk mitigation procedures, the Project Team identified
several broader approaches that have been implemented across the 13 record linkage implementations
that serve to manage re-identification risk. These include:
• Using a controlled or enclave data access model, and requiring approval from a data access
committee, steering committee, or some other governing body prior to accessing the linked
data
• Setting a clear definition of de-identified fo
r all data shared from a repository or for all data
shared in each access tier (e.g., the Genomic Data Sharing Policy currently requires adherence to
the Common Rule and HIPAA Safe Harbor definition of de-identified, PEDSnet has specific
masking rules, All of Us uses different thresholds for each of its tiers, N3C requires additional
rules for HIPAA-limited and PPRL-linked data, and other implementations require removal of the
18 HIPAA identifiers)
• Prohibiting
re-identification through Data Use Agreements or terms of access
• Where feasible, performing a broader risk assessment to determine which (if any) data elements
require more data access controls than others
8.1.7
Key consideration 7: All PPRL tools assessed for this Project meet a basic set of
capability requirements, but vary on certain desirable features
A PPRL implementation for the CARING for Children with COVID studies requires certain essential
capabilities in the PPRL tool selected: (1) the PPRL tools must accommodate a broad set of PII (see
Section 8.1.3) and be scalable, (2) the vendors should have no rights to the data (including PII elements,
associated metadata), and (3) the tool must have appropriate protections for the source data/PII
elements. All seven vendors/organizations assessed met these essential capabilities.
The PPRL tool selected must be able to accommodate hashed codes/tokens from many participants and
sites because there are already at least 150 sites contributing data to the CARING for Children with
COVID studies. As new pediatric COVID studies are launched
63
, it would be valuable to link records with
existing CARING for Children with COVID studies that have been enrolling participants since the start of
the pandemic, to increase data richness and track data collected over time.
The PPRL Technology Assessment conducted in this project examined the largest use case with real
world data as a proxy for the scalability of each product. All tools supported scaling in upwards of a
million records, which is more than sufficient for the number of records generated in CARING for
Children with COVID studies. For many of these tools, the limiting factor on how many records can be
processed is not the software itself but the computational power available, such as memory, central
processing unit (CPU) and random access memory (RAM), so scalability will be dependent on the set up
of the application server/environment.
Most tools use proprietary algorithms for their PPRL products, which raises a concern about whether the
vendors have any right to the PII elements, the hashes/tokens generated from the PII elements, or other
data that may be sent to or used by the software (e.g., metadata or derivative software data).
Vendors/organizations for all assessed tools indicated in their survey that they do not retain rights to
these data or metadata. Additionally, all the vendors/organizations indicated the source data never
leave the data originator’s computer/server; therefore, no unhashed PII elements are ever accessible to
anyone other than the data originator. Several of the vendors surveyed do offer services that allow data
originators to link their data to external commercial datasets in a “data-as-a-service marketplace”
NICHD Office of Data Science and Sharing (ODSS) Page 49 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
model. This assessment did not review the governance models for these arrangements and the topic
merits further review.
All seven tools assessed meet the required criteria for the CARING for Children with COVID studies. The
Project Team identified additional features that are desirable, but not required, that differentiate the
tools assessed. These features include pre-processing/data cleaning, tuneability of matching algorithm,
ease of use, and federal security certification.
Prior to generating the hashes, the data cleaning and/or pre-processing of PII elements step
standardizes and recodes the PII to reduce data entry errors and generate more consistent hashes. This
preprocessing can improve the ability to resolve potential linkage matches and substitution of
nicknames (e.g., “Jim” to “James"). While manipulation of PII elements must be consistently applied
across all CARING for Children with COVID studies for the best quality linkage
38
, such pre-processing and
standardization could be performed prior to using the PPRL software; hence, it is not a required criterion
for a PPRL tool.
Soundex is a type of phonetic encoding that occurs during PII pre-processing and was considered to be a
desirable characteristic to accommodate name misspellings in children. Only some tools have this
feature, and it may introduce risk of high false positive matching rates
64
. For example, some PPRL
implementations, such as PEDSnet, indicated that Soundex yields an inflated number of false positives
when performing linkage that includes families and its use should therefore be considered carefully.
Tunable matching criteria/algorithms (e.g., comparison/classification parameters that can be
configured) for entity resolution is another capability that is a “nice to have,” as PII elements may
contain fields with potential errors and such errors can be accommodated by accepting certain
imperfect matches. Examples of adjusting hash matching criteria include choosing a matching design
with either a higher false positive rate or a higher false negative rate and changing the number of match
parameters. The ability to adjust the linkage schema and associated weights of each combination of PII
elements provides the flexibility needed to improve the ability to detect true matches based on the
availability of certain PII elements when using pediatric data. There may also be certain use cases for
which a different ratio of false negatives to false positive is desirable. For example, a conservative
approach (reducing the number of false positives) better supports the data generation and analysis
goals of the CARING for Children with COVID use cases, while a higher rate of false positives could be
tolerated in use cases that involve contacting sites to potentially recruit participants to a new study (this
approach is currently being explored by N3C
65
). Some tools allow tuning/configuration of their linkage
algorithm for a specific PPRL implementation. Tuning the algorithm and adjusting the linkage schema
does impact all linkages in a given implementation, so the final matching algorithm configuration should
be based on an assessment utilizing a gold standard dataset that closely matches the data sources for
the given implementation.
Similarly, making linkage performance reports available to the data originator (e.g., number of matches,
number of possible matches, number of duplicates) that describe the performance of a software also
helps adjust for and identify errors in the data, such as name misspellings and other reasons potential
linkages should be identified and reviewed. For example, a PPRL tool may provide the data originator
the option to create matches with the option of indicating “With Close Match Checking” (provides some
fault tolerance for PII entry errors) or “No Close Match Checking" (only matches the PII entered) but do
not otherwise have the flexibility to adjust the matching threshold.

NICHD Office of Data Science and Sharing (ODSS) Page 50 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Six of the seven tools the Project Team surveyed generate linkage performance reports. While useful for
informing real-time linkage decisions, these reports should not be used as proxies for rigorous data
linkage quality assessments for a given hashing/matching protocol.
FISMA/FedRAMP certifications are rigorous federal authorization processes that are tailored to ensure
an Information Technology system has an adequate plan for security, clear security responsibilities,
periodic review, and authorization to operate in a Federal IT environment. Although a FISMA/FedRAMP
certification is not a requirement for a PPRL tool for use with the CARING for Children with COVID
studies, these certifications demonstrate compliance with federally approved system security controls
for managing sensitive data. Some tools the Project Team assessed either have or are in the process of
achieving FISMA and/or FedRAMP status. All PPRL tools the Project Team assessed have basic security
features (e.g., encrypted data transfer, database encryption) and security certifications (such as FISMA
certification, SOC 2) in place and are considered truly “privacy preserving” in the sense that PII does not
leave the data originator’s environment.
If PPRL technology is to be implemented at study sites, the ease of use of a product is a “nice-to have”
characteristic for the CARING for Children with COVID study investigators who are generally constrained
for resources and time. Therefore, product usability features such as the presence of a GUI may be a
priority. Having a GUI to enter/upload the PII elements for hashing, instead of requiring the study
sites/researchers to programmatically ingest the data, simplifies the PPRL process for users. Most of the
tools have GUIs, whereas implementing those PPRL products that do not have GUIs would require
additional development efforts, which would result in increased budget and prolonged timelines.
Researchers with more technical experience/knowledge of PPRL have indicated that they would
appreciate the ability to use web services and scripts to automate token generation. All the tools the
Project Team assessed include this feature of automated token generation.
8.1.8
Key consideration 8: Certain PPRL tool features better serve robust implementation
approaches and sustainability
Although all PPRL tools assessed for this project meet a basic set of capability requirements (see Section
8.1.7),
tool features such as the ability to persist hashes over time, software ownership, the cost of
maintenance, and potential interoperability with other PPRL tools impact a tool’s utility for long-term
implementation of record linkage for NIH pediatric studies.
Some of the CARING for Children with COVID studies are longitudinal, and as new studies are rolled out
in the future, it would be ideal to utilize a PPRL tool that can persist hashes and the associated metadata
such that new hashes can be matched with existing hashes without needing to regenerate them from PII
elements. Regenerating hashes may not be feasible for past data submissions (e.g., the investigator may
no longer be engaged or have access to the PII) and could result in additional run time and effort for
entity resolution, increased budget, and other resource considerations when implementing PPRL for
longitudinal data collection. Six of the seven tools the Project Team assessed are able to persist hashes.
Once a PPRL tool is selected for a record linkage implementation, the program/initiative is generally
“locked-in”
ll
to using that product for the duration of the record linkage implementation. A few
commercial PPRL vendors self-reported that they are developing interoperability solutions, but their
methodologies have not been published nor implemented at the time of this assessment. Although
ll
Locked-in, as used here, refers to the operational algorithms and configurations that are distinct and are not interoperable.
NICHD Office of Data Science and Sharing (ODSS) Page 51 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
BRICS is not interoperable with other vendors, BRICS does have a robust network of NIH institutes that
use its Centralized GUID Server approach for PPRL linkages. As long as the proper approvals are in place
(authorizations, research plan approvals, data use agreements, etc.), BRICS is able to link data from 250+
studies across NIH institutes.
The overall cost to NIH to maintain a PPRL implementation over multiple years factors into the
sustainability of the PPRL solution. The tools the Project Team assessed use different cost models. These
cost models include an annual licensing fee in addition to pricing based on the number of sites
generating tokens, a one-time configuration fee plus a licensing fee per site, a fee based on the number
of sites and the scale of the linkage, a fee based solely on the number of records ingested, and only
upfront costs relating to initial setup and any desired customizations to the product. The differences in
these cost models emphasize the differences between commercial vendors (Crossix, Datavant,
HealthVerity, Senzing, and IQVIA) and the government-owned PPRL product (BRICS) and the university-
owned product (NHash):
• The commercial PPRL products have per site or per record costs in addition to some products
with annual licensing fees.
• The government/university owned products only charge for initial configuration.
While assessing the overall implementation cost of these tools was not within the scope of this Project,
these variations in cost models impact consideration for long-term sustainability. Over time, cost models
with only initial set-up costs could be more cost effective than the annual, per site, and/or per record
commercial cost models.
Additionally, commercial vendors are subject to fluctuations in the private market. During the analysis
performed by NCI, the number of candidate software products shrank from eleven to eight as
companies merged or went out of business and products were deprecated.
The NCI report assessed Anonlink, which is an open source PPRL tool used by CODI for pediatric linkage.
This assessment does not include Anonlink as they did not respond to Project Team inquiries. Since
Anonlink has been adopted by CDC’s CODI, further evaluation may be warranted for the tool.
In order to generate a direct comparison of overall costs for each of the PPRL tools in this report, the
CARING for Children with COVID studies would need to finalize technical details of the PPRL
implementation model (requirements for customization, the number of study sites, estimated number
of records generated in the CARING for Children with COVID studies, and specifications/desired
architecture for a GUID server or honest broker).
8.2 Considerations for CARING for Children with COVID
Prior to moving forward with record linkage across the CARING for Children with COVID studies, the
collaborators should agree on an overall approach and the planned scope for the record linkage. A
three-party approach, where approved researchers link the data themselves, should be adopted to
maintain separation between the entity resolver and the data repositories that share the data and
associated linkage information (e.g., GUIDs and/or linkage maps). This approach will enable maintaining
provenance of individual datasets (e.g., tracking which data are from PRISM versus MUSIC, attribution,
which datasets are modified over time) and will allow data access and use requirements (e.g., consent-
based data use limitations
52
) to “travel” with individual datasets. The three-party approach could use
either an NIH GUID tool-based approach (where the GUID server is the entity resolver) or leverage a
NICHD Office of Data Science and Sharing (ODSS) Page 52 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
commercial or open source tool and identify a separate party to play the role of the honest broker and
create linkage maps.
A linked database model is likely the most sustainable and reasonable approach to foster reproducibility
and could encompass many datasets across multiple repositories as long as the same PPRL tool and
entity resolver are used by all repositories. The collaborators should define up front the general scope of
datasets to be included in the implementation, so the linkage (and associated benefits and risks) can be
communicated during the informed consent process to participants and/or data submitters. A relatively
broad scope is required given that CARING for Children with COVID data spans multiple programs
(including RADx) and data repositories (RADx Data Hub, BioData Catalyst, the Kids First Data Resource,
and ImmPort), and there could be a desire to include in the PPRL implementation additional pediatric
COVID studies funded by other programs. One way to establish a clear but broad scope of linkages is to
define the scope of linkage to data associated with NIH-designated or NIH-controlled repositories and/or
other NIH studies. An NIH-defined scope could be too limiting for certain research use cases but would
address the current CARING for Children with COVID use cases.
The CARING for Children with COVID initiative plans to perform whole genome sequencing to
understand inherited risks associated with SARS-CoV-2 infection in children; therefore, these studies are
subject to the requirements of the NIH GDS Policy. Under the current GDS Policy, it may be possible to
link to data from typically “unconsented” sources such as administrative datasets if explicit consent for
linkage and sharing the linked data is obtained in the context of the research studies (thereby changing
the status of the administrative data to “consented”). However, certain administrative datasets (e.g.,
Census) may prohibit re-distribution of their data through other repositories.
If the outcome of the GDS RFI determines that explicit consent is not required for linking consented
genomic data to unconsented data sources, and re-consent is not feasible, thoughtful consideration and
possibly further governance analysis should go into determining whether linkage with unconsented data
sources is appropriate for the CARING for Children with COVID studies. If this type of linkage is agreed
upon by the collaborators, it should be explicitly communicated to all other data contributors who
participate in the linkage scope. Additionally, the collaborators could consider an approach where data
contributors can “opt in” or “opt out” of linkage with a specific unconsented dataset (similar to what has
been done with administrative datasets that are “external” to but linked with N3C EHR data).
The current GDS definition of “de-identified” prohibits linkage with HIPAA limited datasets; however,
adhering to both the Common Rule definition of “de-identified” and HIPAA Safe Harbor, which excludes
certain dates and location information, helps mitigate some re-identification risk that may be introduced
when linking across multiple studies and data types. If the outcomes of the GDS RFI changes the
definition of de-identified under the GDS Policy, additional de-identification standards (e.g., expert
determination
18
) or re-identification risk assessments (e.g., those done by All of Us) could also be
considered.
CARING for Children with COVID study datasets vary in terms of controlled and registered tier data
access requirements. Linkage between datasets of any tier introduces potential re-identification risk,
and this risk can be mitigated through an NIH approval process. For a CARING for Children with COVID
record linkage implementation, the linkage information (e.g., GUIDs, linkage maps) should be
maintained in a controlled access tier, which incorporates an NIH review process and institutional
oversight
66
. With this approach, the original tier status of all unlinked datasets need not change, and
researchers who obtain approval to access the controlled data and linkage information will be able to

NICHD Office of Data Science and Sharing (ODSS) Page 53 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
deduplicate participant data in their analyses. This proposed implementation is similar to the approach
N3C is planning for the EHR-EHR linkage, where level 2 (de-identified) data will not include the PPRL-
generated linkage maps, but users can update their data use request and provide the required LOD from
their IRB to access the level 3 (HIPAA limited dataset), which includes the linkage maps across all
participating EHR datasets.
For the CARING for Children with COVID studies, this approach will require determining which system(s)
will be responsible for maintaining and distributing the linkage information (created by the GUID server
or honest broker) and how it will be operationalized. This approach should be feasible with minimal
development work, given the current authentication/authorization framework of the data repositories
involved. Its implementation will also require determining which data access committees should provide
oversight over the approval to access the linkage maps and how that oversight will be incorporated into
the dbGaP data access request process used by the Kids First Data Resource, BioData Catalyst, and the
RADx Data Hub for managing controlled access data.
Researchers should be able to receive linkage maps that encompass all datasets they are authorized to
access, in line with all of the three-party models reviewed in the assessment. However, for datasets that
are available only in the registered tier and are not associated with a controlled access dataset, a
mechanism needs to be established for users to request that portion of the linkage map as well. To
reduce unnecessary data linkage access requests, each of the repositories could display whether a given
study has common participants with other CARING for Children with COVID studies and list those
studies. This approach should be reviewed by the CARING for Children with COVID program leadership
to ensure there are no edge cases that would introduce reidentification risk to study participants.
Only two CARING for Children with COVID studies currently address record linkage in their informed
consent forms. MUSIC includes language that implies broad linkage with “other research studies” and
one of the PreVAIL kIds has already consented for broad use of a “GUID” approach (Appendix Table 1
).
While the consents from the other studies do not prohibit linkage activities, it would be ideal to explore
the feasibility of re-consenting participants to include explicit record linkage information (drawing from
the example language developed in this report) or seeking institutional approval for linkage, when re-
consent is not feasible.
Since these studies use the GDS Institutional Certifications to certify that the data sharing is appropriate
and consistent with the consent and as record linkage is not addressed in these documents, it is
appropriate to provide submitting institutions the opportunity to certify whether the planned linkage is
appropriate, with input from their IRB and/or equivalent Privacy Board. As part of this certification
process, information should be provided outlining the planned scope of the linkage and how the linked
data will be shared through the data repositories (i.e., linkage information will be available to “approved
researchers” via controlled access only).
There may be data collected from tribal and/or international participants for the CARING for Children
with COVID studies. If there are tribal or international laws, regulations, or policies that conflict with the
proposed process or outcomes of a record linkage implementation and cannot be addressed by
additional agreements, certain participants may need to be excluded from the PPRL implementations.
Additionally, it may be appropriate to consider sharing pediatric COVID data associated with tribal
nations through the forthcoming Tribal Data Repository
67
.
NICHD Office of Data Science and Sharing (ODSS) Page 54 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
The selection and implementation of a specific PPRL tool could constrain the scope of linkage to a
particular set of data originators that are able to use that tool with their studies. This assessment
demonstrates that nearly all PPRL tools can support the basic requirements for pediatric record linkage,
including a three-party linked database model, as well as the proposed governance considerations. The
primary technological limitation is the minimal common subset of PII elements collected across the
CARING for Children with COVID studies. However, as additional PII is likely needed to yield high-quality
linkages with pediatric data (e.g., standardized location information, even new elements like mother’s
maiden name), essentially any PPRL tool in this assessment would be acceptable. Using an NIH-owned
software for a long-term and large-scale implementation strategy may preclude the need for new
vendor contracts, avoid vendor-associated costs, and reduce risk of vendor business model
modifications (e.g., mergers/acquisitions, bankruptcy) that could adversely affect tool maintenance and
longitudinal linkage.
Finally, a long-term pediatric-wide approach to record linkage may look different than responding to the
urgent needs of the pandemic, CARING for Children with COVID, and other pediatric COVID use cases. If
possible, it may be appropriate to leverage existing record linkage implementations that already include
some pediatric COVID studies and expand the scope to encompass the CARING for Children with COVID
and other relevant studies. Further discussions would be needed to ensure the generation of high-
quality linkages for children and that a robust governance approach is configured to share the linkage
information across multiple repositories charged with sharing pediatric COVID data in the federated NIH
data ecosystem.
8.3 Limitations of this Assessment & Future Directions
This assessment represents a snapshot of the landscape of record linkage to support biomedical
research. The findings and considerations in this assessment will serve as a useful guidepost for the
design and implementation of new PPRL implementations. The Project Team identified and prioritized
several additional topics that merit further investigation as they were either outside the scope of this
report or were not uncovered in the specific implementations assessed, including:
• Linkage quality assessments must be performed prior to selecting and implementing any PPRL
tool. These assessments should be based on combinations of PII input elements, matching
algorithms that are relevant to the features of a given record linkage implementation, and
acceptable error rates based on the purpose of a given implementation. In parallel, the
feasibility of collecting relevant PII elements across data submitters should be assessed. In this
assessment, the Project Team surfaced challenges associated with PII elements currently used
by various implementations but uncovered the potential opportunity for using additional and
novel elements such as mother’s maiden name. The Project Team also collected some anecdotal
information from interviews and some quality-relevant information from vendors; however,
performing a linkage quality assessment requires rigorous testing against a relevant gold
standard dataset, the development of which is time and cost intensive. Such efforts are
currently being taken on by research teams within NCI, ONC
68
, and N3C. Ideally, these efforts
should make available gold standard datasets for future quality assessments.
• While this report highlights the importance of transparent yet flexible consent, participant
attitudes towards explicitly consenting for linkage, transparency in how data are used in future
research, and linkage with unconsented data were out of scope for this project and should be
assessed further. The discussion from the NIH Policy and Ethics of Record Linkage Workshop
25
NICHD Office of Data Science and Sharing (ODSS) Page 55 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
which was convened by the NIH Office of Data Science and Sharing, should be the starting point
for these assessments.
• While many vendors publicly share some cost information, actual costs of PPRL are nearly
impossible to determine without first making key decisions for a new record linkage
implementation, including whether an honest broker will be needed, the duration of the desired
linkage, the PII combinations or “tokens” that will be used, the number of sites, and the number
of linkages.
• Multiple PPRL vendors purport to solve the lock-in challenge through development of
interoperability methods such as the “token bridging” approach. This solution appears to be
nascent technology that warrants rigorous analysis before any PPRL implementation includes
this feature as a dependency.
• Finally, as technology rapidly evolves, there could be unforeseen nuances not captured or fully
addressed in this report.
Researchers and other stakeholders can learn from existing record linkage implementations, as well as
the key considerations and observations documented in this report, taking into account the specific
requirements for a given implementation. While technology continues to advance rapidly, there is
nearly an endless number of ways it can be used and it is critical for policy and governance approaches
to be planned in advance of and inherently incorporated in its implementation, especially when sharing
data from human participants.

NICHD Office of Data Science and Sharing (ODSS) Page 56 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
9 GLOSSARY
Table 13: Glossary of Terms
Term Definition
Aggregate data Summary statistics compiled from multiple sources of individual-level data. (NIH)
Authorization The function of specifying access rights/privileges to resources. (HHS)
Authorized users Any appropriately provisioned individual with a requirement to access an information
system. (NIST)
Blocking (or indexing) Blocking or indexing “splits each database into smaller blocks according to some
blocking criteria.” It identifies a smaller set of potential candidate pairs without having to
compare every single pair in the full comparison space. (NCHS)
Bloom filters A data structure that can be used for efficiently checking membership to a set and
whether two sets approximately match. (Schnell, R., Bachteler, T., Reiher, J.: Privacy-
preserving record linkage using Bloom filters. BMC Med Inform Decis Mak 9(1) (2009)
DOI: 10.1186/1472-6947-9-41)
Common data model (CDM) A CDM standardizes the definition, format and model content of data across participating
data partners so that standardized applications, tools and methods can be applied.
(PCORnet)
Controlled access Application and eligibility requirements need to be met and approved (e.g., by a data
access committee) to gain access. (NIH)
“Controlled access” and “access controls” refer to measures such as requiring data
requesters to verify their identity and the appropriateness of their proposed research use
to access protected data. (NIH)
[see also data access model]
Clear text Information that is not encrypted. (NIST)
Data access committee (DAC) The DAC is responsible for reviewing all requests for access to datasets from external
requestors and is composed of individuals with expertise in science, policy, or
bioinformatics resources. (NIH)
Data access models Four types of data repository access models (NIH):
o Controlled access: Application and eligibility requirements need to be met to gain
access
o Registration required: Open to all, but users need to be signed in or registered
with the resource to access
o Open access: No access restrictions or registration required to access
o Mixed: Has both controlled and open access.
Data coordinating center (DCC)
The DCC is an organization that coordinates large multi-site clinical research
programs/trials and can provide common questionnaires, data collection forms and data
management; statistical analysis; overall study training, coordination and quality
assurance; support of ancillary study activities; support of websites or online resources
for the program; and more. (NIH)
Data linkage model Describes the scope of the datasets that are part of the linkage implementation. As defined
in this Report:
o Linked database model—where the linkage information that is created and/or
provided encompasses all datasets in a given database
o Study-specific model—where linkage information is created and/or provided for
the purposes of a specific study
NICHD Office of Data Science and Sharing (ODSS) Page 57 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Term Definition
Data originator/
contributor/submitter
Institutions/organizations/researchers that collect data from patients or study participants
or that collect administrative data; they may also be the party to submit the data to a
repository for sharing.
Data steward A formal position or an assigned accountability with responsibility for the following areas:
o Adherence to an appropriately determined set of privacy and confidentiality
principles and practices
o Appropriate use of information from the standpoint of good statistical practices
(such as by not implying cause and effect when the data only point to correlation)
o Limits on use, disclosure, and retention
o Identification of the purpose for a specific use of the data
o Application of “minimum necessary” principles
o Verification of receipt by the correct recipient, wherever possible
o Data de-identification (HIPAA-defined and beyond)
o Data quality, including integrity, accuracy, timeliness, and completeness
(NCVHS
)
Data use agreement (DUA)
A DUA establishes who is permitted to use and receive data, and the permitted uses and
disclosures of such information by the recipient. (HHS modified)
Data user A person who is authorized by the Data Access Committee (DAC) or equivalent to
access and/or analyze data. (N3C)
Disclosure Disclosure relates to inappropriate attribution of information to a data subject, whether
an individual or an organization. Disclosure occurs when a data subject is identified from
a released file (identity disclosure), sensitive information about a data subject is revealed
through the released file (attribute disclosure), or the released data make it possible to
determine the value of some characteristic of an individual more accurately than
otherwise would have been possible (inferential disclosure).(HHS)
De-duplication The process of removing redundant patient records from a database. (CDC)
De-identification De-identified patient data is patient information that has had personally identifiable
information (PII; e.g., a person’s name, email address, or SSN), including protected
health information (PHI; e.g., medical history, test results, and insurance information)
removed. This is normally performed when sharing the data from a registry or clinical
study to prevent a participant from being directly or indirectly identified. (NIH)
Dictionary attack An attack where an attacker uses pre-computed tables to reverse engineer the inputs to
the hash. For example, if some possible contents are known to be contained in the
inputs to the hashing function (such as name, date of birth, etc.), an attacker can
construct rainbow lookup tables by producing hashes from the complete set of
potentially valid input values. (Dong X, Randolph DA, Rajanna SK. Enabling Privacy
Preserving Record Linkage Systems Using Asymmetric Key Cryptography. AMIA Annu
Symp Proc. 2020 Mar 4;2019:380-388. PMID: 32308831; PMCID: PMC7153159)
Electronic health records (EHRs)
EHRs are electronic versions of the paper charts in your doctor’s or other health care
provider’s office. An EHR may include your medical history, notes, and other information
about your health including your symptoms, diagnoses, medications, lab results, vital
signs, immunizations, and reports from diagnostic tests such as x-rays. (HHS)
Enclave A data enclave is a secure network through which confidential data, such as identifiable
information from census data, can be stored and disseminated. In a virtual data enclave,
a researcher can access the data from their own computer but cannot download or
remove it from the remote server. Higher security data can be accessed through a
physical data enclave where a researcher is required to access the data from a
monitored room where the data is stored on non-network computers. (NNLM)
[see also data access model]

NICHD Office of Data Science and Sharing (ODSS) Page 58 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Term Definition
Encoding/Hashing Encoding: Using a system of symbols to represent information, which might originally
have some other representation. Example: Morse code, phonetic encoding, hashing.
(NIST)
Hashing: A method of calculating a relatively unique output (called a hash digest) for an
input of nearly any size (a file, text, image, etc.) by applying a cryptographic hash
function to the input data. (NIST
)
Encryption Cryptographic transformation of data (called “plaintext”) into a form (called “ciphertext”)
that conceals the data’s original meaning to prevent it from being known or used. If the
transformation is reversible, the corresponding reversal process is called “decryption,”
which is a transformation that restores encrypted data to its original state. (NIST)
Encryption key A cryptographic key that has been encrypted using an approved cryptographic algorithm
in order to disguise the value of the underlying plaintext key. (NIST)
Entity Resolution Process of joining or matching records from one data source with another that describe
the same entity. (Census)
In PPRL, hash codes/tokens are used to match individual records without using PII/PHI.
(N3C)
Federal Information Security
Modernization Act (FISMA)
The Federal Information Security Modernization Act of 2014 requires that agencies
maintain programs that provide adequate security for all information collected,
processed, transmitted, stored, or disseminated in general support systems and major
applications. FISMA requires an annual independent evaluation to determine
effectiveness of information security programs. (HHS)
Federal Risk and Authorization
Management Program
(FedRAMP)
FedRAMP is a government-wide program that provides a standardized approach to
security assessment, authorization, and continuous monitoring for cloud products and
services. (HHS)
Geocoding The process of inputting an address and receiving back latitude/longitude coordinates
calculated along an address range. (Census)
Global Unique Identifier (GUID) The GUID is a subject ID that allows researchers to share data specific to a study
participant without exposing personally identifiable information (PII). The GUID is made
up of random alpha-numeric characters and is NOT generated from PII/PHI. (
NIH)
Governance Governance, as defined in this Report, comprises of the policies, processes, and
controls that address ethics, privacy protections, compliance, risk management, or other
requirements for a given record linkage or privacy preserving record linkage (PPRL)
implementation.
Hash codes/tokens An encrypted value created by an irreversible conversion algorithm and any underlying
Protected Health Information that has been de-identified using the expert determination
method as described under HIPAA regulations at 45 CFR 164.515(b)(1). (N3C)
The string of bits which is the output of a hash function. A hash function maps a bit string
of arbitrary length to a fixed-length bit string using an algorithm that computes a
numerical value (called the hash value) on a data file or electronic message that is used
to represent that file or message and depends on the entire contents of the file or
message. A hash function can be considered to be a fingerprint of the file or message.
(NIST
)

NICHD Office of Data Science and Sharing (ODSS) Page 59 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Term Definition
Health Insurance Portability and
Accountability Act (HIPAA)
Privacy Rule
The Standards for Privacy of Individually Identifiable Health Information are codified in
45 C.F.R. Parts 160 and 164 promulgated by the U.S. Department of Health and Human
Services under the Health Insurance Portability and Accountability Act (HIPAA) of 1996.
The HIPAA Privacy Rule establishes national standards to protect individuals' medical
records and other individually identifiable health information (collectively defined as
“protected health information”) and applies to health plans, health care clearinghouses,
and those health care providers that conduct certain health care transactions
electronically. The Rule requires appropriate safeguards to protect the privacy of
protected health information and sets limits and conditions on the uses and disclosures
that may be made of such information without an individual’s authorization. The Rule
also gives individuals rights over their protected health information, including rights to
examine and obtain a copy of their health records, to direct a covered entity to transmit
to a third party an electronic copy of their protected health information in an electronic
health record, and to request corrections. (
HHS Health Information Privacy)
Honest broker A party that holds de-identified tokens (“hashes”) and operates a service that matches
tokens generated across disparate datasets to formulate a single Match ID for a specific
use case. (N3C)
Individual-level de-identified data Health information that is not individually identifiable (if it does not identify an individual
and if the covered entity has no reasonable basis to believe it can be used to identify an
individual). (HHS)
Institutional Review Board (IRB) An IRB is the institutional entity charged with providing ethical and regulatory oversight
of research involving human subjects, typically at the site of the research study. (NIH)
An Institutional Review Board is an appropriately constituted group that has been
formally designated to review and monitor biomedical research involving human
subjects. An IRB has the authority to approve, require modifications in (to secure
approval), or disapprove research. This group review serves an important role in the
protection of the rights and welfare of human research subjects. (
FDA)
Interoperability According to section 4003 of the 21st Century Cures Act, the term 'interoperability,' with
respect to health information technology, means such health information technology
that— "(A) enables the secure exchange of electronic health information with, and use of
electronic health information from, other health information technology without special
effort on the part of the user; "(B) allows for complete access, exchange, and use of all
electronically accessible health information for authorized use under applicable State or
Federal law; and "(C) does not constitute information blocking as defined in section
3022(a)."
(HIT)
Key escrow The system responsible for storing and providing a mechanism for obtaining copies of
private keys associated with encryption certificates, which are necessary for the
recovery of encrypted data. (NIST)
Letter of determination (LOD) An LOD documents an IRB decision on the status of research. (HHS)
Limited dataset (LDS) Datasets containing protected health information but excludes the following 16 HIPAA
direct identifiers: names, postal address information, other than town or city, State, and
ZIP Code, telephone numbers, fax numbers, electronic mail addresses, SSNs, medical
record numbers, health-plan beneficiary numbers, account numbers, certificate and
license numbers, vehicle identifiers and serial numbers, including license plate numbers,
device identifiers, and serial numbers
Web Universal Resource Locators (URLs), Internet Protocol (IP) address numbers,
Biometric identifies including fingerprints and voice prints, Full-face photographic images
and any comparable image. (HHS)
Linkage Map As defined in this Report:
A linkage map is a crosswalk between participant-level IDs across disparate datasets
and repositories.

NICHD Office of Data Science and Sharing (ODSS) Page 60 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Term Definition
Metadata Information describing the characteristics of data including, for example, structural
metadata describing data structures (e.g., data format, syntax, and semantics) and
descriptive metadata describing data contents (e.g., information security labels). (NIST)
Non-technical controls Non-technical security and privacy controls and activities include such actions and things
as: administrative controls (policies, training, risk management, risk assessment,
workforce security and privacy) and physical controls (facility access controls,
maintenance records, work area use, work are security and privacy, and contingency
operations) (NIST)
Open Access Data within this category presents minimal risk of participant identification. Access to
these data does not require user certification, and researchers may explore data content
without restriction. (NCI)
No access restrictions or registration required to access (NIH)
[see also data access model]
Patient Identifier Unique data used to represent a person’s identity and associated attributes. (NIST)
Personally identifiable information
(PII)
Any information that can be used to distinguish or trace an individual's identity, either
alone or when combined with other information that is linked or linkable to a specific
individual. (NIST and CODI)
Privacy preserving record linkage
(PPRL)
A technique identifying and linking records that correspond to the same entity across
several data sources held by different parties without revealing any sensitive information
about these entities (
UK Office for National Statistics)
Protected Health Information
(PHI)
Individually identifiable health information that is transmitted or maintained in any form or
medium (electronic, oral, or paper) by a covered entity or its business associates,
excluding certain educational and employment records. (NIH)
Pseudo Global Unique Identifier A Pseudo-GUID is a unique ID that is not based on PII. This is a random ID that can be
used as a placeholder when PII is not available, and "promoted" to a real GUID when
the information is obtained at a future date.(NDA)
Record Linkage Combining information from a variety of data sources for the same individual. (AHRQ)
Registered Tier Registration required; open to all, but users need to be signed in or registered with the
resource to access the system, tool or data. Sometimes requires a “click-through”
agreement first. (
NIH)
[see also data access model]
Repository A repository is a place to store and make available data that includes research results,
publications, and scientific data for usage. (NIH)
Salt
Random data used to modify the input of a hash function to guarantee a unique output.
This adds another hashing layer on top of an encryption algorithm, increasing the
difficulty of reversing the encryption of the data. (UK Office for National Statistics)
Secret key
A cryptographic key, used with a secret key cryptographic algorithm, that is uniquely
associated with one or more entities and should not be made public.(NIST)
Soundex Soundex is a phonetic index/encoding that codes names based on the way a name
sounds rather than on how it is spelled. (NIST)
Study_ID An arbitrary, study-specific, site-agnostic, unique identifier that identifies a patient in a
Project dataset. (CODI)
Subject ID As defined in the Report:
Refers to a de-identified subject/participant identifier that can be generated by hashing
or non-hashing methods. If hashing is used, it is different from a hash code/token
(hashed ID) generated using a PPRL tool (as in RADx-rad PreVAIL kIds projects and
PEDSnet).

NICHD Office of Data Science and Sharing (ODSS) Page 61 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Term Definition
Technical controls The security controls for an information system are primarily implemented and executed
by the information system through mechanisms contained in the hardware, software, or
firmware components of the system. (NIST)

NICHD Office of Data Science and Sharing (ODSS) Page 62 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
10 ACRONYMS
Table 14: Acronyms
Acronym
Expansion
ABCD
Adolescent Brain Cognitive Development
ACF
Administration for Children and Families
ACS
American Community Service
ACS
American Community Survey
ADHHS
Alaska Department of Health and Social Services
AES
Advanced Encryption Standard
AHRQ
Agency for Healthcare Research and Quality
ALCAN
Alaska Longitudinal Child Abuse and Neglect Linkage
AMIA
American Medical Informatics Association
AoU
All Of Us
API
Application Programming Interface
BAM
Binary Alignment Map
BEA
Bureau of Economic Analysis
BRICS
Biomedical Research Informatics Computing System
CAPS
Committee on Access, Privacy, Security
CARING for Children
with COVID
Collaboration to Assess Rick and Identify LoNG-term outcomes for Children with COVID
CDC
Centers for Disease Control and Prevention
CDR
Curated Data Repository
cdRNS
Common Data Repository for Nursing Science
CHORDS
Colorado Health Observation Regional Data Service
CIT
Center for Information Technology
CMI
Child Maltreatment Incidence
CMS
Centers for Medicare & Medicaid Services
CODI
Childhood Obesity Data Initiative
CPU
Central Processing Unit
DA
Disclosure Avoidance
DAC
Data Access Committee
dbGAP
Database of Genotypes and Phenotypes
DBMS
Database Management Systems
DCC
Data Coordinating Center
DHDN
Distributed Health Data Network
DMR
Data Management Resource
DOD
Department Of Defense
DRC
Data and Research Center
DSA
Data Sharing Agreement
DSC
Down Syndrome Connect

NICHD Office of Data Science and Sharing (ODSS) Page 63 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Acronym Expansion
DSDR Data Sharing for Demographic Research
DUA Data User Agreement
DUCC Data User on Code Conduct
DUR Data Use Request
EHR Electronic Health Record
EMR Electronic Medical Record
EPA Environmental Protection Agency
ERB Ethics Review Board
ETL Extract Transformation and Load tool
FedRAMP Federal Risk and Authorization Management Program
FISMA Federal Information Security Management Act
FITBIR Federal Interagency Traumatic Brain Injury Research Informatics System
FSDRC Federal Statistical Research Data Center
GDS Genomic Data Sharing
GIID Government Issued ID
GRAF Genetic Relationship and Fingerprinting
GRDR Global Rare Diseases Data Repository
GUI Graphical User Interface
GUID Global Unique Identifier
GWAS Genome Wide Association Studies
HHS Health and Human Services
HIPAA Health Insurance Portability and Accountability Act
HPO Healthcare Provider Organizations
ICF Informed Consent Form
ICS Integrated Client Services
ID Identifier
IHQ Immunization History Questionnaire
IKDR International Kawasaki Disease Registry
IRB Institutional Review Board
IT Information Technology
JAAMH Joint Addiction, Aging, and Mental Health
JDBC Java Database Connectivity
JSON Javascript Object Notation
JWT JSON Web Token
KD Kawasaki Disease
KUMC University of Kansas Medical Center
LHB Linkage Honest Broker
LOD Letter of Determination
LSOA Longitudinal Study of Aging

NICHD Office of Data Science and Sharing (ODSS) Page 64 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Acronym Expansion
MAS Macrophage Activation Syndrome
MIDRC Medical Imaging and Data Resource Center
MIS Multisystem Inflammatory Syndrome
MIS-A Multisystem Inflammatory Syndrome in Adults
MIS-C Multisystem Inflammatory Syndrome in Children
MRN Medical Record Number
MSUA Master Sharing and Use Agreement
MUSIC Multisystem Inflammatory Syndrome In Children
N3C National COVID Cohort Collaborative
NCATS National Center for Advancing Translational Studies
NCHS National Center for Health Statistics
NCI National Cancer Institute
NCSES National Center for Science and Engineering Statistics
NDA National Institute of Mental Health Data Archive
NDI National Death Index
NEI National Eye Institute
NHANES Continuous National Health and Nutrition Examination Survey
NHEFS NHANES 1 Epidemiologic Follow-Up Study
NHGRI National Human Genome Research Institute
NHHCS National Home and Hospice Care Survey
NHIS National Health Interview Survey
NHLBI National Heart, Lung, and Blood Institute
NIA National Institute of Aging
NIAAA National Institute on Alcohol Abuse and Alcoholism Data Archive
NIAID National Institute of Allergy and Infectious Diseases
NICHD National Institute of Child Health and Development
NIDA National Institute on Drug Abuse
NIH National Institute of Health
NIH ODSS National Institute of Health Office of Data Science Strategy
NIMH National Institute of Mental Health
NINDS National Institute of Neurological Disorders and Stroke
NINR National Institute of Nursing Research
NIST National Institute of Standards and Technology
NNHS National Nursing Home Survey
NSCAW National Survey of Child and Adolescent Well Being
OAI Osteoarthritis Initiative
ODBC Open Database Connectivity
ODSS Office of Data Science and Sharing
OHA Oregon Health Authority

NICHD Office of Data Science and Sharing (ODSS) Page 65 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Acronym Expansion
OHRP Office for Human Research Protections
OHSU Oregon Health Sciences University
OMB Office of Management and Budget
PCORnet The National Patient-Centered Clinical Research Network
PDBP Parkinson’s Disease Biomarkers Discovery
PHI Personal Health Information
PID Participant Identifier
PII Personally Identifiable Information
PIK Personal Identification Key
PO Project Officer
POP02 or POPS Pharmacokinetics, Pharmacodynamics, and Safety Profile of Understudied Drugs Administered to
Children per Standard of Care
PPI Patient Provided Information
PPRL Privacy Preserving Record Linkage
PRAMS Pregnancy Risk Assessment Monitoring System
PreVAIL kIds Predicting Viral-Associated Inflammatory Disease Severity in Children with Laboratory Diagnostics and
Artificial Intelligence
PRISM Pediatric Research Immune Network on SARS-CoV-2 and MIS-C
PTSC Participant Technology System Center
PVS Person Identification Validation System
RAB Resource Access Board
RADx Rapid Acceleration of Diagnostics
RADx-ATP Rapid Acceleration of Diagnostics Advanced Technology Platforms
RADx-rad Rapid Acceleration of Diagnostics Radical
RADx-UP Rapid Acceleration of Diagnostics Underserved Populations
RAM Random Access Memory
RDC Research Data Committee
RDR Raw Data Repository
RECOVER Researching COVID to Enhance Recovery
RFI Request for Information
SC Subject Consent
SDI Self Determination Inventory from the Self-Determination Inventory System (SDIS) Data Dashboard
SDOH Social Determinants of Health
SHA-2 Secure Hash Algorithm 2
SIPP Survey of Income and Program Participation
SOA Supplement on Aging
SOC 2 Service Organization Control 2
SRA Short Read Archive
SSA Social Security Administration
SSM Subject Sample Matching
NICHD Office of Data Science and Sharing (ODSS) Page 66 of 130
PPRL for Pediatric COVID Studies – Final Report (September 2022)
Acronym Expansion
SSN Social Security Number
SSTR Sample Status Telemetry Report
TSS Toxic Shock Syndrome
U.K. United Kingdom
VCF Variant Cell Formula

Page 67 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11 APPENDIX
11.1 CARING for Children with COVID Studies – Supplemental Information
Appendix Table 1: Informed Consent Forms from Pediatric COVID Studies Selected for the Project
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
POP02 “If your child is seen at
another location, we may
ask you to sign a form to
allow us to get those
records. Examples
include medical history,
physical exam, recent
laboratory test results,
and evaluations over the
course of your child’s
hospitalization or clinic
visits. If these evaluations
are not already noted in
the medical record, they
may be performed for this
study and will be recorded
as study data.
Additionally, we will
collect whether your child
has or is participating in
other research studies”
Not available Linked data sharing: Not included in the consent form
General data sharing: “What we learn in this study will be
put in a database run by the National Institutes of Health
(NIH) to be shared for future research. This information will
not include anything that identifies your child.
All participants’ de-identified study data and any remaining
de-identified study samples will be submitted to a NIH-
designated storage location, such as the NICHD Data and
Specimen Hub or DASH (https://dash.nichd.nih.gov) or the
NIH database of Genotypes and Phenotypes or dbGaP
(https://www.ncbi.nlm.nih.gov/gap/) from which the data will
be shared with other researchers. Individual level genomic
data will be managed in a controlled-access manner.
Controlled access means that only researchers who apply
for and get permission to use the information and samples
for a specific research project will be able to access the
information. Your child’s genetic data, study samples and
health information, stored in these databases, will not be
labeled with your child’s name or other information that
could be used to identify them. Researchers approved to
access information in these databases will agree not to
attempt to identify your child. De-identified samples may
also be used by other researchers in the future to conduct
tests separate from those being done in the current study.
These researchers may conduct whole genome
sequencing; by doing WGS, these researchers may have
information that is unique to your child.
The purpose of sharing this information is to make more
research possible that may improve children’s health. This
will be done without obtaining additional permission from
you.
“You have the right to stop this
Authorization at any time. Your decision to
stop your Authorization will not involve any
penalty or loss of access to treatment or
other benefits to which you/ your child is
otherwise entitled. If you decide you no
longer want your child to participate in this
study, but do not stop your Authorization,
new health information may be collected
until this study ends.
To stop this Authorization, you should
inform the site investigator, as named on
the first page of this form, of your decision
in writing. Stopping your Authorization will
prevent sharing of PHI in the future but will
not affect any PHI that has already been
gathered or shared.”
“I have been told that if I become
an adult while enrolled in this
study, I will be asked to sign the
consent form.”
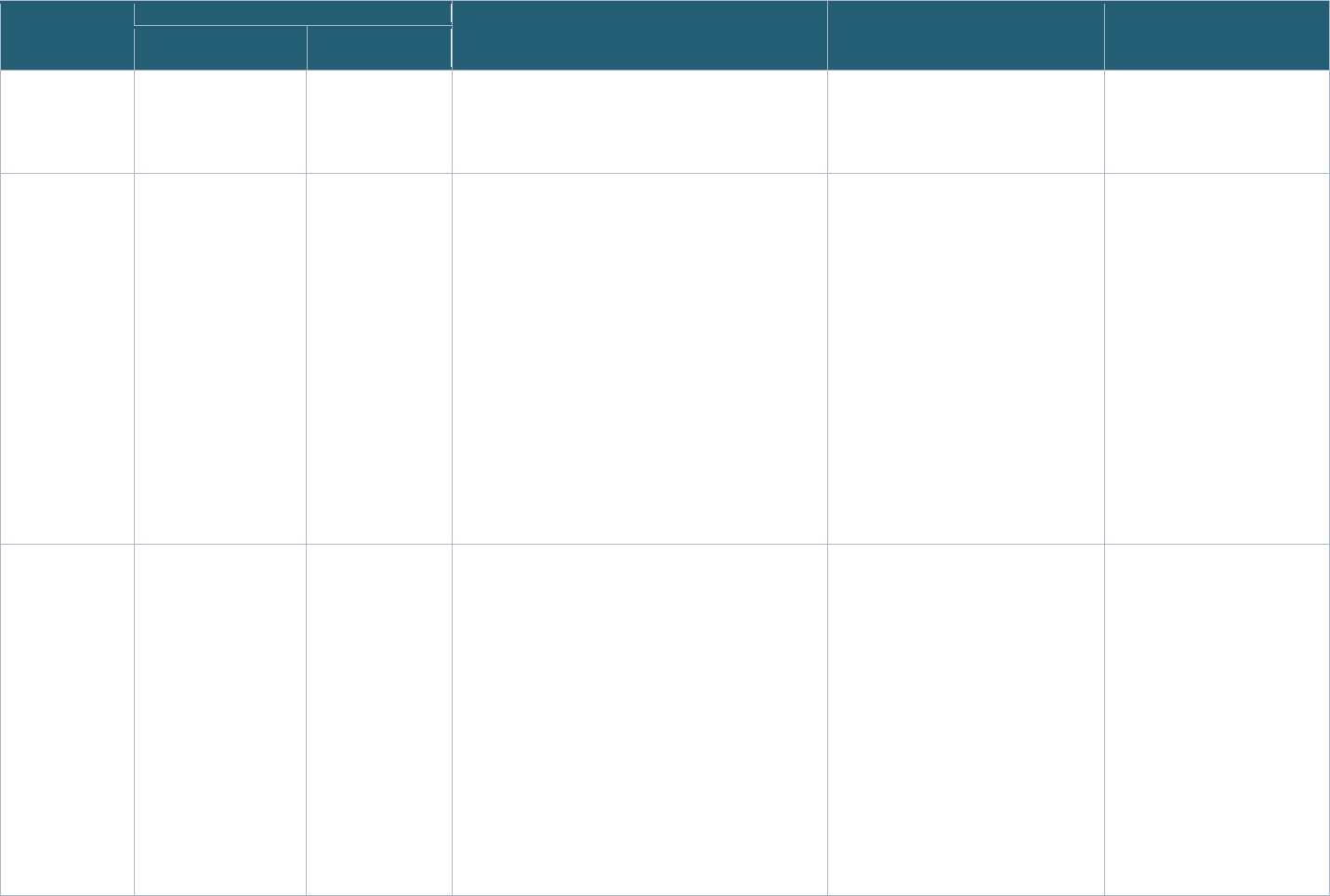
Page 68 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
The data and samples
collected in this study may be kept
forever. We may publish the results of this study. However,
we will not include your or your child’s name or any other
identifying information.”
MUSIC “Study information sent
outside of this institution
will be linked to your
[name] through a study
identification (ID) number.
The link between your
name and this study ID
number will be
kept in a locked, secure
area that only the study
team can get to.”
“If you participated in
other research
studies, we may
collect and share
information and lab
data between studies
to ease the burden
on research staff and
participants alike.
Data and samples
will be available to
researchers within
the PHN as well as
researchers from
other national and
international
institutions for the
study of MIS-C and
other diseases.”
Linked data sharing: Not included in the consent form
General data sharing: “I agree to have my data and
samples shared in a central biobank after PHN funding
ends for future studies in heart disease and other diseases.
‘Biobanking’ is st
oring health information and/or blood,
saliva, or tissue for future research studies. A ‘bank’ is the
place where it is stored. We would like your permission to
bank your blood (or saliva) sample for future research.
In the future, your
data (clinical and genetic) and sample
might be placed in a central data or biobank to make it
easier for researchers to use your samples and data for
research.”
“If you choose to provide a blood (or
saliva) sample for the biobank and later
want to withdraw your sample from the
biobank, it is important that you contact
study staff and tell them in-person or in
writing. You will have the choice to have
the study ID number removed from the
sample and leave the sample and existing
data in the biobank for future research. Or,
you may ask to have the unused samples
and data destroyed. If your sample is
placed in a central biobank, your personal
information was already removed so it
cannot be identified as you and therefore
cannot be removed. We will also not be
able to destroy samples and data that have
already been used or distributed for
research.
“If your child turns 18 years old
while taking part in this study,
he/she will be asked to review and
sign an informed consent form as
an adult if he/she wants to
continue to be in the study”
PRISM “Information about
you/your child and
samples collected for this
study may be shared with
other PRISM researchers.
We will not ask you for
additional permission
before sharing the
information with other
PRISM researchers.”
Not available Linked data sharing: Not included in the consent form
General data sharing: “
Once the data and samples have
been stripped of all personal information identifying
you/your child, they can be shared without your consent.
As a NIH-sponsor
ed clinical study, we serve the public by
sharing research information (data) that is collected during
a study with the scientific community to advance science
and health. We will keep the information resulting from this
study in a safe place, controlled by the NIH, called a central
data repository. The scientific information will be available
for future studies that may help future patients.
Your/your child’s
data may be stored for a very long time.
The information in the central data repository will not
contain personal information that would identify you/your
child, such as name, birthdate, address, etc. We will not
Once samples have been stripped of all
identifiers, or have been sent to another
laboratory, or have been used for a
research test, you will not be able to
change your mind about giving permission
for their use.”
Not available

Page 69 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
ask you for additional permission before sharing the
information in the central data repository.”
RADx-rad PreVAIL kIds Studies
AICORE-ids:
Artificial
Intelligence
COVID-19 Risk
AssEssment for
kids
(PI: Dr. Ananth
Annapragada)
Not available Not available Linked data sharing: Not included in the consent form
General data sharing: “Information that identifies you may
be removed from your identifiable private information
collected as part of this research, and after such removal,
your information may be used for future research studies or
distributed to another investigator for future research
studies without additional consent/authorization from you.
Sharing and Future Research Studies with Identifiable
Biospecimens Information that identifies you may be
removed from your identifiable biospecimens collected as
part of this research, and after such removal, your
biospecimens may be used for future research studies or
distributed to another investigator for future research
studies without additional consent/authorization from you.”
“Even after you have signed this form, you
may change your mind at any time. Please
contact the study staff if you decide to stop
taking part in this study. If you choose not
to take part in the research or if you decide
to stop taking part later, your benefits and
services will stay the same as before this
study was discussed with you. You will not
lose these benefits, services, or rights.”
Not available
Diagnosing and
Predicting Risk in
Children with
SARS-CoV-2
Related Illness
(PI: Dr. Jane
Burns)
Not available Not available Linked data sharing: Not included in the consent form
General data sharing: “Your c
hild's blood sample, body
fluids, or swabs may be used in additional research related
to Kawasaki disease to be conducted by the University of
California personnel for an unlimited period of time. These
samples may be shared with other investigators at other
institutions.”
“If you decide later that you do not want
the specimens collected from your child to
be used for future research, you may tell
this to Dr. Burns, who will use her best
efforts to stop any additional studies.
However, in some cases it may be
impossible to locate and stop such future
research once the materials have been
shared with other researchers.”
“When your child turns 18 years of
age, he /she will not be re-
consented for the continued use of
banked specimens.”
Discovery and
Clinical Validation
of Host Biomarkers
of Disease Severity
and Multi-System
Inflammatory
Syndrome in
Children (MIS-C)
with COVID-19
(PI: Dr. Charles
Yen Chiu)
Not available Not available Linked data sharing: Not included in the consent form
General data sharing: “Your information, blood, and saliva
will be sent to scientists in several laboratories. After our
study is over, your information and any leftover blood and
saliva will be sent to a central data repository at the
National Institute for Health (NIH) and will be available for
other scientists who are studying COVID to use in their
research. Any information that could be used to identify
you from your medical records or clinical samples will be
removed before it is given to scientists outside of UCSF.”
“If you decide later that you do not want
your sample and information to be used for
future research, you can tell us, and we will
destroy any remaining identifiable sample
and information if it is no longer needed for
your care.”
Not available
COVID-19 Network
of Networks
Expanding Clinical
“The research team may
use or share your or your
child’s information
Not available Linked data sharing: Not included in the consent form “You may also withdraw your
consent/permission for the use of data
already collected about you or your child,
Not available

Page 70 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
and Translational
Approaches to
Predict Severe
Illness in Children
(CONNECT to
Predict Sick
Children)
(PI: Dr. Lawrence
Kleinman)
collected or created for
this study with the
following people and
institutions: Rutgers
University Investigators
involved in the Study,
Non-Rutgers Investigators
on the Study Team:
National Institute of
Health, our study
sponsor; Yale University,
partner study sites; New
York Medical College,
partner study sites.
Those persons or
organizations that receive
your or your child’s
information may not be
required by Federal
privacy laws to protect it
and may share your
information with others
without your permission, if
permitted by the laws
governing them.”
General data sharing: “The research team is interested in
advancing the science to improve the diagnosis and
treatment of COVID for children, adolescents, and young
adults. Therefore, we may share remaining data and
biospecimens with other collaborators, including companies
after removal of all personal identifiers that could be linked
to your identity without obtaining additional informed
consent from you.”
“We may share your or your child’s health and genetic
information through databases at the National Institutes of
Health (NIH), including the database of Genotypes and
Phenotypes (dbGaP). By sharing this information, the hope
is to maximize the chance for researchers to use and learn
from your or your child’s information to better understand
the effects of the coronavirus that causes COVID-19 in
children, adolescents, and young adults. The NIH team will
work with Dr. Kleinman and the study team from Rutgers to
coordinate the secure transfer, storage, and access of your
or your child’s information. The NIH team will make sure
that this information cannot be used to identify you or your
child and that it remains password-protected and available
only to qualified researchers studying the coronavirus that
causes COVID-19.”
but you must do this in writing to Lawrence
Kleinman, M.D., M.P.H. at Children’s
Health Institute, 89 French Street, Room
1338, New Brunswick, NJ 08901, 732-235-
7906.
Any data that has already been sent to NIH
or its designee, such as another
organization that integrates data from
across studies at several institutions (also
known as a Data Coordinating Center) or
data that has been published cannot be
withdrawn because there may not be any
identifiers with the data.”
“You may change your mind and not allow
the continued use of your or your child’s
information (and to stop taking part in the
study) at any time. If you take away
permission, your information will no longer
be used or shared in the study, but we will
not be able to take back information that
has already been used or shared with
others. If you say yes now but change your
mind later for use of your or your child’s
information in the research, you must write
to the researcher and tell him or her your
decision.”
A Data Science
Approach to
Identify and
Manage
Multisystem
Inflammatory
Syndrome in
Children (MIS-C)
Associated with
SARS-CoV-2
Infection and
Kawasaki Disease
(KD) in Pediatric
Patients
“De-identified study data
(information that does not
allow anyone to
determine your child’s
identity by removing all
names, contact
information, medical
record number and any
other information that can
be linked directly to
you/you child) will be
combined with data from
all other sites participating
in this study. All IKDR
“As part of this study,
a Global Unique
Identifier (GUID) will
be created. A Global
Unique Identifier
(GUID) is a secure
one-of-a-kind code
that is assigned to
your child so we can
securely track your
child participating in
multiple projects and
databases.
Linked data sharing: Not included in the consent form
General data sharing: “External researchers can get data
directly from the IKDR. They will first obtain REB approval
and sign an agreement with the Hospital for Sick Children
(where the IKDR data coordinating center is located).
These agreements will control how your child’s study data
will be used. They will not be permitted to disclose or to
transfer study data to anyone else. They will also not be
permitted to use study data for purposes other than those
included in the agreements. Researchers will also agree
that they will not attempt to re-identify your child from their
study data. The information from this registry will be
“It is your choice, and your child’s, to
decide to take part in this study, and
participation is voluntary. You and your
child can change your mind at any time
during the research study.
The study team may
ask why you are
withdrawing your child for reporting
purposes, but you do not need to give a
reason to withdraw your child from the
study if you do not want to.
Withdrawal from the study will not have
any effect on the care your child or your
Not available

Page 71 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
(PI: Dr. Cedric
Manlhiot)
investigators are required
to have an agreement
with the Hospital for Sick
Children in Toronto to
participate in this study
and will be allowed to
access de-identified data.
Data might be accessed
by IKDR investigators for
approved research or to
participate in IKDR
research activity (e.g.,
helping with result
interpretation).”
Please let the study
team know if you
have any questions
or if you do not want
multiple studies that
your child is
participating in to be
linked through a
GUID.”
available only to researchers who have received REB
approval for their research.
Data will be deposit
ed in an approved National Institutes of
Health (NIH) data registry in the United States after they
undergo a process called anonymization where some
information such as dates are modified to further reduce
the possibility of re-identification. The information will be
merged with the information from other patients across the
world. The NIH has a strict process to ensure the security
and privacy of research data it makes available through
NIH-approved registries. Researchers will be required to
enter into an agreement with the NIH to obtain access to
the data. This agreement stipulates that researchers will not
be permitted to disclose or to transfer study data to anyone
else. They will also not be permitted to use study data for
purposes other than those included in the agreements.
Researchers will also agree that they will not attempt to re-
identify your child from their study data. Only researchers
from academic institutions may obtain access to data
through this mechanism.”
family will receive at SickKids. If you
decide to have your child leave the study,
you can contact a member of the study
team to let them know. If you no longer
want your child’s study information to be
used in this research, you can request your
child’s data to be withdrawn and
destroyed. Please note that any study data
that has been included as part of the
analysis or that has been shared cannot be
withdrawn.”
Diagnosis of MIS-C
in Febrile Children
(PI: Dr. Audrey
Odom John)
Not available Not available Linked data sharing: Not included in the consent form
General data sharing: “We will use and may share data
and/or specimens for future research. They may be shared
with researchers/institutions outside of CHOP. This could
include for profit companies. We will not ask for your
consent before using or sharing them. We will remove
identifiers from your data and/or specimens, which means
that nobody who works with them for future research will
know who you are.
The NIH repository stores genetic information and
phenotypic data from many studies. The NIH then shares
that information with researchers. We will send the
information about you and the other participants to a
repository at the NIH. The information will be de-identified
(no names or other direct information about you will be
included). The NIH will not be able to re-identify you or any
other individual.
The NIH intends to share the collected information with
other researchers. The researchers who receive data must
“You may change your mind and withdraw
your permission to use and disclose your
health information at any time. To take
back your permission, it is preferred that
you inform the investigator in writing.
In the letter, state that you changed your
mind and do not want any more of your
personal information collected. The
personal information that has been
collected already will be used if necessary
for the research. No new information will
be collected. If you withdraw your
permission to use your personal health
information, you will be withdrawn from the
study.”
Not available

Page 72 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
promise to keep the data confidential and to use it only for
the purpose approved by NIH. They must also promise to
not try to re-identify anyone.”
Identifying
Biomarker
Signatures of
Prognostic Value
for Multisystem
Inflammatory
Syndrome in
Children (MIS-C)
(PI: Dr. Juan
Salazar)
Not available Not available Linked data sharing: Not included in the consent form
General data sharing: “At the end of the study, your
information, survey answers, and any leftover blood and
saliva will be shared with the National Institute of Health’s
Rapid Acceleration of Diagnostics (RADx) Data
Coordinating Center. The RADx is a NIH program that
funds studies researching ways to use technology for
COVID testing. All studies that are funded will share their
data and samples with the Data Coordinating Center to
create a research resource for everyone to use. This study
is funded by the RADx program.
All data and samples that are shared with the Data
Coordinating Center will be completely de-identified by
removing all of your private information.
The Data Coordinating Center can store your information
and samples indefinitely.”
Genomic data sharing: “In order to allow researchers to
share results, the NIH has developed special sample/data
“banks” that collect the results and analyze samples/data
from research studies, including genetic studies. For
example, there is a database called The NIH Database of
Genotypes and Phenotypes (DbGaP). Some of your
genetic, genomic or health information might be placed
into one or more of these banks so other qualified and
approved researchers can do more studies. We do not
think that there will be further risks to your privacy and
confidentiality by sharing your health information, samples
and/or genetic information with these banks. However, we
cannot predict how genetic information will be used in the
future. The samples and data will be sent with only your
research code number attached. Your name or other
directly identifiable information will not be given to these
central banks. There are many safeguards in place to
protect your privacy.”
“You can leave any time after you start.
To leave, email Dr. Salazar at:
The fil
e linking your code to your priva
te
information will be destroyed.
Any remaining blood or sal
iva s
amples will
be destroyed.
All of your information will be kept and still
used, but it won’t have any of your private
information.”
“If the research subject reaches
the age of 18 prior to the close of
the study, we will attempt to
contact them and re-consent them
as adults. If we are unable to
contact them, they will not be
discontinued from study. However,
their data and specimens will be
completely de-identified including
the destruction of any link to
identifiers from coded data.”
Page 73 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Study Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
Across Sites Within a
Study
Across Studies
Severity Predictors
Integrating Salivary
Transcriptomics
and Proteomics
with Multi Neural
Network
Intelligence in
SARS-CoV2
Infection in
Children (SPITS
MISC)
(PI: Dr. Usha
Sethuraman)
Not available Not available Linked data sharing: Not included in the consent form
General data sharing: Your identifiable information in the
research study records may also be shared with, used by or
seen by collaborating researchers, the sponsor of the
research study (National Institutes of Health), the sponsor’s
representatives including the Data Coordinating Center
assigned by the NIH, and certain employees of the Central
Michigan University and Pittsburgh University if needed to
oversee the research study.”
“However, you can withdraw your
permission to allow the research team to
review your medical records in writing to
Dr. Usha Sethuraman, Children’s Hospital
of Michigan, Central Michigan University at
any time. Any identifiable research or
medical information recorded for, or
resulting from, your participation in the
study prior to the date that you formally
withdrew your consent may continue to be
used and disclosed for the purposes
described above.
You may also withdraw your permission at
any time through a written request. Any
identifiable research or medical information
recorded for, or resulting from, your
participation in the study prior to the date
that you formally withdrew your consent
may continue to be used and disclosed for
the purposes described above.”
Reconsent form language:
“I understand that I am currently
participating in a research study. I
further understand that consent for
my participation in this research
study was initially obtained from
my authorized representative
since I was unable to provide
direct consent at the time that this
initial consent was requested. I
have now turned age 18 and I am
able to provide direct consent for
continued participation in this
research study.”

Page 74 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2 Governance Assessment Supplemental Information
11.2.1
Governance Summary For Record Linkage Implementations Using PPRL
Appendix Table 2: Governance Summary for Record Linkage Implementations Using PPRL
Record Linkage
Implementation
Datasets Linked Authorization (Consent/IRB/Other) for PPRL
Tool
PII Elements
Used for PPRL
Entity
Resolution
(Matching)
Performed by
Data Linking
Performed
by
Data
Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing
Linked Data
1 The Biomedical
Research Informatics
Computing System
(BRICS) Instances:
o NINDS/Parkinson’
s Disease
Biomarker
Program/NIA
o NEI
o NCATS/Global
Rare Diseases
Data Repository
(GRDR
)
o NINR/ Common
Data Repository
for Nursing
Science (cdRNS
)
o The Federal
Interagency
Traumatic Brain
Injury Research
(FITBIR
)
Data within BRICS
instances: clinical,
genetic,
phenotypic,
specimen, and
medical imaging
User and their
institution agree to
the terms of data
submission, which
specifically
addresses linkage
via use of the GUID;
no explicit language
for linking or use of
GUID in the
informed consent
User and their
institution agree to
the terms of data
submission, which
specifically
addresses linkage
via use of the
GUID; no explicit
language for
linking or use of
GUID in the
informed consent
Data Access
Committee
approval; all
approvals include
access to the
GUIDs
BRICS
GUID tool
o Complete
legal given
(first) name of
the subject at
birth
o Middle name
(if available)
o Complete
legal family
(last) name of
subject at
birth
o Day of birth
o Month of birth
o Year of birth
o Name of city/
municipality in
which subject
was born
o Country of
birth
[Optional PII
elements in
Section
11.2.1.1]
GUID server
for each BRICS
instance or a
BRICS
instance can
connect to the
Centralized
GUID server.
Presently
BRICS
instances:
PDBP, NEI,
NCATS,
NINDS, NIA,
cdRNS connect
to the
centralized
GUID server.
Researchers
who have
approval to
access the
data
Linked
database
model
Controlled Section
11.2.1.1
2 NIMH Data Archive
(NDA) Repository
Phenotypic,
clinical, genomic/
pedigree,
neuroimaging, and
other neurosignal
recordings data
o Example consent
language
describes GUID
but use of
language in
studies is not
verified by NDA
Data submitters
agree that
informed consent
from participants
aligns to “broad
data use” by
signing the NDA
Data Access
Committee
approval; all
approvals include
access to the
GUIDs
NDA
GUID
tool
o First Name
o Middle Name
o Last Name
o Sex
o Date of Birth
o City/
Municipality
of Birth
NDA GUID
server
Researchers
who have
approval to
access the
data
Linked
database
model
Controlled Section
11.2.1.2
mm
Two types of linkage models: Linked database model, where the linkage information that is created and/or provided encompasses all datasets in a given database; Study-specific model, where linkage information is
created and/or provided for the purposes of a specific study
nn
Data access models: Open access = no access restrictions or registration required to access; Registration required = open to all, but users need to be signed in or registered with the resource to access; Controlled access =
application and eligibility requirements need to be met to gain access (e.g., by a data access committee); Enclave = data cannot leave a specific system boundary (e.g., cannot be downloaded)

Page 75 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementation
Datasets Linked Authorization (Consent/IRB/Other) for PPRL
Tool
PII Elements
Used for PPRL
Entity
Resolution
(Matching)
Performed by
Data Linking
Performed
by
Data
Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing
Linked Data
o Data submitters
agree to data
linkage by signing
the NDA Data
Submission
Agreement.
Data Submission
Agreement
3 National COVID
Cohort Collaborative
(N3C) EHR data
linkage
[Linked data not yet
available to users]
EHR data of
COVID-19
patients from N3C
participating
institutions (=data
partners) who
have opted in for
PPRL
o Data partners
must agree to
linking data by
signing the
Linkage Honest
Broker Agreement
o Informed consent
not collected from
patients
o NCATS obtained
a waiver of
consent from NIH
IRB
o Informed
consent not
collected from
patients
o NCATS
obtained a
waiver of
consent from
NIH IRB
o Local IRB letter
of determination
for HIPAA
limited dataset
o N3C Data
Access
Committee
approval for
data use, which
includes access
to linkage map
with MATCH_ID
and Pseudo IDs
(generated by
the Honest
Broker for the
specific use
case)
Datavant Combinations of
PII elements to
generate 18
tokens per
record (last
name, first
name, DOB,
gender, SSN,
email, zip5/9,
cell phone)
Third-party
honest broker:
Regenstrief
N3C Enclave Linked
database
model
Enclave Section
11.2.1.3
4 N3C – Class 0: N3C
EHR data linkage with
data from an external
enclave
Example: Linkage
with data from the
Medical Imaging and
Data Resource
Center (MIDRC
)
[Linked data not yet
available to users]
o EHR data from
N3C data
partners
o Data stored in
an external
enclave —
imaging data
(currently)
o N3C data
partners must
agree to linking
their EHR data by
signing the
Linkage Honest
Broker
Agreement, and
provide
permission to link
EHR data with
individual external
datasets
o Informed consent
not collected from
N3C patients;
NCATS obtained
a waiver of
consent from NIH
IRB
o Interconnect
Agreement
established with
o Informed
consent not
collected from
patients;
NCATS
obtained a
waiver of
consent from
NIH IRB
o Interconnect
Agreement
established with
N3C and the
external
enclave (the
interconnect
agreement is
being
developed and
not yet live)
o Local IRB letter
of determination
o N3C Data
Access
Committee
approval for
data use, which
includes access
to linkage map
with MATCH_ID
and Pseudo IDs
(generated by
the Honest
Broker for the
specific use
case)
Datavant Combinations of
PII elements to
generate 18
tokens per
record (last
name, first
name, DOB,
gender, SSN,
email, zip5/9,
cell phone)
Third-party
honest broker:
Regenstrief
N3C Enclave Linked
database
model
Enclave;
Ephemeral
work bench
(a temporary
extension of
the N3C
enclave)
Section
11.2.1.4

Page 76 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementation
Datasets Linked Authorization (Consent/IRB/Other) for PPRL
Tool
PII Elements
Used for PPRL
Entity
Resolution
(Matching)
Performed by
Data Linking
Performed
by
Data
Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing
Linked Data
N3C and the
external enclave
(the interconnect
agreement is
being developed
and not yet live)
5 N3C – Class 2:
N3C EHR data
linkage with external
datasets
Examples: Linkages
with viral variant data
and mortality data
ingested into N3C
EHR data from
N3C data partners
linked to external
data for COVID-19
patients; current
sets of external
data linkages
include:
o Viral variant
data (submitted
to NCBI
repository by
data partners)
o Mortality data
(from
government
mortality
sources and
obituary sites )
o Data partners
must agree to
linking their EHR
data by signing
the Linkage
Honest Broker
Agreement, and
must provide
permission to link
EHR data with
external data
o Informed consent
not collected from
patients
o NCATS obtained
a waiver of
consent from NIH
IRB
o Mortality data
sources – no
authorization
required as the
data are
purchased by
N3C from various
private sources
Informed consent
not collected from
N3C patients;
NCATS obtained a
waiver of consent
from NIH IRB
o Local IRB letter
of determination
o N3C Data
Access
Committee
approval for
data use, which
includes access
to linkage map
with MATCH_ID
and Pseudo IDs
(generated by
the Honest
Broker for the
specific use
case)
Datavant Combinations of
PII elements to
generate 18
tokens per
record (last
name, first
name, DOB,
gender, SSN,
email, zip5/9,
cell phone)
Third-party
honest broker:
Regenstrief
N3C Enclave Linked
database
model
Enclave
(private user
workspace
within the
N3C enclave)
Section
11.2.1.5
6 PEDSnet EHR data from the
11 participating
PEDSnet
institutions,
including
demographic data,
outpatient
encounters,
inpatient
admissions, ER
encounters,
anthropometrics,
vital signs,
providers,
diagnoses,
o Large
observational
studies operate
under waiver of
consent
o Consent for data
linking is
embedded in the
broad study
consent
o PEDSnet Data
and Steering
Committees
approve linkage
for a study;
o Broad study
consent
o PEDSnet
Steering
Committee
PEDSnet Data
Committee
Datavant o Last name
o First name
o First initial of
first name
o Sex
o DOB
o Zip3
o Soundex of
first name
o Soundex of
last name
PEDSnet Data
Coordinating
Center (DCC)
as the honest
broker
The honest
broker may be
an external
entity for out-
of-network
linkages,
depending on
the study
PEDSnet
DCC
Study-
specific
linkage
model
Enclave Section
11.2.1.6
Page 77 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementation
Datasets Linked Authorization (Consent/IRB/Other) for PPRL
Tool
PII Elements
Used for PPRL
Entity
Resolution
(Matching)
Performed by
Data Linking
Performed
by
Data
Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing
Linked Data
treatments, visit
payer, lab test
results, and
medications.
Several linkages
have been done
with health plans,
disease specific
registries, vital
statistics, and
geocoded data.
PEDSnet sites
decide whether to
participate in
linkage on a
study-by-study
basis.
7 CDC/The Childhood
Obesity Data Initiative
(CODI)
Colorado child
health data from:
o Denver Public
Health &
Hospital
Authority
o Children’s
Hospital
Colorado
o Kaiser
Permanente
Colorado
o Girls on the
Run of the
Rockies
o Hunger Free
Colorado
CODI expanded in
2020 to include a
North Carolina
cohort including
clinical and
community data in
the Triangle region
from health care,
local/state health
departments, and
community-based
organizations in
North Carolina.
o Data partners
agree to linking
data within the
CODI network
via
the Master
Sharing and Use
Agreement
(MSUA)
o IRB approval
o CHORDS
Rese
arch Council
approves dat
a
linkage study
request.
o Data par
tners
agree to share
linked data
within CODI
network v
ia the
Master Sharing
and Use
Agreement
(MSUA)
o Master Sharing
and Use
Agreement
(MSUA)
AnonLink o First name
o Last name
o Date of birth
o Sex
o Phone
number
o Household
street
address
o Zip code
CODI Data
Coordinating
Center (DCC)
CODI Data
Coordinating
Center
(DCC)
Study-
specific
linkage
model
Controlled Section
11.2.1.7

Page 78 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.1.1
The Biomedical Research Informatics Computing System (BRICS) Instances: NINDS/Parkinson’s Disease Biomarker Program, NIA, NEI, NCATS/GRDR,
NINR/cdRNS, FITBIR
• Description
69
: The NIH Biomedical Research Informatics Computing System (BRICS) is a collaborative and customizable bioinformatics repository
designed to efficiently collect, validate, harmonize, and analyze research datasets. BRICS was designed to address the wide-ranging needs of several
biomedical research programs
70
. The overall concept was to develop services that could be integrated and deployed as instances for individual research
programs. De-identification of each patient within a research study is supported by the use of a Global Unique Identifier (GUID). It is a de-identification
tool developed for researchers to use prior to submission of data to a specific BRICS instance. With the implementation of the BRICS GUID tool
71
,
researchers can have access to data across studies, without revealing personally identifiable information (PII), while correlating study data across
different studies and uniquely sorting across distinct and redundant datasets. At this time, BRICS does not link with external genomic data repositories
such as the database of Genotype and Phenotype (dbGaP), but it does link with the NINDS biorepository, Biospecimen Exchange for Neurological
Disorders (BioSEND).
• Data sources
72
: Data sources depend on the instances.
o In the BRICS Centralized GUID server solution
71,73
used by multiple instances below, the source of data is from studies conducted by the
respective institutes/program:
The National Institute of Neurological Disorders & Stroke (NINDS), Parkinson’s Disease Biomarkers Program (PDBP), and the National
Institute of Aging (NIA)
74
The National Eye Institute (NEI)
75
The Global Rare Diseases (Patient Registry) Data Repository (GRDR) at the National Center for Advancing Translational Sciences (NCATS)
76
The Common Data Repository for Nursing Science (cdRNS) at National Institute of Nursing Research (NINR)
77
o In the Federal Interagency Traumatic Brain Injury Research (FITBIR) instance
78
, the source of data is from the traumatic brain injury (TBI) studies
funded by the DoD and NIH.
• Data types
69
: Variety of data from BRICS instances which includes clinical, genetic, phenotypic, biospecimen, and medical imaging data.
• Linkage agreements:
o Submission: Each data submitter must agree to linkage via the use of GUID as part of the data submission requirements of each of the respective
repository. For example: the NINR cdRNS
79
and the FITBIR
80
data sharing policies states the GUID is a unique code that allows linkage of all
submitted information on a single participant, giving researchers access to information that may have been collected elsewhere. Both programs
also expect that an Institutional Review Board (IRB) and/or Privacy Board has verified that the submission is consistent with informed consent,
that the data are de-identified according to the respective repository standards, risk to the study population has been considered, and the data
were collected in a manner consistent with NIH/DOD regulations, but the IRB is not asked to sign the Submission Request.
o Access/Use: Linkage of data between the data sources are performed by the researchers who are interested in using the data
81
. They must first
obtain approval from the Data Access Committee (DAC) of the respective study/program to access the data, and then they perform the linkage
using the subject-level GUID
72
.
Page 79 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
• Entity resolution: Performed by each instance’s GUID server.
o Standardization/pre-processing of PIIs
82
: Instructions regarding required PII elements to ensure that a valid GUID is created include:
–
The "Last Name" fie
ld must contain the family name given at birth, prior to legal name change, or marriage. If there is any doubt as to the
original legal name at birth, refer to the information on the birth certificate. Name suffixes such as "Jr.," "Sr.," “III,” etc. should be ignored.
–
If the partici
pant’s "First Name" is a compound name, such as Anne Marie, or Jose- Luis, it may be unclear whether the second part of the
compound is a first name or a middle name. In such cases, use the first name as you would report it on other records, such as school
transcripts, or credit card billing statements. If in doubt, refer to the birth certificate.
–
If the partici
pant does not have a "Middle Name" (known not to have a middle name at birth), leave this field blank and respond “No” when
asked if the individual has a middle name. The GUID Software has a selection to accommodate this possibility. If in doubt, refer to the birth
certificate.
–
If the "City or
Municipality of Birth” has undergone a name change during a participant's lifetime, use the name of the city at the time of the
participant’s birth. Examples of this are Peking / Beijing, or Bombay / Mumbai. Again, if there is any doubt, refer to the birth certificate.
The following PII fields are optional:
–
Physical sex of su
bje
ct at b
irth
–
Government-iss
ued or National ID (i.e., SSN) (No one has provided government issued ID to date.)
–
Country issui
ng government-issued or National ID
– Organization/cohort association (for example, PDBP)
o GUID generation process
73
: Each instance of BRICS is connected to a BRICS GUID server to generate GUIDs. An instance of BRICS either uses a
centralized GUID server called the Multi-tenant GUID Solution (example: NINDS) or has its own GUID server (example: FITBIR). Each instance of
the GUID server has a different salt. The centralized GUID server salt is consistent across studies within that instance.
–
Researchers w
ithin BRICS instances log into the GUID client on their local computers and enter in subject PII.
– The GUID client (on local computers) uses PII to derive a series of one-way hashes which securely encrypt PII information.
–
The one-way hashes
are sent to the GUID server for reference and storage. No PII is sent to the server.
– The GUID server returns a GUID identifier. If the one-way hashes match an existing GUID identifier, an existing GUID will be returned. If this
is a new subject, a new GUID will be returned.
o GUID quality assessment
73
: Some hash codes leave out one PII element which allows the GUID system to determine if there is a close match. If
the hashes are the same other than the one PII element, the system can notify the user to check if the PII was entered incorrectly. The quality is
also validated during the data entry at the client site, which includes practices such as checking that the months are between 1-12 and day is
between 1-31. In the GUID interface, data has to be entered twice to confirm validation. Currently, data pre-processing in BRICS does not include
breaking out syllables within names prior to hashing to facilitate “fuzzy matching” of names (e.g., Soundex).
o Hash/GUID storage and destruction procedures
73
: The one-way hashes are sent from the BRICS instances to the NIH GUID server for reference
and storage. The hashes are stored in an encrypted database. The hashes are not shared and cannot be accessed except potentially by BRICS
developers. Withdrawal of consent leads to the destruction of hashes and GUIDs. If subjects no longer consent to their data being shared, the
Page 80 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
GUID and the associated hashes are removed from system. The study or program officers must inform BRICS to remove the GUIDS associated
with the patient and the associated hashes will be removed.
•
Data Linkage
73,75
: BRICS facilitates the linkage of data by adding the GUID to the records associated with each subject, which allows researchers to
distinguish between unique subjects and recurrent subjects across datasets. BRICS databases can support data queries (cohort discovery) and link data
with GUIDs both within a study (by linking the data from different forms/datasets) or between different studies (such as between different FITBIR
studies). The BRICS system does not track all queries since they are often refined. However, all downloads of data from the query tool are audited and
recorded (e.g., Visualization | FITBIR (nih.gov)
; see figure below). In this example usernames are masked but the System Administrators can identify
actual users as well as
metrics including what data was download from each study. In addition, each owner of a study can download a report of who has
accessed their study data.
Presently, the inst
ances d
o not directly communicate with each other; a GUI/interface can only query within an instance of BRICS. However, when a
common GUID server is used between two instances, a user could manually see if participants match. They would first have to get DAC approval and
download the data from the two different instances. There are programmatic ways of doing this, but presently the GUI/interface does not provide the

Page 81 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
ability to query across different instances. However, the foundational infrastructure has already been developed (query and download APIs) and is
included in the BRICS system. Extending the BRICS query tool to query across instances is very possible and is not a massive level-of-effort.
o Process:
1.
Once the research
er has a GUID, they may submit data associated with this subject. No data may be submitted to the system without a
GUID.
2.
Once data are i
n the database, a researcher may query this data. The GUID allows researchers to distinguish between unique subjects and
recurrent subjects across all datasets (i.e., studies).
3.
Researchers acces
s data across different studies from within their instances, without revealing PII, while correlating study data across
different studies and uniquely sorting between distinct and redundant datasets.
•
Sharing and access
ing linked data—authorizations/agreements/approvals:
o De-identification status of shared data: As per NINR/cdRNS policy
79
and FITBIR
80
policy, data submitted to BRICS/NINR or FITBIR will be de-
identified such that the identities of data subjects cannot be readily ascertained or otherwise associated with the data by the BRICS/cdRNS or
FITBIR staff or secondary data users.
o Authorization for sharing linked data
72
: User and their institution agree to the terms of data submission, which specifically addresses linkage via
use of the GUID; no explicit language for linking or use of GUID in the informed consent.
o Deductive disclosure review
81
: Deductive disclosure review of linked data is not currently done by the programs. The researcher links the
datasets after obtaining approval for access from the respective program’s data access committees. The programs do not track the linkage.
However, each program requires that approved users agree to a Data Use Certification (DUC), which explicitly prohibits re-identification of study
participants. For example, the FITBIR DUC
83
states, “Recipient agrees that data will not be used, either alone or in conjunction with any other
information, in any effort whatsoever to establish the individual identities of any of the subjects from whom data were obtained.” Also, the DUC
states the potential requirement for an IRB approval from the user’s institution: “If Recipients request access to data on individuals for whom
they themselves have previously submitted data to FITBIR, they may gain access to more data about an individual participant than they
themselves collected. Consequently, these research activities may be considered “human subjects research” and may require that they obtain
institutional IRB approval of their Research Project.”
o Approvals/agreements for accessing linked data
72
: Approval for access from the respective program’s data access committees.
11.2.1.2
National Institute of Mental Health (NIMH) Data Archive (NDA)
• Description
84
: The National Institute of Mental Health Data Archive (NDA) provides infrastructure for sharing human subjects research data, tools,
methods, and analyses, enabling collaborative science and discovery. The NDA is a collection of research data repositories including the NIMH Data
Archive (NDA), the Osteoarthritis Initiative (OAI), the Adolescent Brain Cognitive Development (ABCD) data repository, and the National Institute on
Alcohol Abuse and Alcoholism Data Archive (NIAAA
DA
). The NDA infrastructure was established initially to support autism research but has grown into an
informatics platform that facilitates data sharing across all of mental health and other research communities, making data available from each of these
repositories combined into a single resource with a single process for gaining access to all shared data.

Page 82 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
• Data sources
85
: NDA accepts human subjects research data related to mental health and other NIH-funded scientific domains. All NIMH-funded
researchers performing human subjects research are required to submit data to NDA as per the 2019 NIMH Data Sharing Policy
86
. In addition, any
researchers who have acquired high-quality research data appropriate to an NDA-supported research cluster may request to submit data, regardless of
funding source and location.
• Data types
85
: Clinical, phenotypic, genomic/pedigree, neuroimaging, and other neurosignal recordings data on human subjects.
•
Linkage agreeme
nts
87
: All data submitters to the NIMH Data Archive must agree to the following as per the NIMH Data Archive Data Submission
Agreement:
o “Submitter agrees to collect the information required to generate a Global Unique Identifier (GUID) for all research participants, using software
provided by the NIMH Data Archive (https://nda.nih.gov/s/guid/nda-guid.html
),”
o “Submitter may use the NIMH Data Archive GUID Tool to generate pseudoGUIDs if their IRB determines that the information required to create
a GUID may not be collected from research participants. Submitter agrees to submit all subject data to the NIMH Data Archive with a GUID or
pseudoGUID.”
o “Submitter acknowledges that data are submitted to the NIMH Data Archive in accordance with informed consent of research participants
and/or with the approval of the IRB.”
o The Agreement specifies linkage via GUID: “NDA GUID allows the NIMH Data Archive to link together all submitted information on a single
participant, giving authorized researchers access to information even if the data were collected at different locations or through different
studies”.
o “Submitter agrees that data and Supporting Documentation submitted to the NIMH Data Archive may be accessed and used broadly by
approved users for research and other activities as authorized by and consistent with law.”
o The Policy
88
for the NIMH Data Archive (NDA) also expects that the data submission is consistent with consent where feasible:
–
“For prospect
ive studies, in which data sharing through the NDA is conceived within the study design at the time research participants
provide their consent, the NIMH expects specific discussion within the informed consent process and documentation that participant’s data
will be shared for research purposes through the NDA. For retrospectively collected data, the NIMH anticipates considerable variation in the
extent to which data sharing and future research have been addressed within the informed consent documents; therefore, the NIMH
expects the submitting institution to determine whether a retrospective study is appropriate for submission to the NDA (including an IRB
and/or Privacy Board review of specific study elements, such as participant consent).”
•
Entity reso
lution:
o Standardization/pre-processing of PIIs
89
: The following information is required to create a GUID and should be recorded exactly as it appears on
the birth certificate, to ensure that it does not change over the course of the participant’s life:
– First Name
–
Middle Name
–
Last Name
–
Sex

Page 83 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
– Date of Birth
–
City/Municipality o
f Birth
If adequate information is not available to fully create a valid GUID, a pseudoGUID
90
can be created. This is a random ID that can be used as a
placeholder where this information is not available, and “promoted” to a real GUID when the information is obtained at a future date.
o GUID generation process
89
: Performed by the GUID system at NDA:
1.
An authorized me
mber of the research project team (user) with an active Data Submission Agreement may request access to and then
download the NDA GUID Tool on a local computer
91
. Third-party NDA GUID Tool requests must be submitted by an NDA user with an active
NDA account and an authorized Signing Official (SO) from that user’s institution and are reviewed on a case-by-case basis. All NDA GUID Tool
users must agree to the terms of use for the tool.
2.
The user enter
s participant PII into the tool.
3. The tool generates a series of one-way hash codes based on the PII entered, without the PII ever leaving the computer.
4.
The one-way hash
codes are encrypted and securely sent to the GUID system at NDA.
5. If the hash codes match an existing hash code, the GUID associated with that existing hash code is sent back to the researcher. The GUID is
an alphanumeric code that is randomly and persistently linked to the hash codes within the secure NDA GUID system and cannot be traced
back to the PII entered by the research project team member.
6.
If the hash cod
es do not match an existing hash code in the NDA GUID system, a new GUID is created and sent back.
o GUID quality assessment
92
: The hash codes generated by the algorithm depend on the spelling of the words (PII) entered, so there’s no way to
account for/predict typos or data entry errors. To address possible data entry errors, it is possible to reconcile GUIDs at a later date
93
.
Occasionally, the submitted hashes will closely (but not exactly) match the hashes of an existing GUID subject. This situation primarily occurs
when working with twin research subjects who have differing first names beyond the first letter. When this happens, the tool will prompt the
user (data originator) to indicate whether it should use the existing GUID or create an entirely new GUID. Generally, unless the user knows for
certain that the subject is truly a different person, e.g., as in the twin example, they should choose to “Create New GUID.”
92
o Hash/GUID storage and destruction procedures
89
: The hashing algorithm is run inside the NDA GUID Tool software that researchers must run on
their own computer. The PII input into the NDA GUID Tool never leaves the researcher’s local environment. Instead, one-way hashes are
securely transmitted to the NDA cloud database, which uses the same secure transmission protocol (via a secure web service) to return random
alphanumeric strings called GUIDs. GUIDs and their matched hash codes are stored in the secure database maintained by NDA’s security team in
the Amazon cloud. Additional information is available at GUID Tool Terms of Use
.
• Data linkage
94
: NDA facilitates the linkage of data by adding the GUID to the records associated with each subject, which allows the NDA to associate a
single research participant's genomic, imaging, clinical assessment, and other information even if the data were collected at different locations or
through different studies. The GUID is then used as the primary participant identifier in data submissions. The GUID allows researchers to distinguish
between unique subjects and recurrent subjects across all datasets. Linkage of data is performed by researchers who have approval from the NDA Data
Access Committee to access the datasets.
Page 84 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
• Sharing and accessing linked data—authorizations/agreements/approvals:
o De-identification status of shared data
88
: As per NDA policy, data submitted to NDA will be de-identified such that the identities of data subjects
cannot be readily ascertained or otherwise associated with the data by the NDA staff or secondary data users.
o Authorization for sharing linked data
87
: Based on the required data submission agreement provided by the submitter: “Submitter agrees that
data and Supporting Documentation submitted to the NIMH Data Archive may be accessed and used broadly by approved users for research and
other activities as authorized by and consistent with law.” Example informed consent language that addresses the use of the GUID is available;
however, it is not confirmed whether such language is used as part of the data submission.
o Deductive disclose review: Deductive disclosure review of linked data is not currently done by the program. The researcher links the datasets
after they obtain approval for access from NDA. The approved users must agree to a Data Use Certification (DUC)
95
, which explicitly prohibits re-
identification of study participants. The DUC states, “Recipient agrees that data will not be used, either alone or in conjunction with any other
information, in any effort whatsoever to establish the individual identities of any of the subjects from whom data were obtained that data will
not be used to attempt to establish the individual identities of any of the study participants from whom data were obtained (or their relatives).”
o Approvals/agreements for accessing linked data
93,95
: Access to linked data require approval from the Data Access Committee and may require
IRB approval of the Research DUC. The NDA DUC prohibits users from re-identifying the study participants from whom data were obtained.
Researchers who submitted data to NDA may have access to personal identifying information for research participants in the original study at
their institution and therefore their use of NDA data from other studies with the same participants may be considered “human subjects
research” and subject to IRB approval. The NDA DUC states: “If Recipients access data on individuals for whom they, themselves, have previously
submitted data to the NIMH Data Archive, Recipients may gain access to more data about an individual participant than they, themselves,
collected. Consequently, these research activities may be considered “human subjects research” within the scope of 45 C.F.R. 46. Recipients
must comply with the requirements contained in 45 C.F.R. 46, as applicable, which may require IRB approval of the Research Data Use
Statement.”
11.2.1.3
National COVID Cohort Collaborative (N3C) EHR Data Linkage
[Linked data from this implementation is not yet available to users]
• Description
96
: The N3C Data Enclave is a centralized, secure, national clinical data resource with powerful analytics capabilities that the research
community can use to study COVID-19, including potential risk factors, protective factors, and long-term health consequences. The N3C collects data
derived from the electronic health records (EHRs) of people who were tested for COVID-19 or who had related symptoms, as well as data from
individuals infected with pathogens that can support comparative studies, such as SARS1, MERS and H1N1. EHR to EHR data linkages are not yet
available to users.
• Data sources
96
: Participating institutions (also called “N3C data partners”) release EHR data to N3C under the HIPAA Privacy Rule that allows medical
and health care institutions to release data for research without obtaining an individual’s authorization if direct identifying information is removed and
appropriate oversight and agreements are in place. Under the HIPAA Privacy Rule requirements, these institutions can release what is called a limited
data set to N3C. As of September 2022, the N3C enclave hosts EHRs from over 15.5 million patients.

Page 85 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
• Data types
97
: The EHR data in N3C include demographics, symptoms, lab test results, procedures, medications, medical conditions, physical
measurements.
•
Linkage agreements
65
: Participation in PPRL is voluntary for N3C data partners. Data partners who choose to participate in PPRL must do the following:
o Agree and sign the Datavant software license agreement
o Agree and sign the Linkage Honest Broker (LHB) Agreement (LHBA)
98
. The LHB for N3C is Regenstrief and the LHBA provides terms and
conditions for data linkage and outlines the general terms of data use. Institutions do not need to sign the LHBA (i.e., participate in PPRL) in
order to contribute data.
o Send the de-identified tokens and Pseudo ID
oo
to the LHB and the Pseudo ID to the N3C along with data payload.
•
Hashed token gen
eration by data partners using Datavant tool:
o Standardization/pre-processing of PIIs
99
: Data partners collect data in various data models (OMOP, TRINETX, PCORnet, ACT, FHIR etc.); hence,
the PII elements collected by individual data partners are standardized.
o Hash/token generation
100
: Performed by N3C data partners who have signed the LHBA and installed Datavant at their institutions. Datavant
tokens are generated using cryptographic hash functions, specifically with SHA-256, one-way, irreversible cryptographic hash function from
identifiable information. Datavant tokens are certified under the Expert Determination Standard in the HIPAA Privacy Rule.
o Token storage and destruction procedures
65
: The N3C de-identified tokens are held separately from data residing within the N3C data enclave.
The LHB is a neutral entity located outside of the N3C enclave that serves as an escrow for the cryptographic hash codes (tokens). Token storage
and destruction is defined by the agreement between LHB and N3C
101
. The destruction agreement assures a standard process for receiving,
reviewing, authorizing, executing, and confirming a token data destruction request for the LHB system. The requesting party must submit a
written request to an authorized NCATS official who will first verify that the request is appropriately scoped to the token data pertaining to the
requesting party and will then honor the party’s request and will submit an LHB Token Destruction Form to the LHB to execute the destruction.
Once destroyed by the LHB, the authorized NCATS official will inform the requesting party.
•
Entity resol
ution & data linkage
100
: Entity resolution is performed by the LHB, and data linkage is done within N3C enclave by requesting researchers.
Entity resolution/deduplication across all participating EHR datasets is a requirement for any institution that participates in the LHBA because of its
importance to the data quality of the N3C data enclave and its scientific mission. However, access to deduplicated EHR records across N3C is not yet
available.
o Process
100
: N3C performs entity resolution builds on a weekly basis.
1.
Data partner
s send a randomly generated Pseudo ID (=PAT_ID = Source ID) and the cryptographic hashes/tokens to LHB (Regenstrief).
2. N3C enclave receives EHR data payload with the Pseudo IDs (=PAT_ID = Source ID) from data partners.
3.
When the N3C e
nclave receives the data payload from a data partner, an N3C_ID is assigned.
4. The N3C_ID together with the site’s randomly generated ID for a specific record (=Pseudo ID = PAT_ID = Source ID) are sent by the N3C data
enclave to LHB.
oo
Pseudo ID is the de-identified subject ID used in the data by the data partners for sharing. It is also referred to as PAT_ID or Source ID.

Page 86 of 130
5. LHB runs Datavant matching algorithms and generates a MATCH_ID which represents the records that should be linked (linkage map).
pp
6. When a user is approved for access to the Limited Data Set level, the N3C enclave provides the linkage map (with MATCH_ID, N3C_ID, Site
ID, a
nd Pseudo ID) and data in
the researcher’s private workspace in the N3C enclave. The user accesses the datasets for an individual
participant based on the linkage map. [Note: N2C provides all
of the EHR data available in N3C to the users along with the linkage map; no
removal of duplicate records across sites is p
erformed.]
o Linkage/matching quality assessment
101
: A linkage quality assessment report is being prepared by N3C. In general, the linkage quality measures
will depend on the linkage use case—for example: the linkage quality for EHR data linkage is stringent to support academic research but could be
less stringent for recruitment use cases (what N3C calls “cohort discovery”)
• Sharing and accessing linked data—authorizations/agreements/approvals:
o De-identification status of shared data
102
: PPRL-linked data are Level 3 (limited dataset), where 16 of 18 HIPAA identifier have been removed;
data retains dates and zip codes.
o Authorization for sharing linked data
103
: N3C participating institutions do not obtain consent from individual patients for the data they send to
the N3C. The 1996 HIPAA Privacy Rule allows medical and health care institutions to release data for research without obtaining an individual’s
authorization if direct identifying information is removed and appropriate oversight and agreements are in place. Under the HIPAA Privacy Rule
requirements, data partners can release what is called a limited dataset. This is what participating health sites send to the N3C.
o Deductive disclosure review currently not being done at N3C
101
; however, access to PPRL-linked EHR data will be made available for users who
have approval for accessing HIPAA Limited Data Set (which is a Level 3). Accessing the HIPAA limited dataset requires a letter of determination
from the data requestor’s IRB in addition to and other requirements for access (see below).
o Agreements/approvals for accessing linked data
102
:
– Technical data privacy controls: Linked data is provisioned in a private workspace within the N3C enclave
– Non-technical data privacy controls: The following non-technical controls are in place for accessing the linked level 3 data:
Data Use Request (DUR) with justification to access the linked data, approved by N3C Data Access Committee
Signed institutional Data Use Agreement (DUA) executed with NCATS
Institutional IRB Letter of determination (LOD)
Attestation to: N3C Code of Conduct, IT security training, and human subjects protection training
11.2.1.4
N3C Class 0—External Data Linkage
[Linked data from this implementation is not yet available to users]
• Description
96
: The N3C Data Enclave is a centralized, secure, national clinical data resource with powerful analytics capabilities that the research
community can use to study COVID-19, including potential risk factors, protective factors and long-term health consequences. The N3C collects data
derived from the EHRs of people who were tested for COVID-19 or who had related symptoms, as well as data from individuals infected with pathogens
pp
Also referred to
as N3C_GUID, N3C_HDB_MATCH_ID, Linked_ID
NICHD Office of Data Science a
nd Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report (September
2022)

Page 87 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
that can support comparative studies, such as SARS 1, MERS and H1N1. N3C facilitates the linking of external datasets with EHR data available in the N3C
enclave using PPRL to generate a richer dataset that can answer new questions about COVID-19. The details below specifically pertain to PPRL linkage
with external Class 0 datasets that originate from different enclaves and allows for a temporary extension of the N3C Data Enclave in the form of an
ephemeral workbench to accommodate this requirement
65
. A current example of a Class 0 linkage is with imaging data from the Medical Imaging and
Data Resource Center (MIDRC). Class 0 data linkage is not yet available to users.
• Data sources
97
: EHR data in the N3C enclave (submitted by N3C data partners) and imaging data from external data repositories, such as imaging data in
MIDRC
•
Data types
97
: EHR data in N3C (including demographics, symptoms, lab test results, procedures, medications, medical conditions, physical
measurements) and external individual-level data, such as imaging data.
•
Linkage agreements
65
: Authentication of Class 0 data linking is at the NIH level. An interconnect agreement is required between N3C and the enclave
from where the external datasets are used for Class 0 linkage. For N3C data partners—participation in PPRL is voluntary, and linkage of EHR data with
external datasets is limited to only those N3C data partners who are participating in PPRL
100
. Each PPRL participating data partner has the option to
choose which type of data linkage they want to participate in—that is, they can choose whether or not to permit linking their EHR data with external
data such as imaging data. Participating sites can, at any time, update their participation in linkage of multiple datasets by changing their setting of
institutional parameters within N3C via a web interface dashboard
100
. Individual data partners can only see their linkage permissions in the dashboard.
N3C data partners who choose to participate in PPRL with external datasets must comply with the following:
o Agree and sign the Datavant software license agreement
o Agree and sign the Linkage Honest Broker (LHB) Agreement (LHBA)
o Send the Pseudo_ID to the LHB and N3C (along with the data payload)
o Agree to link to the given external dataset via the PPRL Site Permissions Dashboard
•
Hashed token generati
on:
o Standardization/pre-processing of PIIs
99
: N3C data partners collect data in various data models (OMOP, TRINETX, PCORnet, ACT, FHIR etc.);
hence, the PII elements collected by individual data partners are standardized.
o Hash/token generation
100
: Both the N3C data partners (who are participating in PPRL) and external data stewards, such as MIRDC, generate
hashed tokens using the Datavant tool installed on a local server. Datavant tokens are generated using cryptographic hash functions, specifically
with SHA-256, one-way, irreversible cryptographic hash function from identifiable information. Datavant tokens are certified under the Expert
Determination Standard in the HIPAA Privacy Rule.
o Token storage and destruction procedures
65
: The N3C de-identified tokens are held separately from data residing within the N3C data enclave.
The LHB is a neutral entity located outside of the N3C enclave that serves as an escrow for the cryptographic hash codes (tokens). Token storage
and destruction is defined by the agreement between LHB and N3C. The destruction agreement assures a standard process for receiving,
reviewing, authorizing, executing, and confirming a token data destruction request for the LHB system. The requesting party must submit a
written request to an authorized NCATS official who will first verify that the request is appropriately scoped to the token data pertaining to the

Page 88 of 130
requesting party and will then honor the party’s request and will submit an LHB Token Destruction Form to the LHB to execute the destruction.
Once destroyed by the LHB, the authorized NCATS official will inform the requesting party.
• Entity resolution/data linkage
100
:
o Entity resolution performed by the LHB, based on tokens received from the N3C data partners and external data stewards, and when requested
by N3C enclave based on two criteria:
– The data partner whose EHR data are being linked has provided the permission to link with imaging data
– The ‘linkage requesting researcher’ has been approved by N3C enclave to access the linked data
o Linked database model: Data linkage performed by N3C enclave encompasses all participating datasets.
o Process
101
:
1. Data partners send the Pseudo ID and the hashed tokens to LHB for EHR data
2. External data steward (MIRDC) also sends their MIRDC Pseudo ID and the hashed tokens to the LHB
3. When the N3C enclave receives the EHR data payload from a data partner, an N3C_ID is assigned
4. The N3C_ID and the Pseudo IDs for the EHR data and the Class 0 data are sent by the N3C enclave to LHB. The LHB runs Datavant matching
algorithms and generates a MATCH_ID
qq
(unique de-duplicated ID) that maps to N3C Pseudo ID and external (MIRDC) Pseudo ID; the linkage
map represents the records that should be linked.
5. When a user is approved for access to the Limited Data Set level, the N3C enclave provides the linkage map (with MATCH_ID, N3C_ID, Site
ID, and Pseudo IDs) and the respective data in a secure ephemeral workbench, which is a temporary virtual workspace. The workbench is
ephemeral because it is short-lived for a specific task and then destroyed when the user’s work is completed. [Note: N3C provides all of the
EHR and Class 0 data available in N3C to the users along with the linkage map; no removal of duplicate records across sites is performed.]
o Linkage/match quality assessment: A linkage quality assessment report is being prepared by N3C.
• Sharing and accessing linked data—authorizations/agreements/approvals:
o De-identification status of linked data
102
: PPRL linked N3C and external data such as imaging data are classified by N3C as Level 3 limited dataset
(LDS), which consists of patient data that retain the dates of service and patient ZIP code.
o Authorization for sharing linked data
103
: Participating institutions do not obtain consent from individual patients for the data they send to the
N3C. The 1996 HIPAA Privacy Rule allows medical and health care institutions to release data for research without obtaining an individual’s
authorization if direct identifying information is removed and appropriate oversight and agreements are in place. Under the HIPAA Privacy Rule
requirements, data partners can release what is called a limited data set. This is what participating health sites send to the N3C. For external
Class ) data, an interconnect agreement must be established for (linking and) sharing data.
o Deductive disclosure review
104
: Prior to approving each external dataset, re-identification risk is assessed during the technical integration call by
the Tools and Resources Review Committee comprised of federal employees from NCATS, the N3C community, and the Information Systems
Security Officer.
qq
Also referred to as N3C_GUID,
N3C_HDB_MATCH_ID, Linked_ID
NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Page 89 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o Agreements/Approvals for accessing linked data
100
:
– Technical da
ta privacy controls: Linked data are provisioned in a temporary ephemeral workbench within N3C
–
Non-technical
data privacy controls: The following non-technical controls are in place for accessing the linked level 3 data:
DUR with justific
ation to access the linked data, approved by N3C Data Access Committee
Signed institutional DUA
Institutional I
RB Letter of determination (LOD)
Attestation
to: N3C Code of Conduct, IT security training, and human subjects protection training
Interconnect a
greement between N3C and the external data system
11.2.1.5
N3C Class 2 External Data Linkage
• Description
96
: The N3C Data Enclave is a centralized, secure, national clinical data resource with powerful analytics capabilities that the research
community can use to study COVID-19, including potential risk factors, protective factors and long-term health consequences. The N3C collects data
derived from the EHRs of people who were tested for COVID-19 or who had related symptoms, as well as data from individuals infected with pathogens
that can support comparative studies, such as SARS 1, MERS and H1N1. N3C facilitates the linking of external datasets with EHR data available in the N3C
enclave using PPRL to generate a richer dataset that can answer new questions about COVID-19. The details below specifically pertain to PPRL linkage
with two examples of external data linkages that fall under Class 2 and are currently performed within N3C—viral variant data and mortality data
100
.
• Data sources
100
: EHR data in the N3C enclave (submitted by N3C data partners) and the following external datasets:
o Viral variant data: Collected by N3C data partners but stored in NIH’s National Center for Biotechnology Information (NCBI) Sequence Read
Archive (SRA); currently only viral variant summary data provided by the N3C data partners are linked. When the viral variant sequence data use
case is ready, it will be imported into N3C.
o Mortality data: The mortality data sources include government mortality sources (death certificates and person-reporting), ObituaryData.com,
and private obituary sites
•
Data types
100
: EHR data in N3C (including demographics, symptoms, lab test results, procedures, medications, medical conditions, physical
measurements), viral variant summary data (currently; sequence data will be available later), and mortality/death date
•
Linkage agreeme
nts
65
: N3C authorizes the linkage of Class 2 datasets. For N3C data partners, participation in PPRL is voluntary, and linkage of EHR data
with external datasets is limited to only those N3C data partners who are participating in PPRL
65
. Each PPRL participating data partner has the option to
choose which Class 2 datasets their EHR data can link to—for example: they can choose to permit linking their EHR data with viral variant and/or
mortality data. Participating sites at any time can update their participation in linkage of multiple datasets by changing their setting of institutional
parameters within N3C via a web interface dashboard
100
. Individual data partners can only see their linkage permissions in the dashboard. Data partners
who choose to participate in PPRL with external datasets must agree to the following:
o Agree and sign the Datavant software license agreement
o Agree and sign the Linkage Honest Broker (LHB) Agreement (LHBA)

Page 90 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o Agree to send the de-identified tokens and Pseudo ID
rr
to the LHB and the Pseudo-ID along with the EHR data to N3C
o Agree to link to the given external dataset PPRL Site Permissions Dashboard
• Hashed token generation:
1.
For viral va
riant summary data linkage
105
: Because the viral variant summary data are collected by N3C data partners for their cohort of COVID-
19 patients, hashing is performed by the N3C data partners who are participating in PPRL using the Datavant tool. The details are, therefore,
similar to what is described above in N3C EHR data linkage section (11.2.1.3
).
2. For mortality data linkage: N3C data partners who are participating in PPRL generate the hashed tokens for EHR data as described in N3C EHR
data linkage section (11.2.1.3). An ext
ern
al party engaged by N3C aggregates mortality data from government and private sources and generates
hashed tokens using PII elements.
•
Entity resol
ution
100
:
o Process: Performed by LHB based on (1) tokens received from the N3C data partners for viral variant summary data linking and from N3C for
mortality data linking, and (2) when requested by N3C enclave based on two criteria:
–
The data part
ner whose EHR data are being linked has provided the permission to link with viral variant and/or mortality data
–
The linkage-re
questing researcher has been approved by N3C enclave to access the linked data
o Linkage/match quality assessment: A linkage quality assessment report is being prepared by N3C.
•
Data Linkage
100
: Linked database model encompassing all participating datasets. Performed by N3C enclave; below is the process:
1.
Data partners send
Pseudo ID along with their EHR data payload to N3C and set permissions for EHR data linkages with external datasets within
N3C dashboard (web interface).
2.
When the N3C e
nclave receives the EHR data payload from a data partner, an N3C_ID
is assigned.
3.
For viral variant dat
a: Data partners send the following to N3C for viral variant data
105
: a crosswalk between the local specimen ID and the
N3C_ID, and the viral variant summary data.
4.
N3C data part
ners send the Pseudo ID and the hashed tokens for the EHR data to the LHB. For mortality data: The external party engaged by N3C
sends the hashed tokens to the LHB.
5.
N3C enclave sen
ds the N3C_ID and the Pseudo ID for the EHR data to LHB.
6.
LHB runs Data
vant matching algorithms and generates a MATCH_ID that maps to Pseudo IDs; the linkage map represents the records that should
be linked
7.
When a user is app
roved for access to the Limited Data Set level, the N3C enclave provides the linkage map (with MATCH_ID, N3C_ID, Pseudo ID,
and Site ID) and the respective data in a secure private workspace. [Note: N3C provides all of the EHR and Class 2 data available in N3C to the
users along with the linkage map; no removal of duplicate records across sites is performed.]
rr
Pseudo ID is the de-identified subject ID used in the data by the data partners for sharing. It is also referred to as PAT_ID or Source ID.

Page 91 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
• Sharing and accessing linked data—authorizations/agreements/approvals:
o De-identification status of the shared data
102
: Linked viral variant data and mortality data are classified as Level 3 limited dataset, which consists
of patient data that retain the dates of service and patient ZIP code.
o Authorization for sharing linked data
103
: Participating institutions do not obtain consent from individual patients for the data they send to the
N3C. The 1996 HIPAA Privacy Rule allows medical and health care institutions to release data for research without obtaining an individual’s
authorization if direct identifying information is removed and appropriate oversight and agreements are in place. Under the HIPAA Privacy Rule
requirements, data partners can release what is called a limited dataset. This is what participating health sites send to the N3C.
o Deductive disclosure review
104
: Prior to accepting Class 2 datasets into the N3C enclave, re-identification risk associated with Class 2 (external)
datasets is assessed during the technical integration call by the Tools and Resources Review Committee comprised of federal employees from
NCATS, the N3C community, and the Information Systems Security Officer. Additional methods are evoked for certain datasets such as using zip3
instead of zip
5 or removal of zip for data from tribal reservations.
o Agreements/approvals for accessing linked data
100
:
– Technical c
ontrol: Linked data are provisioned in the researcher’s private workspace within N3C
–
Non-technica
l controls: The following non-technical controls are in place for accessing the linked level 3 data:
DUR with jus
tification for to access the linked data, approved by N3C Data Access Committee
Signed inst
itutional DUA
Institutional
IRB Letter of Determination (LOD)
Attestatio
n to: N3C Code of Conduct, IT security training, and human subjects protection training
11.2.1.6
PEDSnet
• Description: PEDSnet
106
, a national pediatric learning health system, is a large national community of hospitals and healthcare organizations, researchers
and clinicians, and patients and families. The PEDSnet community works together to identify the most important research questions that can reduce
children's suffering and support their health and well-being.
•
Data sources
107
: EHR data from the 11 PEDSnet participating institutions. The sites include Children’s Hospital of Philadelphia (lead), Ann & Robert H.
Lurie Children's Hospital of Chicago, Children’s National Hospital, Children’s Hospital Colorado, Cincinnati Children’s Hospital Medical Center, Nationwide
Children’s Hospital, Nemours Children’s Health System (both the Delaware and Florida health systems), Riley Children’s Hospital, St. Louis Children’s
Hospital, Seattle Children’s Research Institute, Stanford Children's Health, and Texas Children’s Hospital.
•
Data types
108
: Demographic data, outpatient encounters, inpatient admissions, ER encounters, anthropometrics, vital signs, providers, diagnoses,
treatments, visit payer, lab test results, and medications; access to physician notes and other unstructured data such as operative, imaging, and
pathology reports.
•
Linkage agreem
ents
109
: Linkage is determined by the PEDSnet sites based on the particular use case/research study. When sites sign up to be part of the
PEDSnet network, they must agree to hash their data; however, individual PEDSnet study sites can decide whether to participate in data linkages on a
Page 92 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
study-by-study basis once the PEDSnet Steering Committee approves the data linkage as part of a specific study research plan. PEDSnet also uses
Datavant technology to create deduplicated reports of their data on an annual basis.
• Hash/token generation
109
: Hashes are only generated when a patient is enrolled in a research study conducted by two or more PEDSnet institutions
(they are not generated for routine patient care, and they are not stored in a persistent fashion).
o Standardization/pre-processing of PIIs: Study sites collect data in the PEDSnet Common Data Model which is harmonized to the PCORnet
Common Data Model; therefore, the PIIs collected by individual data partners are standardized.
o Hash/token generation: Performed by the 11 participating institutions using Datavant.
o Hash/token generation success rate: Hashing success rate is calculated by the Data Coordinating Center (DCC). The success rate is dependent on
the PII elements used. The tokens that use last name, first name, sex, and DOB generate the best match with over 93% success rate.
o Hash/token storage and destruction procedures: Hash codes/ tokens are stored at the DCC and destroyed at the end of each specific study; the
study sites never access them.
• Entity reso
lution and data linkage
109,110
: Performed by the PEDSnet DCC for in-network study linkages and annual reports.
o Process:
1.
PEDSnet stu
dy sites install Datavant software which uses various PII elements to generate de-identified cryptographic hash codes for each
patient.
2.
Study sites
send the hash codes and site-level patient IDs to the DCC along with the limited dataset (with dates, zip codes and Census block
groups). The DCC never accesses the PII/PHI.
3.
The DCC uses
the hash codes to perform entity resolution and generates a Master Patient Index (MPI) for the specific study.
4. The DCC links data based on the MPI and distributes the linked dataset to the study team using another DCC-assigned patient identifier that
is unique for that specific study only; study researchers access the linked data within the PEDSnet enclave.
5.
The MPI is de
stroyed after the study is completed; there is no persistent PEDSnet wide MPI.
o Linkage quality assessment
109: For smaller projects: The DCC has validated the linkage in the past by identifying the relevant patient for the
study, assembling the case report forms from multiple sites, and sending the patient and case report form information to the respective PEDSnet
sites, who will then perform a chart review to ensure the patient is the same. For larger projects: chart review is not feasible; so, linkages must
be done more stringently (such as stringent hashing parameters) to ensure quality.
•
Sharing and acc
essing linked data—authorizations/agreements/approvals
34,46
:
o De-identified status of shared data: PII elements are eliminated from data prior to linking. The sites send the hashed codes generated from PII
elements to the DCC which securely stores the hashed codes and performs entity resolution. The DCC then assigns a study specific study ID to
the participant which is only known by the DCC and the study. The study researchers never see the hashed code. The study handles a de-
identified dataset in which the participants are assigned a unique study specific ID.
o Sharing/accessing linked data
109
: PEDSnet Steering Committee approval is required for linking data based on review of the study research plan.
Much of PEDSnet’s work involves large observational studies and operates under waiver of consent, but when consent is obtained, the consent
language is broad and addresses sharing linked data.
Page 93 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o Deductive disclosure review
109
: A risk review is done for each proposed study, but no separate deductive disclosure review of the linked data is
performed. Data use agreement prohibits reidentification and re-use for other studies. All members must agree to the PEDSnet standard policies
when joining the Network; additional terms of data use are stipulated on a case-by-case basis. Policy also requires masking cell counts <11 in
report and manuscripts. However, smaller cell counts can be reported if five or more institutions contributed to the dataset and institutions
agree to allow the small cell sizes to be revealed.
o Agreements/approvals for accessing linked data: Investigators, data analysts, and statisticians who need to access the database will first apply to
be an authorized user. Approval for access is required from the PEDSnet Data Committee. For approved PEDSnet studies, the Data Coordinating
Center will set up a workspace within the PEDSnet database environment and transfer the minimum necessary data for the research project to
the workspace. The workspace will support database and statistical applications allowing the team to conduct data analyses.
11.2.1.7
CDC/The Childhood Obesity Data Initiative (CODI)
• Description
111,112
: The Centers for Disease Control and Prevention (CDC) designed CODI to integrate clinical and community longitudinal data for
childhood obesity research, evaluation, and surveillance. The goals of the initiative were to demonstrate enhanced data capacity to conduct childhood
obesity research and surveillance within an existing distributed health data network (DHDN), and to develop and share reusable tools/resources to
encourage similar work. CODI brings together data stored across different sectors and organizations to create individual-level, linked longitudinal records
that include SDOH, clinical and community interventions, and health outcomes. CODI also creates longitudinal household records, linking individual
longitudinal records within the same household. Five Colorado-based organizations collaborated to expand an existing DHDN, Colorado Health
Observation Regional Data Service (CHORDS) network, to include community-generated data and assemble longitudinal patient records for research.
(CODI expanded in 2020 to include a North Carolina cohort for individual and household level data. Data linkage and standardization and infrastructure
optimization are currently in progress. CODI is a demonstration project of the CHORDS network and operates within the CHORDS governance
framework. The CHORDS Governance Committee, which meets regularly and represents all CHORDS participants, is the decision-making body for both
the CODI project and the CHORDS network.)
•
Data sources
112
: CODI Data Partners share de-identified linked data outside their institutional boundaries based on HIPAA Privacy Rule 45 C.F.R. 164.514
(b). Current data sources include:
o Colorado child health-related data from:
Three health sy
stems (Denver Public Health & Hospital Authority, Children’s Hospital Colorado, and Kaiser Permanente Colorado)
Two communit
y-based organizations (Girls on the Run of the Rockies and Hunger Free Colorado)
o North Carolina:
Clinical and
community data in the Triangle region from health care, local and state health departments, and community-based
organizations
•
Data types
112
: Social determinants of health data, clinical interventions (EHR), community interventions, health outcomes, and geographic data
• Linkage agreements
111
:
Page 94 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o The Master Sharing and Use Agreement (MSUA) is a multiparty reciprocal agreement which defines the roles of each party, general network
functionality relationship between DCC and Data Partners, and tasks the DCC is permitted to carry out for the CODI network. The MSUA
designates and empowers the DCC to conduct PPRL activities, create and distribute queries for data, process and aggregate site-specific
datasets, share study datasets with data users, and delegate authority to the DCC to sign DUAs on behalf of Data Partners.
• Hashed token generation
111
: Anonlink, an open source PPRL software, modified such that data from more than two organizations could be matched at
the same time.
o PII elements
113
: Given name, family name, date of birth, sex, phone number, household street address, and household ZIP Code
o Anonlink schema
114
: Name-sex-DOB-phone, name-sex-DOB-ZIP, name-sex-DOB-parents, name-sex-DOB-address
•
Entity reso
lution
115
: Performed by the DCC, which the MSUA designated as the University of Colorado.
o Process:
1.
The DCC shar
es with each Data Partner configuration information which contains information on how to normalize PII before hashing.
This ensures a consistent representation of PII which serve as the inputs for hashing.
2. Data Partners input PII along with a randomly generated encryption key to compute specific hash values for each individual. Because a
given individual corresponds to multiple hash values, the Data Partner also includes a HASHEDID, which is the hash of that site’s arbitrary
patient identifier from the CODI warehouse (this ID is not PII).
3.
The Data Partners
send the hashed values and the HASHEDID to the DCC.
4. After receiving the collections of hashed values from Data Partners, the DCC builds a hash index from these values to identify any
matches. By combining hash values and the HASHEDIDs, the DCC can determine all of the hashes that correspond to the same individual.
The DCC then assigns a unique network wide ID, known as the LINK_ID, for each unique individual.
5.
The DCC then s
ends the mapping between the HASHEDID and the globally unique LINK_ID to the Data Partner. Each Data Partner stores
its patients’ LINK_IDs in its local CODI warehouse for use in future research queries. Sharing of the LINK_ID is prohibited with any
research dataset.
•
Data linkage
115
: Study-specific linkage model; the DCC performs the data linkage.
o Process:
1.
Researcher
s query data across multiple CODI sites after an agreement has been established with the DCC.
2.
Once the agreement
is in place, the DCC distributes queries to Data Partners. Data Partners execute the agreed upon query and review
site-specific results before returning results to the DCC.
3.
The DCC then
combines site-specific results to build a longitudinal record in accordance with the applicable protocol and returns the
research dataset to the researcher/data requester. The DCC replaces the LINK_ID with a study-specific STUDY_ID, unique to the query,
before distributing the data to the requester.
•
Sharing and acc
essing linked data—authorizations/agreements/approvals
111
:
o Governance s
tructure:
–
CODI DCC work
s with data requesters to develop and submit local IRB protocols for approval.
Page 95 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
– CHORDS Research Council oversees research reviews and makes recommended approvals for studies to the Governance Committee.
–
CHORDS Gove
rnance Committee and Research Council handle the decision-making, creating guiding principles, and maintaining the
governance plan which documents policies and procedures for data request initiation, Data Partner approval and participation, regulatory
requirements, security, privacy and confidentiality, and publication and presentation guidelines.
– Multi-organizational DCC-merged datasets with site identifiers removed is owned by the DCC, who then acts as the data steward.
o
De-identificatio
n status of shared data: Longitudinal records are stripped of site and patient identifiers (i.e., the unique ID resulting from PPRL)
and are temporarily
provisioned for analysis to researchers on a study-specific basis. For recipients of longitudinal records who are CODI Data
Partners, stripping identifiers ensures that patients from the receiving Data Partner cannot be reidentified. Policies prohibit data users from
reidentifying patients.
o Authorization for data sharing:
–
Researchers co
mplete a CHORDS Project Intake Form which is then sent to the data partners via email by the Research Project Manager.
–
The CHORDS R
esearch Council provides a recommendation to the CHORDS Governance Committee. At the subsequent CHORDS Governance
Committee meeting, the study is presented, and the Committee votes on formal approval for the study to proceed. Once approved, Data
Partner participation is formally requested, and written participation responses are required.
–
CODI Data Use and T
ransfer Agreement requirements pertaining to receipt of a limited dataset or individual-level de-identified dataset
differ, dependent upon the data user:
For a study l
ed by a Non-Data-Partner Data User, a study-specific Data Use and Transfer Agreement is required.
For a study l
ed by a Data-Partner Data User, a study-specific Data Use and Transfer Agreement is not required.
–
The MSUA is a m
ultiparty reciprocal agreement which defines the parameters of data exchange, approved uses of CODI data, and
expectations of end-users. The reciprocal nature of the MSUA allows it to act as a DUA for a project initiated by a CODI Data Partner. The
MSUA also allows the DCC to create and distribute queries, process, and aggregate site-specific datasets, share study datasets with data
users, and delegated authority to the DCC to sign DUAs on behalf of Data Partners.
–
All CODI st
udies require approval or designation as non-human subjects research by the requesting researcher’s IRB.
o Deductive disclosure review: Deductive disclosure review is not performed for datasets that are linked for CODI studies. Additionally, the
Working Group determined that garbled information (PPRL identifiers) did not require added governance protections to maintain privacy. Data
Partners would be notified of any unapproved use or breach of garbled information or the unique identifier. The governance plan establishes
guidelines for data destruction at a study’s conclusion.
o Agreement/approvals for accessing linked data:
–
The MSUA inc
ludes a reciprocity provision that allows sharing of limited or de-identified datasets among CODI Data Partners without an
additional DUA. For researchers from organizations not participating in CODI, a study specific DUA is required. MSUA appendices include a
template DUA approved by CODI Data Partners for use in research studies and a Responsible Use of Data Agreement that defines the
responsibilities of researchers receiving CODI longitudinal records.
Page 96 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
– CODI data users are required to sign a Responsible Use of CODI Data Agreement and return the signed agreement to the DCC. The
Responsible Use of CODI Data Agreement defines the expectations of a data user, as a recipient of a CODI research dataset from DCC, and
the limitations on the use of that research dataset. When a data user receives a research dataset containing individual-level data from the
DCC, the Responsible Use of CODI Data Agreement functions in concert with the specific terms detailed in the Data Use and Transfer
Agreement.
Page 97 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.2
Governance Summary for Record Linkage Implementations Not Using PPRL
Appendix Table 3: Governance Summary for Record Linkage Implementations Not Using PPRL
Record Linkage
Implementation
Datasets Linked Authorization for Linkage
Methodology
PII Elements
Used for
Linking
Entity
Resolution
(Matching)
Performed
by
Data Linking
Performed
by
Data Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing Linked
Data
1 Database of
Genotypes and
Phenotypes
(dbGaP)
Genotype data,
phenotype data,
genome wide
association (GWAS)
data, Short Read
Archive (SRA) data,
expression data, image
data, etc.
Data Submitter Data
Submitter
NIH Data Access
Committee
approval
Genetic
Relationship and
Fingerprinting
(GRAF) analysis
within studies and
Subject ID
(string) matching
across studies
Not applicable dbGaP Data
Curation
Team
Requesting
investigators
Linked database
model for
matches across
studies and
study-specific
linkage model for
within study
Controlled
data but
public dbGaP
IDs
Section
11.2.2.1
2 All of Us (AoU)
o EHR data, genomic
data via
biospecimens,
physical
measurements,
patient provided
information (PPI) via
surveys (topics
include
sociodemographic,
overall health,
lifestyle, and health
care access and
utilization), data from
mobile devices,
biosamples
o Future external
datasets: mortality
data, EPA, USDA,
health registry,
pharmacy data, etc.
Consent from
participants and
IRB approval to
use PII for
linkages with
external datasets
Consent from
participants
Approval from the
AoU Resource
Access Board
(RAB) for access to
individual level data
via the registered
and controlled tiers
Internal AoU data
linkages: using a
common
Participant ID
(PID, a unique
random 10-
character string)
External data
linkages: two
current pilot/
assessment
projects via
Datavant
Internal AoU
data linkages:
Not applicable
For two
external data
linking
projects:
Unknown
Internal AoU
data
linkages:
AoU Data
and
Research
Center (DRC)
For two
external data
linking
projects: third
party
AoU Data and
Research
Center (DRC)
Internal AoU
data linkages:
Linked database
model
For two external
data linking
projects: Study-
specific linkage
model
o Public Tier
o Registered
Tier
(Enclave)
o Controlled
Tier
(Enclave)
Section
11.2.2.2
Page 98 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementation
Datasets Linked Authorization for Linkage
Methodology
PII Elements
Used for
Linking
Entity
Resolution
(Matching)
Performed
by
Data Linking
Performed
by
Data Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing Linked
Data
3 PCORnet-DS
Connect/DS-
DETERMINED
Study
o EHR data of
PCORnet patients
with Down Syndrome
who were recruited to
DS-DETERMINED
for linkage with
external data which
include:
o DS-Connect demo-
graphic and Initial
Health Questionnaire
(IHQ) data
o Self Determination
Inventory (SDI)
survey data
Informed consent Informed
consent for
sharing with
study team
and with a NIH
designated
data
repository
Access
requirements to be
defined based on
the NIH designated
repository used for
sharing the data.
Linked using
unique patient
specific referral
code, which is a
combination of
de-identified
pat_id generated
at the PCORnet
sites and the
study_id
generated by
REDCap for the
study
Study Referral
Code
(=study id+
pat_id)
Study Team Study Team Study-specific
linkage model
Controlled Section
11.2.2.
3
4 Georgetown
Federal Statistical
Research Data
Center (FSRDC)
o Survey data from
American Community
Survey (ACS),
Population Survey,
Survey of Income
and Program
Participation (SIPP)
o Administrative data
from Social Security
Administration (SSA)
and the Centers for
Medicaid and Medi-
care Services (CMS)
o Other types of data
from 5 agencies:
Agency for
Healthcare Research
and Quality (AHRQ),
Bureau of Economic
Analysis (BEA),
Bureau of Labor
Statistics (BLS),
National Center for
Health Statistics
(NCHS), and
National Center for
Science and
Engineering Statistics
(NCSES)
o Data provided by
researchers
o Data owners
(per their data
collection
authorities
and/or IRBs)
and Census
requirements
o For statistical
purposes only
o Data
owners (per
their data
collection
authorities
and/or
IRBs) and
Census
requirement
o For
statistical
purposes
only
o Data owners and
Census
requirements,
including Special
Sworn Status to
access the
FSRDC data
enclave
o For statistical
purposes only
Probabilistic
matching
software (Multi-
Match) to
generate a
Protected
Identification Key
(PIK)
o SSN
o Date of birth
o Name
o Gender
o Address/es
Census Census Study-specific
linkage model
Enclave Section
11.2.2.4
Page 99 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementation
Datasets Linked Authorization for Linkage
Methodology
PII Elements
Used for
Linking
Entity
Resolution
(Matching)
Performed
by
Data Linking
Performed
by
Data Linkage
Model
mm
Data Access
Model
nn
Additional
Information
Link
Record Linkage Sharing
Linked Data
Accessing Linked
Data
5 National Center
for Health
Statistics (NCHS)
with National
Death Index (NDI)
o National Center for
Health Statistics
(NCHS) survey data
o Mortality data from
the National Death
Index
o Consent from
participants
o NCHS
Research
Ethics Review
Board (ERB)
o Consent
from
participants
o Public
version has
been
cleared for
public
distribution
by CDC.
o Access to
restricted-use
files is reviewed
by the NCHS
Research Data
Center (RDC)
o Combination of
probabilistic
and
deterministic
data linkage.
o First name
o Middle initial
o Last name
o Date of birth
o State of birth
o State of
residence
o Race
o Sex
o Marital
status
o SSN9 or
SSN4
Data Linkage
Program at
NCHS
Data Linkage
Program at
NCHS
Linked database
model
Enclave Section
11.2.2.5
6 The Child
Maltreatment
Incidence (CMI)
Data Linkages
project,
Alaska
Department of
Health and Social
Services/ Oregon
Health Sciences
University
(ADHHS/ OHSU)
Project
o 2009-2011
Pregnancy Risk
Assessment
Monitoring System
(PRAMS) survey
data from Oregon
Health Authority
(OHA), Oregon
Public Health
Division
o 2009 birth and death
Records from OHA,
Center for Health
Statistics
o Child welfare data
from Oregon
Department of
Human Services
IRB approvals
from OHA and
Oregon Health
and Science
University
IRB approvals
from OHA and
Oregon Health
and Science
University
Access is restricted
to staff listed in the
DUA and IRB
protocol
Combination of
probabilistic and
deterministic
matching method
that leveraged
ALCANLink
technology, which
was created for a
similar previous
project
o First name
o Last name
o Middle
name
o Date of birth
o Birth
certificate
number
Trusted third
party:
Integrated
Client
Services
(ICS)
Trusted third
party:
Integrated
Client
Services
(ICS)
Linked database
model
Controlled Section
11.2.2.6

Page 100 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.2.1
Database of Genotypes and Phenotypes (dbGaP)
• Description
116
: The database of Genotypes and Phenotypes (dbGaP) was developed to archive and distribute the data and results from studies that have
investigated the interaction of genotype and phenotype in humans. When dbGaP was established in 2007, data that were uploaded originated from
studies that used different subjects, but with time, newer studies began submitting data that used subjects from previous studies. To link the same
subjects from the newer studies to the previous studies, dbGaP developed a method to match the study provided subject IDs while generating a unique
dbGaP subject ID, which serves as the primary key for all data in dbGaP
117
.
•
Data sources
118
: Genotype, phenotype, GWAS, SRA, expression, and image data generated by NIH funded studies.
•
Data types
119
: The types of data distributed through the dbGaP include phenotype data, GWAS data, summary level analysis data, SRA data, reference
alignment (BAM) data, VCF (Variant Call Format) data, expression data, imputed genotype data, image data, etc.
•
Linkage authori
zation/agreement/approval
118
: Based on decisions made by the data submitter. The GDS Institutional Certification
34
does not address
record linkage or require explicit consent for record linkage.
• Entity resolution
117
: Linked database model
1.
Study submi
tters are required to provide the following along with the de-identified study data:
–
Subject Co
nsent (SC) file: Comprehensive list of all unique de-identified subject IDs, their assigned consent group, and biological sex value.
[Each subject should be submitted with a single, unique, de-identified subject ID. Subject IDs should be an integer or string value.]
–
Subject Sample
Mapping (SSM) file: File mapping SUBJECT_IDs (consented subjects and their phenotype data) to molecular SAMPLE_IDs. Has
explicit list of samples from each subject. A Sample ID is a biological aliquot from a person and is the primary key for the molecular data.
–
Subject Phenotypes file: Includes any number of measured data and/or descriptive traits per individual and is how the demographic, clinical
and exposure variables for a person are submitted. The primary ID in this file is the SUBJECT_ID. All SUBJECT_IDs must be listed in the SC file.
–
Pedigree file: Li
sts the genealogical relationships of subjects within a study. If there are no known relationships, this file does not need to be
submitted.
–
Sample Attribut
es file: Includes sample level variables such as body site, analyte type, etc. The primary ID in this file is the SAMPLE_ID. All
SAMPLE_IDs in this file must be listed in the SSM file.
–
Data Dictionar
y: Table that defines and describes the variables in the corresponding dataset (DS) file. If the DS file contains coded values, the
code meanings need to be included in the DD file. Each of the above listed files also have a data dictionary defining the variables and coded
values.
2.
Once dbGaP receives
the above files, the dbGaP Curator performs entity resolution within and across studies:
–
Within studies: Genetic Relationship and Fingerprinting (GRAF)
120,ss
analysis is used to assess inconsistencies between molecular data sample
IDs and phenotype sample IDs, phenotypic and genotypic sexes, and self-described races/ethnicities and populations inferred from
ss
Per Mike Feolo, Staff Scientist at NCBI and Team Lead for dbGaP: GRAF has been incorporated into the dbGaP automated curation pipeline to check for relatedness within a study as a data curation and quality measure.
It is not typically used to check for relatedness or similarity across studies. GRAF is efficient and accurate at identifying the same subjects across datasets. It could be useful for data linkage; however, dbGaP policy is that
subject linking across studies using GRAF is not performed unless the PI approves it or unless there is a “good reason” to do so. One example of using GRAF as a data linkage tool is NHLBI’s TopMed project, where GRAF
was used to prove subjects are exact matches in these large cohorts.
Page 101 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
genotypic data, unintended data duplications, incorrect pedigree information, and subject relationships. GRAF ensures that the relatedness
status between samples correspond to what are being reported by submitters, by inferring the sexes, ancestry backgrounds, and
relationships between samples using the fingerprint genotypes.
– Across studies: Incoming subject ID is checked against existing studies using a custom string matching analysis. If the subject ID of the
incoming data potentially matches with existing data, curators notify the submitter to verify the origin of the matched subjects.
If the submitter
wishes to confirm that the incoming subject ID matches with an existing subject ID in dbGaP, the Subject Consent File is
sent back to the submitter to add the existing study’s Subject_Source and Source_Subject_ID and resubmit to dbGaP. (The source is the
source repository, consortium, institute, or other study where the existing subject originated
121
.)
Once resubm
itted, dbGaP will assign the unique dbGaP subject ID for each individual. Submitters will have access to the dbGaP subject
IDs via a Sample Status Telemetry Report (SSTR) provided by dbGaP once the IDs and consents have been loaded into the dbGaP
database.
•
Sharing and acc
essing linked data—authorizations/agreements/approvals:
o Authorization for sharing linked data:
–
Consent
122
: As of 2015, the Genomic Data Sharing Policy requires that data submitted to dbGaP be consented for sharing to a public
database. All subjects must be assigned a single consent group in the Subject Consent files upon submission. Consent groups correspond
with the data use limitations on the Institutional Certification, which include: General research use, Health/Medical/Biomedical, Disease-
Specific, and other. Data use limitation modifiers that can be added, if applicable, are local IRB approval required, Publication required,
Collaboration required, Not-for-profit use only, Methods, and Genetic studies only.
–
Consent withdr
awal
123
: Consent versioning is a dbGaP feature which allows for the termination of distribution of datasets with participant
data who wish to retract their consent for sharing. Since dbGaP is not the primary study site, but instead is an archive and distribution point
for data supplied by primary study sites, withdrawal of data from an individual participant in the original study requires the submission of a
new version of data tables to dbGaP by the primary study site. The dbGaP study accession number (for example phs000001.v1.p1) will
change to account for these versioning changes. If any participants have withdrawn consent or changed into a different consent group, then
the “v” version and the “p” participant set will increment, and only the most recent version of the data will remain accessible.
– Institutional certification
34
: The institution, in consultation with its IRB or equivalent body, assures NIH that the study data being submitted is
consistent with the NIH GDS Policy, and with the informed consent of the original study participants. Data use limitations (and modifiers)
should be based on language in informed consent forms.
o De-identification status of shared data: To comply with the Genomic Data Sharing Policy
24
, data should be de-identified to meet the definition
for de-identified data in the HHS Regulations for Protection of Human Subjects (Common Rule) and also be stripped of the 18 identifiers listed in
the HIPAA Privacy Rule, e.g., names, cities, dates, telephone numbers, social security numbers, and any other potentially identifying information,
characteristic, or code. Additionally, submitter-provided subject IDs must be two steps removed from PII in study records; therefore, the dbGaP
subject IDs are also at least two steps removed from PII.
o Deductive disclosure review: A formal deductive disclosure review is not conducted.

Page 102 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o Agreements/approvals for accessing linked data:
– Consent
34
: The informed consent under which the data or samples were collected determines whether the submitted data should be
available in an unrestricted or a controlled access manner. Controlled access data are available to investigators only after review of their
proposed research use. Unrestricted access data are publicly available to anyone.
–
Data Access Co
mmittee (DAC) approval
124
: Access to individual-level controlled access data is dependent upon approval by the data access
committees. NIH DACs review all requests for access to datasets distributed through dbGaP. Some DACs are specific to a single NIH IC, such
as the National Human Genome Research Institute (NHGRI) and review only Data Access Requests (DARs) funded by and/or submitted to
dbGaP through that IC. Other DACs represent multiple ICs with similar research goals, such as the Joint Addiction, Aging, and Mental Health
(JAAMH) DAC, which includes the National Institute on Drug Abuse (NIDA), National Institute on Alcohol Abuse and Alcoholism (NIAAA),
National Institute on Aging (NIA), and National Institute of Mental Health (NIMH). Users must sign dbGaP's DUC Agreement, including
agreement not to use the requested datasets, either alone or in concert with any other information, to identify or contact individual
participants from whom data and/or samples were collected, unless they have specific IRB approval to do so. Additionally, the NIH GDS
Policy prohibits investigators who download unrestricted-access data from NIH designated data repositories from attempting to identify
individual human research participants from whom the data were obtained
24
.
– IRB approval
125
: Some datasets require additional local IRB approval for use, if required per the Institutional Certification.
11.2.2.2
All of Us (AoU)
• Description
126,127
: The All of Us (AoU) Research Program is a historic effort to collect and study data from one million or more people living in the United
States. The program aims to reflect the diversity of the United States and to include participants from groups that have been underrepresented in health
research in the past. Unlike research studies that focus on one disease or group of people, AoU is building a diverse database that can inform thousands
of studies on a variety of health conditions. The program began national enrollment in 2018 and is expected to last at least 10 years.
•
Data sources:
o AoU
data
128
: From direct volunteers (DV) collected through the AoU Participant Portal or from participants enrolled through participating
healthcare provider organizations (HPO) collected through the Data and Research Center (DRC) at Vanderbilt.
o External data
129
: Sources for linkage of AoU data with external data vary. AoU has linked to Census American Community Survey (ACS) data and
anticipates linking with mortality data, Census, Environmental Protection Agency (EPA), U.S. Department of Agriculture Food Access Research
Atlas, private and public claims data, health registry data, private pharmaceutical data, etc.
o PPRL projects: Thus far, AoU PPRL projects have only been performed for the purposes of evaluation and experimentation, but not for releasing
linked data through the workbench, and include:
–
A collabora
tion among AoU, N3C, and NHLBI BioData Catalyst (a project within the NIH COVID Jumpstart initiative
130
) to identify common
participants across the three programs and internally report results at the aggregate level; no individual-level data was exchanged across or
linked between systems
Page 103 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
– An EHR/claims data linkage project to assess the completeness of the AoU EHR data by linking with external claims data from a claims
aggregator
• Data types
131
: EHR data, genomic data via biospecimens, physical measurements, patient provided information (PPI) via surveys (topics include
sociodemographic, overall health, lifestyle, and health care access and utilization), data from mobile devices, measurements, biosamples.
•
Linkage author
izations/agreements/approvals
132, 133
:
o Consent: Consent for data linkage is collected from participants at the time of enrollment. AoU is not currently enrolling participants under 18
years of age. The informed consent for AoU is modular. Each module requires an electronic signature from the participant. Module 1 (Primary
Consent)—Consent to Join the All of Us Research Program—gives an overview of all program activities. Signing the primary consent indicates
general understanding of the research program and approval to take part in the PPI, Data Linkage, Physical Measurements, Biospecimen
Collection, Biobanking, Biomarker assays, Genomic Testing, and Sensor/Wearable Technology activities if invited. The Consent to Join the All of
Us Research Program has the following language:
–
“If you join
, we will gather data about you. We will combine it with data from other people who join. Researchers will use this data for lots of
studies. By looking for patterns, researchers may learn more about what affects people’s health.”
–
“If you deci
de to join All of Us, we will gather data about you. We will gather some of the data from you directly. We will gather some of the
data from elsewhere.”
–
"Data about you
r health from other sources: We will add data from other sources to the data you give us. For example, environmental data
and pharmacy records. This will give researchers more data about factors that might affect your health.... We will use data that identifies
you like your name and date of birth to add data that is specific to you. For example, we may add data from pharmacy records or health
insurance records. If you have had cancer, we may add data from cancer registries...”
– “These other sources can contain sensitive data. For example, they may tell us about your mental health, or use of alcohol or drugs. They
may contain sexual or infection data, including HIV status. Because of this, we will ask the All of Us ethics committee to review and approve
each data source before we add it.”
o HIPAA Authorization: Participants must specifically consent to AoU accessing and linking to data from their EHR by signing the “All of Us
Research Program HIPAA Authorization for Research EHR/Part 2 Supplement
134
"
– “We will acc
ess your whole EHR. That means we will take a copy of all the tests, results, and images in your EHR. This includes data about
your diagnoses, medications, symptoms, allergies, and treatments.”
–
“We will add
your EHR to your All of Us record. Your record will be part of the All of Us scientific database”
o IRB approval
133
: While the primary consent encompasses data linkage, it is anticipated that prior to linking participant data with external
sources, an amendment will be filed with the IRB for any linkages to “health registries” or “claims data” that require the DRC to share
participant-identifying information to an outside entity. Such submissions would detail the data to be linked and the general methods for doing
so. No additional participant consent will be undertaken. The consent discusses that identifying information may be shared in this process.
• Entity resolution/data linkage: Linked database model
Page 104 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o Internal AoU data linkage
133
: AoU data are linked upon arrival to the raw data repository (RDR) using a common participant ID (PID) to create the
AoU Core Dataset:
–
Participan
ts are assigned a PID, a unique random 10-character string (format P000000000), upon registration with the AoU Program. The PID
generation is not based on PII elements.
– Individual level participant data collected through the Participant Portal (PPI, mobile device data, fitness tracker data, biosample, etc.) and
via the DRC (EHR, physical measurements, biosample) is linked using the common PID. This Core Dataset is stored in the RDR, maintained by
the DRC.
–
New individ
ual-level datasets are merged with existing participant study record in the RDR using PIDs.
o External data linkage
129
: AoU has performed linkages with external data sources using PPRL tools only for evaluation and experimentation and
plans to perform geocoding methods.
–
For the two P
PRL projects, AoU is exploring Datavant’s PPRL tool.
1.
For the col
laboration project among AoU, N3C, and NHLBI BioData Catalyst (the COVID Jumpstart initiative
130
), hashed tokens based on
first name, last name, date of birth, SSN, gender, zip, email, and cell (18 tokens) are generated by each collaborator and matched via a
trusted third-party honest broker to identify how many participants are represented in both programs. No data transfer has occurred
between the organizations.
2.
For the EHR/c
laims data linkage project, AoU DRC has engaged an external party to generate tokens for the participants and identify
matches with the claims data. No data transfer has occurred yet.
–
AoU program is
still deciding which PPRL tool to officially use moving forward. AoU has established a Working Group focused on science,
policy and technology of data linkages; this Working Group is developing criteria and governance for data linkages and will develop criteria
for data linkages in 2022.
–
Geocoding b
ased linkages have not yet occurred. When a participant registers, they provide their address, which will be used for linking their
data using geocoding. AoU would like to perform data linkages based on geospatial location, for example with EPA data, environment
exposure data, or other county level data, but would need to determine the acceptable level of geolocation information to use without the
risk of deductive disclosure. AoU is in discussions with NIEHS to explore geocoding.
o Linkage quality assessment
129
: AoU is still in a very early stage of exploring data linkages and has limited experience with assessing the quality of
the linkages.
–
Internal Ao
U data linkages: AoU evaluates quality for internal data linkages by detecting duplicated data (via PID) to prevent data duplication
in the RDR.
–
External dat
a linkages: AoU is using gold standard external datasets to ensure high quality and trustworthy data sources (for example: the
American Community Survey (ACS)/Census are gold standard sources as Census is collecting data at the geographical level. For the
EHR/claims data linkage project, the team is still determining how to assess the quality of the matches.
•
Sharing and acc
essing linked data—authorizations/agreements/approvals:
Page 105 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o De-identification status of the linked data
135
: AoU does not release data that directly identifies participants (i.e., data that can be linked to
specific individuals by users either directly or indirectly through coding systems), even to users with access to controlled data. These inaccessible
data elements include, but are not limited to, personal names, addresses of residence or employment, medical record numbers, and social
security numbers. Furthermore, these individual-level data will be coded, and authorized users will not be given the key to this code.
o Deductive disclosure review:
–
Internal AoU
data linkages
133
: AoU currently does not have a disclosure review board in place, however, all internal dataset linkages are
reviewed for disclosure risk prior to release. Additionally, direct identifiers are removed from structured and unstructured data streams, and
the data gets organized and curated into a Curated Data Repository (CDR). A de-identified Core Dataset made available as registered tier or
controlled tier via the AoU Research Program Research Portal/Research Workbench. AoU will use a variety of approaches to remove explicit
personal identifiers such as name, email, phone number, street address, medical record number (MRN) and Social Security number (SSN)
from the datasets made available for research purposes, including from free text data sources such as open response fields and EHR notes.
As part of this process, personally identifiable information (PII) is removed from participant data as follows: PII from structured fields (e.g.,
Yes/No questions, selecting a birthdate, rating an experience from 1 to 10) will be replaced with code. The Committee on Access, Privacy,
and Security (CAPS) will evaluate these approaches prior to release and routinely control their quality to minimize the risk of inappropriate
re-identification.
–
External dat
a linkages
129
: AoU currently does not have a disclosure review board in place, however, datasets / assets are reviewed for
disclosure risk prior to release. [Note: the Working Group is expected to develop governance around this type of data linkage.]
o Authorization for sharing linked data
132
: Authorization to share AoU data, including linked data, via the CDR is based on informed consent:
–
“The scient
ific database will have individual-level data and samples. This includes your DNA data. Access to this database will be controlled.
Researchers will have to be approved by All of Us to use this database. They will have to have special training before they can be approved.
Their research may be on nearly any topic. They may look for patterns in DNA. This may help them discover different ways that DNA affects
people. These researchers may be from anywhere in the world. They may work for commercial companies, like drug companies. They may
be citizen or community scientists. Citizen and community scientists are people who do science in their spare time.”
– “The data researchers get from studying your samples and DNA may be added to the All of Us scientific database.”
–
“Except if you w
ithdraw (quit) or there are limits imposed by law, there is no limit on the length of time we will store your samples and data.
Researchers will use your samples and data for research long into the future.”
–
“In order to wo
rk with your health data, researchers must sign a contract stating they will not try to find out who you are.”
–
“Once your i
nformation is shared with All of Us, it may no longer be protected by patient privacy rules (like HIPAA). However, it will still be
protected by other privacy rules. These include the rules that researchers must follow to access the All of Us scientific database.”
o Agreements for accessing linked data
133
: De-identified individual level (linked) data stored in the CDR is made available for querying by the research
community through a dedicated analysis platform, the AoU Research Program Research Portal/Research Workbench, for research. The PID is
converted to a research ID before releasing it for access, and the research ID maintains the linkage of the de-identified data. Qualified researchers
who wish to access the data must agree to not remove data (registered and controlled tier—see definitions below) from the Research Portal without
Page 106 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
approval. The AoU will bring researchers to the data rather than allowing researchers to download data to their own machines. The CAPS serves as
the stewards of the data. The Research Portal has three levels of data access: public tier, registered tier, and controlled tier. The registered tier
includes participant-level data with a number of transformations to protect participant privacy (re-identification risk analysis performed). The
controlled tier contains data elements that may not, in their own right, readily identify individual participants, but may increase the risk of
unapproved re-identification when combined with other data elements; data is pre-processed to minimize risk by domain experts. Data is at the
more granular level (zip 3 vs state) in the controlled tier than in the registered tier. The requirements for access vary between the public vs
registered and controlled tiers (note: access requirements are subject to change):
– Public Tier: No individual participant level information included. It contains only summary statistics and aggregate information (bin size set at
20 individuals). No login required to access public tier data.
– Registered and Controlled Tier: While the two tiers may have different requirements in the future, the current requirements for access
include:
Institutional Data Use Agreement
Registration and eRA Commons Account Validation
Responsible Conduct of Research Training
Signing the Data User Code of Conduct
Approved access by the Research Access Board (RAB)
o Approvals f
or accessing linked data
135
: The research that occurs within the CDR Workbench accessible via the registered or controlled tiers are not
subject to IRB review or approval. Users may be bound by institutional policies governing research, which may include local IRB review. Data user’s
institution must enter into an institutional data use agreement with AoU for an individual to become an authorized user. Review of the research
proposal and approval from the AoU Resource Access Board (RAB) is also required.
11.2.2.3
PCORnet-DS Connect/DS-DETERMINED Study
• Description
136
: The DS-DETERMINED study supports and enhances Down Syndrome research and the DS-Connect® registry by leveraging PCORnet, the
National Patient-Centered Clinical Research Network. This study links PCORnet to the DS-Connect® Registry and tests capability in three dimensions: 1)
increase DS-Connect® enrollment, 2) extract of clinical observations, treatments, and outcomes from PCORnet patients, and 3) conducting cognitive
assessment survey of self-determination in the PCORnet Down Syndrome population.
•
Data sources
137
:
o PCORnet, the National Patient-Centered Clinical Research Network. PCORnet partners of this study include University of Missouri, University of
Kansas Medical Center, Allina Health System, and University of Pittsburgh.
o DS-Connect Registry. A resource for people with Down Syndrome and their families to connect to researchers and health care providers, express
interest in participating in certain clinical studies, and take confidential health-related surveys.
o Self-Determination
Inventory System (SDIS) Data Dashboard, a platform for measures related to self-determination.

Page 107 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
• Data types
138
: EHR data from PCORnet will be linked to demographic and Initial Health Questionnaire (IHQ) data from DS-Connect Registry and the
survey data from the SDIS Data Dashboard. The PCORnet common data model is used to encode and organize the EHR data.
•
Linkage agreemen
ts
137
: PCORnet sites agree to become a recruiting site for the study; DS-DETERMINED does not have a separate agreement with the
SDI website because the site is created and operated by the University of Kansas, which is a PCORnet site. Although DS-Connect does create NDA GUIDs
for participants, the GUIDs are not shared with PCORnet as part of this study.
•
Entity resol
ution/data linkage
137
: Linked database model. Study participants are tracked using a unique referral code, which is a combination of the
study ID and patient ID; the referral code links the participants across PCORnet/REDCap, DS-Connect and the SDI website. The software for DS-
Determined linkage is available on GitHub (https://github.com/kumc-bmi/ds-determined-tools
).
o Process for tracking participants:
1.
Eligible pa
rticipants (with a pat_id) are recruited from PCORnet and are sent a unique trial invite ID
2.
When the pat
ient clicks on the link in the invite, they are directed to REDCap where they log in with their trial invite ID to complete their
consents for the study. Once registered for the study, REDCap generates a unique referral code which is a combination of pat_id + study_id,
and the participant is sent the link to the DS-Connect.
3.
Participants t
hen register at DS-Connect and SDI using the unique referral codes. The referral code is used to link across PCORnet/REDCap,
DS-Connect and the SDI website, while keeping the participant’s PII elements private.
4.
Participant dat
a from DS-Connect and SDI are sent to PCORnet/REDCap for linkage via the referral code. PCORnet sites store the mapping of
trial invite codes to pat_id, which allows for linkage to EHR data.
o Quality assessment
137
: The linkage quality is assessed in terms of maintaining the linkage process rather than the matching rate because linkage
is performed via explicit referral codes that were tracked and matched automatically. The team receives raw files at a weekly cadence from both
SDI and DS-Connect that report on which patient has completed the study and how far they have progressed. The DS-Connect files report how
far the patient has progressed in filling out the DS-Connect surveys. The SDI report only states whether the participant has completed the survey
(yes or no).
• Sharing and acc
essing linked data—authorizations/agreements/approvals:
o De-identification status of shared data
137
: The study is currently in progress, and the data has not been shared to external researchers; however,
when shared it will be shared through the INCLUDE Data Hub and de-identified of 18 HIPAA identifiers.
o Authorization for sharing linked data
138
: DS-DETERMINED Consent Forms include sharing data with the Kansas University Medical Center (KUMC)
research team and with a NIH designated data repository. There is also a disclaimer that de-identified research results will be shared outside of
KUMC for research purposes. Participants may contact Evan Dean, Ph.D., if they would like to have their data removed from future use in the
study.
o Deductive disclosure review
137
: Deductive disclosure review is not conducted on the linked data.
o Agreements/approvals for accessing linked data
137
: NIH will have to determine how the linked data should be shared via the new INCLUDE Data
Hub. The INCLUDE Data Hub has a multi-tiered access approach with controlled and registered tiers. Participants and families have access to the
registered tier data. Access to both registered tier and controlled-access data require agreeing to not attempt to re-identify or contact
Page 108 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
participants represented in INCLUDE Data Hub. The agreements for accessing the registered tier
139
is available through the INCLUDE Data Hub
and for accessing the controlled tier via dbGaP
66
.
11.2.2.4
Georgetown Federal Statistical Research Data Center
• Description
140
: The Census Bureau operates 31 open Federal Statistical Research Data Center (FSRDC) locations, including at the Massive Data Institute
at Georgetown University. The FSRDCs partner with over 50 research organizations including universities, non-profit research institutions, and
government agencies
141
. Federal and state statistical agencies collaborate with the Census Bureau to provide microdata to approved researchers in the
secure FSRDC environment. At FSRDCs, qualified researchers can access restricted-use microdata from a variety of statistical agencies to address
important research questions.
• Data sources
140
: American Community Survey (ACS), Population Survey, and Survey of Income and Program Participation (SIPP), Social Security
Administration (SSA), Centers for Medicare & Medicaid Services (CMS), Agency for Healthcare Research and Quality (AHRQ), Bureau of Economic
Analysis (BEA), Bureau of Labor Statistics (BLS), National Center for Health Statistics (NCHS), National Center for Science and Engineering Statistics
(NCSES) and data provided by researchers
•
Data types
140
: Survey data, administrative data, health data, economic data, U.S. labor/workforce data, science and engineering and technology
workforce data
•
Linkage agreem
ents
142
: Census determines whether datasets they have acquired for agency use can be linked. For linkages involving researcher-
provided data, the researcher must certify and show proof (via the data sharing agreement) from the data owner that they have permission to link data
from a specific agency or multiple agencies with Census data and with each other. Since the linkage is based on PII, an IRB from the organization
contributing the data may need to make a determination regarding whether this use of the data is appropriate. Depending on how the data were
collected, other federal authorities may dictate the data linkage.
•
PIK generation
process
143
: The Person Identification Validation System (PVS) at Census uses probabilistic matching software (Multi-Match) to assign a
unique Census Bureau identifier called Protected Identification Key (PIK) for each person that matches a reference file of government administrative
data.
•
Entity reso
lution/data linking: Study-specific model. Data linkage is done by Census for a fee ($19,000 per linkage/join request; one request can include
linkages across multiple datasets). Linkage is only allowed for statistical purposes.
o Process
142
:
– The PVS match
es incoming files to reference files created with data from the Social Security Administration (SSA) Numerical Identification
file (= SSN master file/Numident) and SSA data with addresses obtained from federal files. The reference files used for the matching includes
federal data for generally the same time period as the incoming files.
– Reference file contains one record for each SSN, and is based on SSN, the Census Bureau assigned PIK, date of birth, name, gender, and
addresses where the person may have resided (within a specified time frame).
– When a linkage can be made between an incoming file and the reference file, the PIK is appended to the incoming file.
–
The PIK serve
s as a person deduplication key within files.

Page 109 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o Linkage quality assessment
142
: The Census attempts to assign a PIK to all files; the success rate of assigning the PIK varies on the quality of the
completeness of the PII in the files. The most robust linkages are with social security number (SSN), name, and date of birth.
–
Data from CMS and S
SA are very high quality and generally produce PIKs that are almost 100%.
–
National chang
e of address files from Postal Service only contains name and address which results in a lower PIK rate (40-60%).
– Parameters defining matches and non-matches affect the PIK rate.
–
Additional
information can improve the match rates (e.g., parent names or place of birth).
•
Sharing and acc
essing linked data—authorizations/agreements/approvals
144
:
o De-identification status of shared data: Users can access read-only, de-identified versions of approved files. Use of data provisioned by Census
through the FSRDCs are governed by laws and policies from agencies that supplied data. For example, Title 13 and 26
of the U.S. Code that
protects the privacy and confidentiality of the subjects and their data. Violation of the laws are federal crimes and punishable by serious
penalties including prison time and fines.
o Deductive disclosure review: Census uses the term “disclosure” as privacy protection—this includes:
–
Redacting t
he PII and adding the PIK before provisioning the data to the researcher in the FSRDC
142
– Prior to rem
oving the analytical results and statistical products from the FSRDC workspaces, applying Disclosure Avoidance (DA) techniques
to ensure that the products do not disclose the confidentiality of the subjects or their data
145
– Approval fr
om the Disclosure Review Board (DRB) prior to disseminating statistical products or publications derived from the analysis
o Authorization for sharing the linked data: FSRDCs receive authorization via the data sharing agreement provided by the researcher when
requesting linkage of Census data with data from other agencies.
o Agreements/approvals for accessing linked data: Linked data can only be access in restricted physical and IT infrastructure (enclave).
Researchers must obtain Special Sworn Status
37
, complete background check and training, and sign an agreement in order to use the FSRDCs. In
those agreements, they vow to:
–
Not attempt to re
identify
–
Only use the
data for statistical purposes
–
Not misuse affi
liation or PIV card
–
Understand
there is no guarantee of privacy when using the FSRDC lab
–
If the resea
rcher is bringing in their own cohort of data, they agree for Census to take possession of that data and be the steward of the
data.
11.2.2.5
National Center for Health Statistics (NCHS) with National Death Index (NDI)
• Description
146
: A linkage between the National Center for Health Statistics (NCHS) survey on health outcome data and health care utilization information
with the National Death Index (NDI) to maximize the scientific value of the NCHS survey data without increasing respondent reporting burden.
•
Data sources
147
: Death record information from the NDI for each person dying in the United States or a U.S. territory from 1979 through 2015. The NCHS
data being used in this linkage were from the following populated-based health surveys and years:
Page 110 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
o National Health Interview Survey (NHIS): 1985-2014
o Continuous National Health and Nutrition Examination Survey (NHANES): 1999-2014
o NHANES III (1988-1994)
o NHANES II (1976-1980)
o NHANES I Epidemiologic Follow-up Study (NHEFS)
o Second Longitudinal Study of Aging (LSOA II)
o Supplement on Aging (SOA)
o National Home and Hospice Care Survey (NHHCS): 2007
o National Nursing Home Survey (NNHS): 1985, 1995, 1997, 2004
•
Data types
147
: The data types were comprised of national population and provider surveys and mortality data from death certificates.
• Linkage authorizations/agreements/approvals
147
:
Linkage authorization
148
: The specific federal laws that authorize the National Health Interview Survey (NHIS) to ask for PII for linkage purposes are the
Section 308(d) of the Public Health Service Act (42 United States Code 242m(d)), the Confidential Information Protection and Statistical Efficiency Act
(Title V of Public Law 107-347), and the Privacy Act of 1974 (5 U.S.C. § 552a). Furthermore, the NCHS survey data are deemed eligible to link based
on whether a survey participant gives consent for data linkage in the survey and whether adequate PII is present for linkage.
• Entity resolution/Data linkage
146
: Linked database model. Performed by the NCHS Data Linkage Program. The linkage algorithm was developed with
custom code (using SAS 9.4) and was tailored to perform these specific linkages.
o The primary identifiers used in the linkages were: SSN9 or SSN4 (depending on the survey year or cycle of the survey), first name, middle initial,
last name or father’s surname, month of birth, day of birth, year of birth, state of birth, state of residence, race, and sex.
o Process:
1.
Participants
with exact SSNs matches were joined via a deterministic linkage. Matches were joined and validated by a comparison of other
identifying fields.
2.
Probabilistic link
age was then conducted to identify likely matches, or links, between all records, including those where SSN was missing. All
deterministic matched pairs (from Step 1) were assigned a probabilistic match probability of 1; other records were linked and scored as
follows:
Pairs were formed v
ia blocking.
Potential matches were scored based on the concurrence of first name, middle initial, last name or father’s surname, year of birth,
month of birth, day of birth, state of birth, state of residence, race, and sex.
Match probabilities were estimated through a model which assigned the estimated probability that pairs are matches.
3.
Pairs with the
highest estimated match probability were selected which were believed to represent the same individual between the data
sources.
o Linkage quality assessment
146
: To account for changes in the data collection process for some NCHS surveys and potential demographic shifts among
survey participants, an enhanced linkage methodology was adopted in order to produce high quality matches with a low degree of linkage error. The
Page 111 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
change in data linkage quality is due to the fact that NHIS traditionally collected full 9-digit Social Security Numbers (SSN9) from survey participants;
however, there was increased refusal to provide SSN and consent for linkage, thus in the 2007 surveys, NHIS began to collect only the last four digits of
SSN (SSN4) and added an explicit question about linkage for those who refused to provide SSN. The linkage algorithm that was previously used to create
the 2015 linked mortality files had to be enhanced to account for the changed PII collected. Specifically pertaining to the NHIS, from 2007-2014 (the
years when the collection of SSNs changed) the 2019 Linked Mortality Files (LMF) captured 83.3% of the previously linked records. However, in the prior
years when SSN9 was collected, 1986-2006 NHIS, the 2019 LMF captured 94.0% of the previously linked records. Of note, for the 1999-2014 NHANES
where the collection of SSN9 continued to be part of the data collection process, the 2019 LMF captured 92.5% of the previously linked records.
•
Sharing and
accessing linked data—authorizations/agreements/approvals
146
:
o De-identification s
tatus of the linked data
146
: NCHS removes all direct personal identifiers from both the restricted-use and the public-use linked
mortality files. Only select geographic variables are available in the restricted-use datasets.
–
Public-use Li
nked Mortality Files (LMFs) are made available for selected surveys and will include a limited set of mortality variables for adult
participants only. The public-use versions of the 2019 LMFs will be subjected to data perturbation techniques including synthetic data
substitution for follow-up time and underlying cause of death for select records to reduce the participant disclosure risk. Current public use
linked files are available for: 1986-2014 NHIS, 1999-2014 NHANES, and NHANES III surveys.
o Authorization for sharing linked data
146
: Authorization is obtained through informed consent. Participation to NCHS surveys is voluntary and
participants consent to data linkages and sharing their de-identified data to health professionals and to the public.
o Deductive disclosure review
150,147
: To prevent the possibility of re-identification, the NCHS-NDI LMFs are not available as public-use files.
Researchers who want to obtain the NCHS-NDI LMFs must submit a research proposal to the NCHS Research Data Center (RDC) to obtain
permission to access the restricted use files. All researchers must submit a research proposal to determine if their project is feasible and to gain
access to these restricted data files. The proposal provides a framework which allows RDC staff to identify potential disclosure risks.
– Restricted-use Linked Mortality Files are accessible only through the NCHS Research Data Center (RDC). Data access is reviewed by the NCHS
Research Data Center (RDC). To avoid risk of participant re-identification researchers are not allowed to use public-use and restricted-use
Linked Mortality Files, together, in the RDC.
–
Additionally,
no outputs will leave RDC facilities without first being reviewed by an RDC Analyst for possible disclosures of confidential
information.
149
o Agreement/appro
vals for accessing linked data
146
: The linked data files are made available in secure facilities for approved research projects at
NCHS RDCs. Researchers must submit a proposal to use the restricted LMFs as well as sign the NCHS Data Use Agreement
150
which states that
the researcher will:
1. Use the data in this dataset for statistical reporting and analysis only.
2.
Make no atte
mpt to learn the identity of any person or establishment included in these data.
3. Not link this dataset with individually identifiable data from other NCHS or non-NCHS datasets.
4.
Not engage i
n any efforts to assess disclosure methodologies applied to protect individuals and establishments or any research on methods
of re-identification of individuals and establishments
Page 112 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.2.6
ACF/The Child Maltreatment Incidence (CMI) Data Linkages—Multiple projects
• Description
151
: Child Maltreatment Incidence Data Linkages (CMI Data Linkages) program identified five research sites with experience linking
administrative data to examine child maltreatment incidence and related risk and protective factors and supported these sites to enhance their
approaches to administrative data linkage through acquisition of new data sources, use of new methods, or replication of existing methods. The
ADHSS/OHSU project was conducted as part of the CMI Data linkages work as a replication study of the Alaska Longitudinal Child Abuse and Neglect
Linkage project (ALCANLink). ALCANLink was developed to examine over time the incidence of maltreatment, predictive and etiologic factors, and
disparities related to child maltreatment in Alaska. ALCANLink partnered with the Oregon Health Authority and Oregon Health Sciences University to
replicate the ALCANLink methods for the state of Oregon.
•
Data sources
151
: Oregon Health Authority (OHA), Oregon Department of Human Services (OHSU), and ALCANLink.
•
Data types
151
:
o Oregon Preg
nancy Risk Assessment Monitoring System (PRAMS) survey data (2009– 2011)
o Oregon birth and death records data for 2009
o Child protective services record data (2009–2018) for children born in 2009 and child protective services record data for children born in 2009–
2011 whose mothers responded to the PRAMS survey
o ALCANLink data which linked Alaskan 2009–2011 PRAMS cohort to vital records and child welfare records
•
Linkage authoriz
ations/agreements/approvals
151
: IRB approval is required from OHA and Oregon Health and Science University
•
Entity reso
lution/data linkage
151
:Linked database model. Performed by Integrated Client Services (ICS), an Oregon State operated data warehouse, as
required by IRB approval. Much of the linking and processing of data was programmed in a software called RedPoint.
o Process: ICS used a combination of probabilistic, deterministic, and manual matching each month to make/maintain the individual-level links.
Each month, each “class” of probabilistic matching components (one class might be Names-DOB, another might be Names-SSN, etc.) went
through iterations in which the matching criteria gradually loosened. With some exceptions, most of the matching components were a mix of
deterministic matching on some fields and probabilistic matching on others. Records went through multiple matching components, and the
highest-scoring match was chosen at the end.
1.
A state agency
, Integrated Client Services (ICS), completed data linkages on behalf of the research team, as per IRB requirements from data
partners. This agency receives and links data from multiple state programs and agencies on a monthly basis. As an intermediary, the unit
provided a structured process for executing data use agreements, accessing data, and completing linkages.
2. Annual data was first processed through an Extract Transformation and Load (ETL) tool called Pentaho®. This tool systematically identifies
and merges in new information for cases that have already been linked using unique project IDs (using ID Keys and Foreign Keys).
3.
Next, a determin
istic match based on the birth certificate number was used to link PRAMS and vital records data.
4.
A probabili
stic match using a weighted Jaro-Winkler edit distance scoring based on names and date of birth was used to link vital records to
CPS data.
Page 113 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
5. Data are stored on secure servers maintained by OHSU's Advanced Computing Center. The servers are physically located at an off-site facility
with emergency power, weekly backup, multiple firewalls, and physical security. Only staff listed on the DUA and in the IRB protocol for this
study have access to the data
o Linkage quality assessment
151
: Involving a separate agency in data linkage meant that the research team was not able to monitor the quality and
completeness of linkages during that process. It was therefore important to establish a high level of confidence and trust in the linkage approach
from the outset. The site team held an in-person meeting with representatives from ICS to discuss the basic approach and linkage flow for each
data source. The team then documented this flow in project materials and its IRB application. Ultimately, it was determined that ICS’s linkage
approach was close enough to the ALCANLink method.
• Sharing and accessing linked data—authorizations/agreements/approvals
151
:
o De-identification status of shared data
151
: The dataset is stripped of all direct identifiers, leaving only the encrypted unique identifier
o Deductive disclosure review: Information not available.
o Authorizations for sharing data:
–
Data Use Agreement fo
r the PRAMS data was approved by Oregon Health Authority (OHA), Oregon Public Health Division, Section of
Maternal and Child Health
–
DUA for vital reco
rd data was approved by the OHA, Center for Health Statistics
–
Data Sharing Agree
ment (DSA) for child welfare data was approved by Oregon Department of Human Services, Children, Adults and Family
Division.
ADHSS/OHSU site had permission to use child welfare data for the CMI Data Linkages project specifically; it is not a broad authorization
–
DSA was appro
ved by Integrated Client Services
o Agreements/approvals for accessing linked data
151
: Data are stored on secure servers maintained by OHSU's Advanced Computing Center. The
servers are physically located at an off-site facility with emergency power, weekly backup, multiple firewalls, and physical security. Only staff
listed on the DUAs and in the IRB protocol for this study have access to the study data.
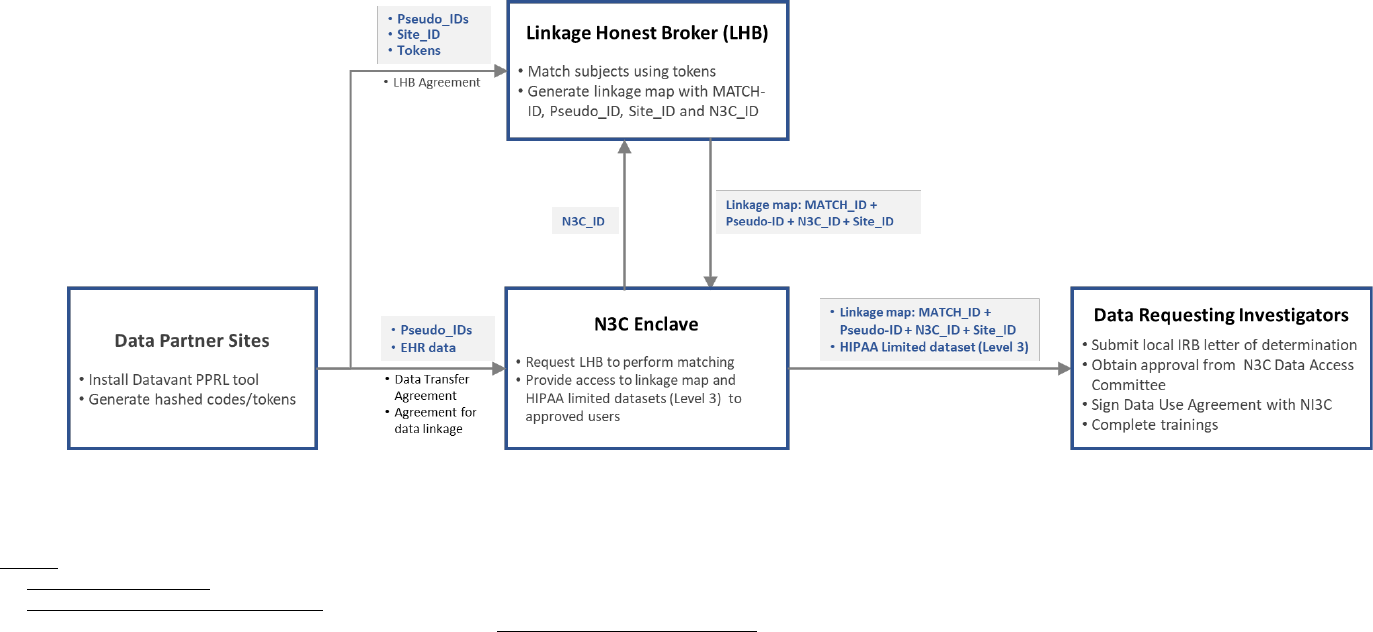
Page 114 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.3
Example Data Flow Schematic for Record Linkage Implementations
Appendix Fi
gure 1: Example Data Flow Schematic for PPRL Linkage: N3C (EHR to EHR Linkage)
Sources:
1. https://covid.cd2h.org/PPRL
2. https://ncats.nih.gov/n3c/about/data-overview
3. N3C Privacy-Preserving Record Linkage and Linked Data Governance: https://doi.org/10.5281/zenodo.5165212
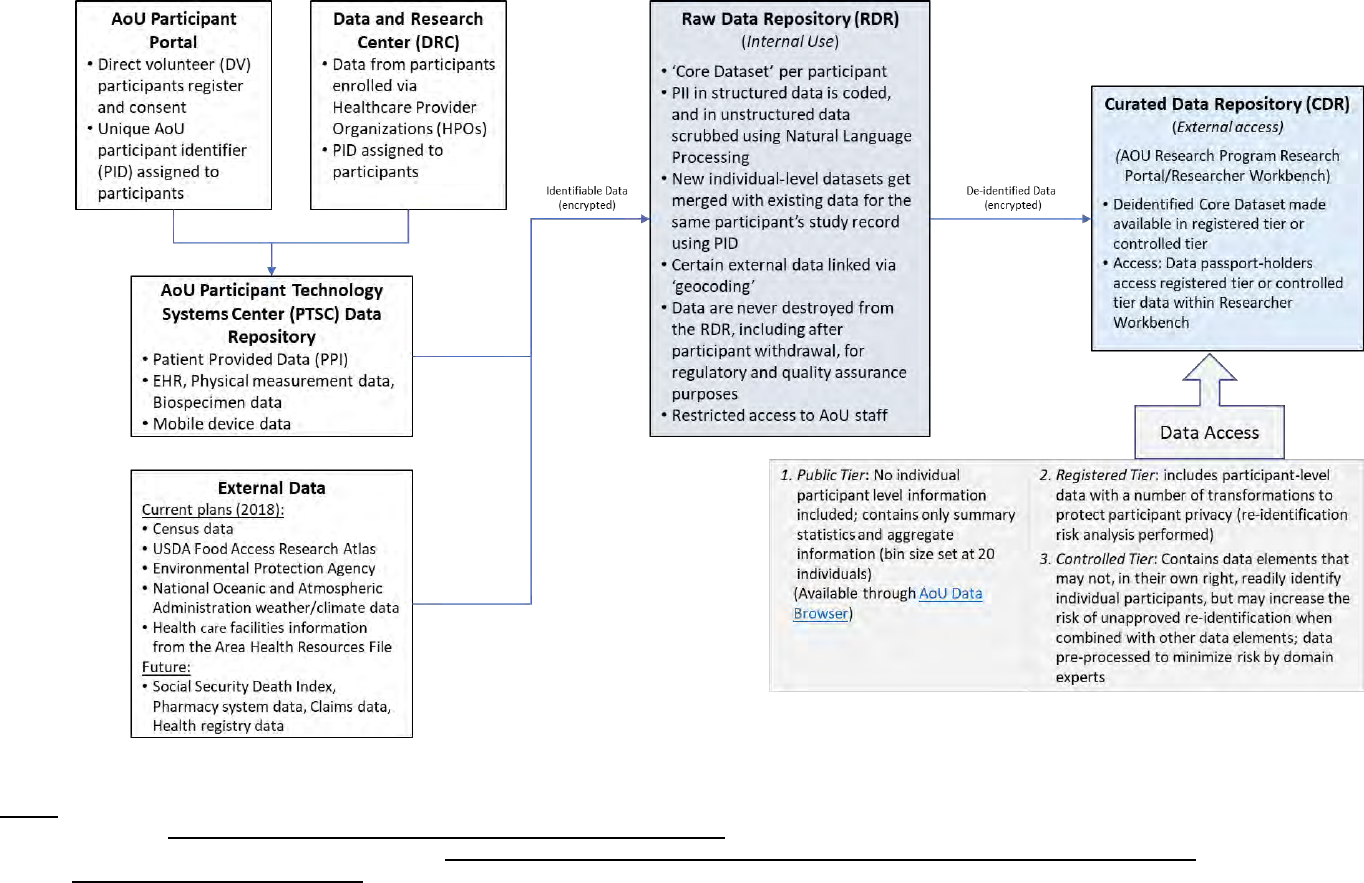
Page 115 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Appendix Fig
ure 2: Example Data Flow Schematic for Non-PPRL Linkage: All of Us
Sources:
1. AoU Protocol: https://allofus.nih.gov/sites/default/files/aou_operational_protocol_v1.7_mar_2018.pdf
2. Framework for Access to All of Us Data Resources v1.1: https://www.researchallofus.org/wp-content/themes/research-hub-wordpress-theme/media/data&tools/data-access-
use/AoU_Data_Access_Framework_508.pdf
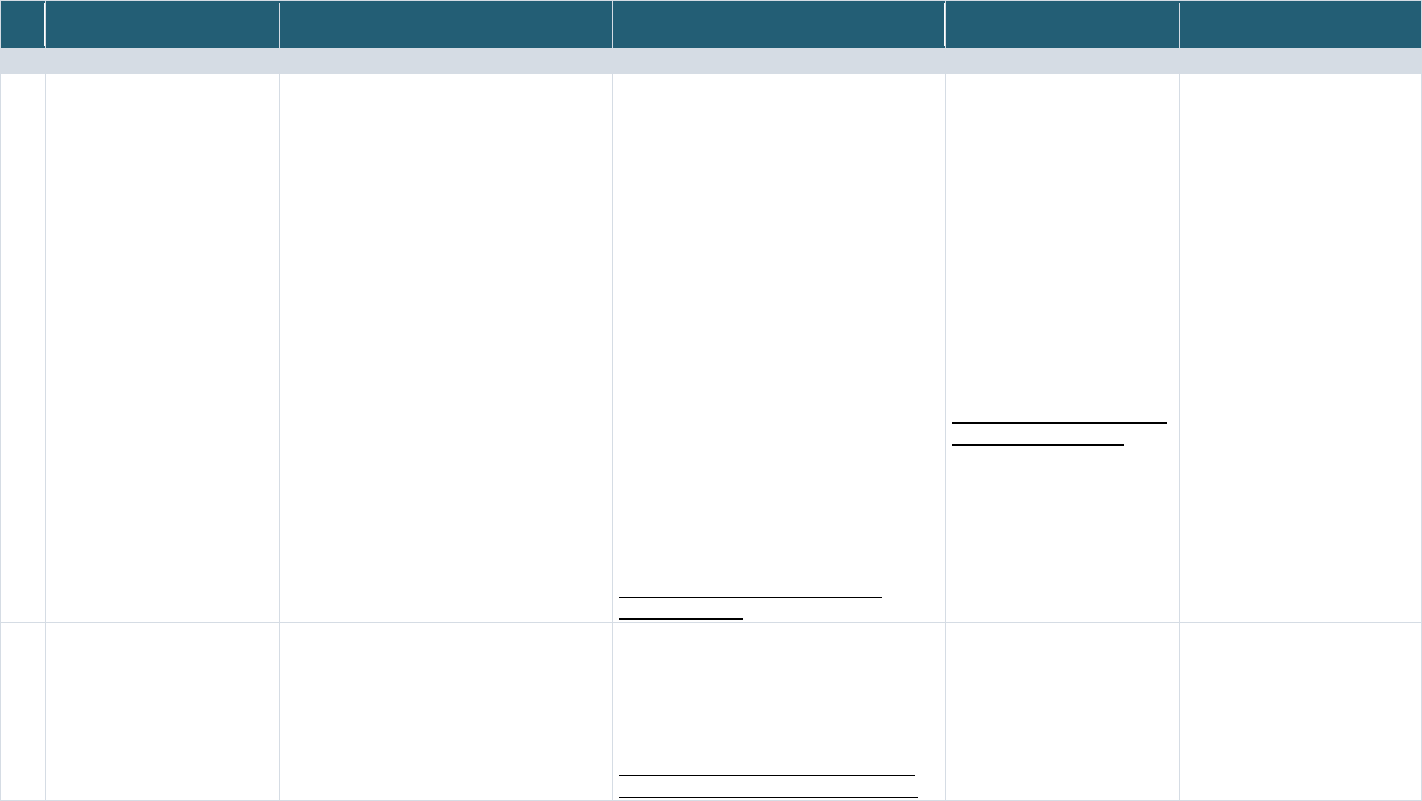
Page 116 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.4
Consent Language for the Record Linkage Implementations Assessed in the Project
Appendix Table 4: Consent Language for the 13 Record Linkage Implementations (If available)
Record Linkage
Implementations
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
PPRL IMPLEMENTATIONS
1A BRICS Instance – NINDS/
PDBP
No explicit language on Data Linkage Data Management Resource (DMR) Consent
Language:
o The data will be submitted to a
database, with all identifying
information removed (it will be
anonymous).
o No personal identifiers will be sent to
the Repository or the database.
o Samples and data will be distributed
to scientists for use in research and
teaching only.
o The sample and unidentified data will
be available to researchers at
hospitals, universities, and
commercial organizations.
o There is a risk that someone could
use information from the sample you
submitted, via DNA, to identify you if it
were matched with another DNA
sample provided by you. However,
any user of this sample must agree
not to use it for that purpose, and the
risk, while real, is small.
https://pdbp.ninds.nih.gov/policy#dmr-
consent-language
DMR Consent Form:
“You have the right to withdraw
from this research project at
any time. If possible, any
samples and data you have
contributed will be discarded if
you request this; however,
because of the sample and
data-masking, we may not
always be able to identify which
samples were donated by you.
Your withdrawal from the study
will in no way affect access to
medical care for which you are
otherwise eligible.”
https://pdbp.ninds.nih.gov/polic
y#dmr-consent-language
No explicit language on
reconsent
1B BRICS Instance –
FITBIR
No explicit language on Data Linkage “All links with your identity will be removed
from the data before they are shared. Only
de-identified data which do not include
anything that might directly identify you will be
shared with FITBIR users and the general
scientific community for research purposes.”
https://fitbir.nih.gov/sites/default/files/inline-
files/FITBIR_Data_Sharing_Policy_final.pdf
No explicit dynamic consent
language
No explicit reconsent language
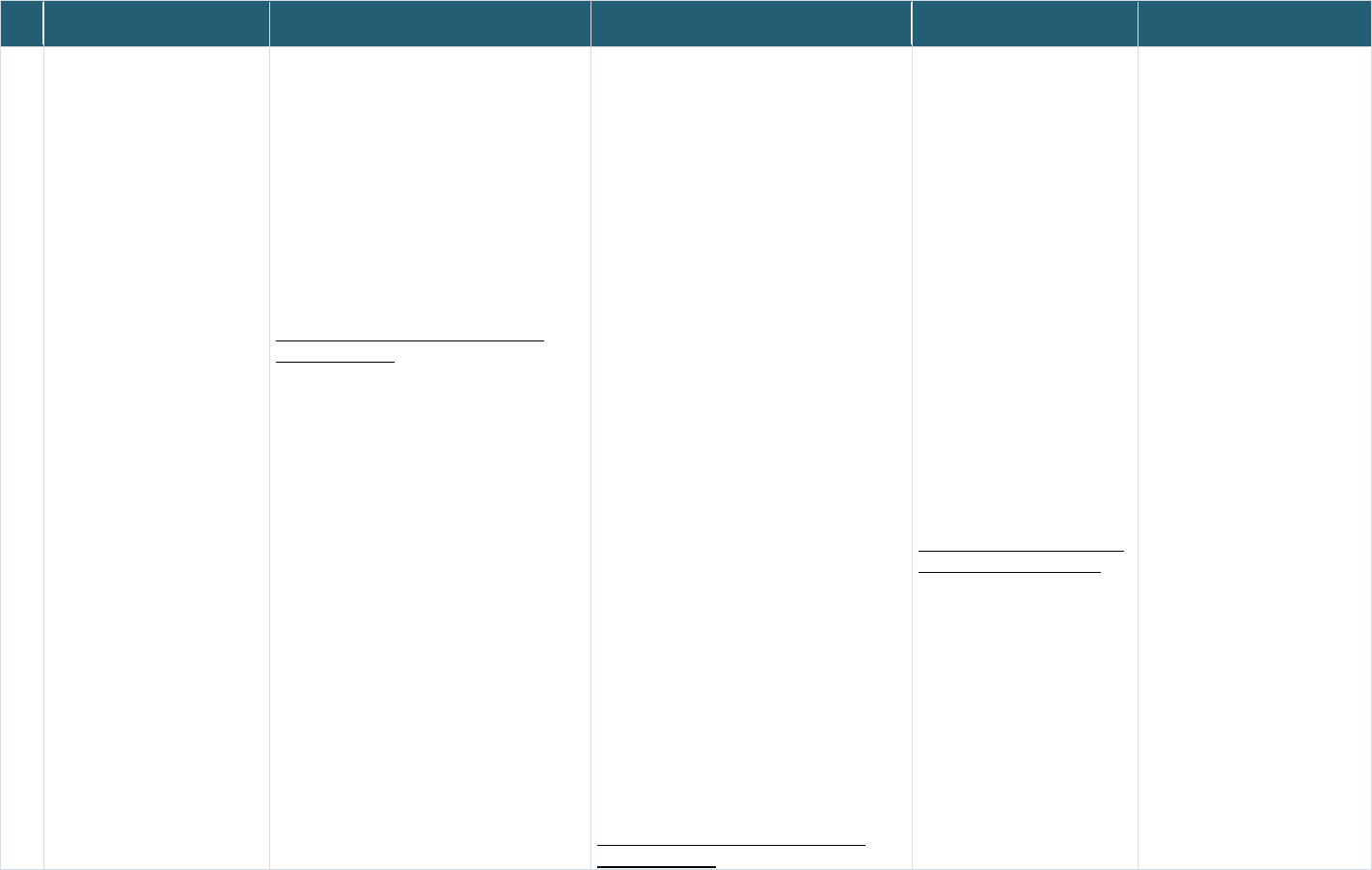
Page 117 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementations
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
2 NIMH NDA Sample consent language provided on the
NDA website (below), but it is not verified by
the NDA repository:
“It is possible that you will participate in more
than one study that sends data to NDA. NDA
can connect your data from different studies
by matching the code number on your
deidentified data from each study. This data
matching helps researchers who use NDA
data to count you only one time. It also helps
researchers who use NDA to better
understand your health and behavior without
knowing who you are.”
https://nda.nih.gov/contribute/contribute-
data.html#infocon
“NDA is a large database where deidentified
study data from many NIH studies are stored
and managed. Sharing your deidentified study
data helps researchers learn new and
important things about brain science more
quickly than before.”
“Deidentified st
udy data means that all
personal information about you (such as
name, address, birthdate and phone number)
is removed and replaced with a code number.
The study researchers will have to collect
your personal information from you in order to
make that code number. The code number
cannot be used to identify you. The study
researchers will never send your personal
information to NDA.”
“During and after the study, the study
researchers will send deidentified study data
about your health and behavior to the NDA.
Other researchers across the world can then
request your deidentified study data for
different research projects. Every researcher
(and the institution to which they belong) who
requests your deidentified study data must
promise to keep your data safe and promise
not to try to learn your identity. Experts at the
NIH who know how to keep your data safe will
review each request carefully to reduce risks
to your privacy. Sharing your study data does
have some risks, although these risks are
rare. Your study data could be accidentally
shared with an unauthorized person who may
attempt to learn your identity. The study
researchers will make every attempt to
protect your identity.”
https://nda.nih.gov/contribute/contribute-
data.html#infocon
“You may decide now or later
that you do not want your study
data to be added to NDA. You
can still participate in this
research study even if you
decide that you do not want
your data to be added to NDA.
If you know now that you do not
want your data in NDA, please
tell the study researcher before
leaving the clinic today. If you
decide any time after today that
you do not want your data to be
added to NDA, call or email the
study staff who conducted this
study, and they will tell NDA to
stop sharing your study data.
Once your data is part of NDA,
the study researchers cannot
take back the study data that
was shared before they were
notified that you changed your
mind.”
https://nda.nih.gov/contribute/c
ontribute-data.html#infocon
No explicit reconsent language
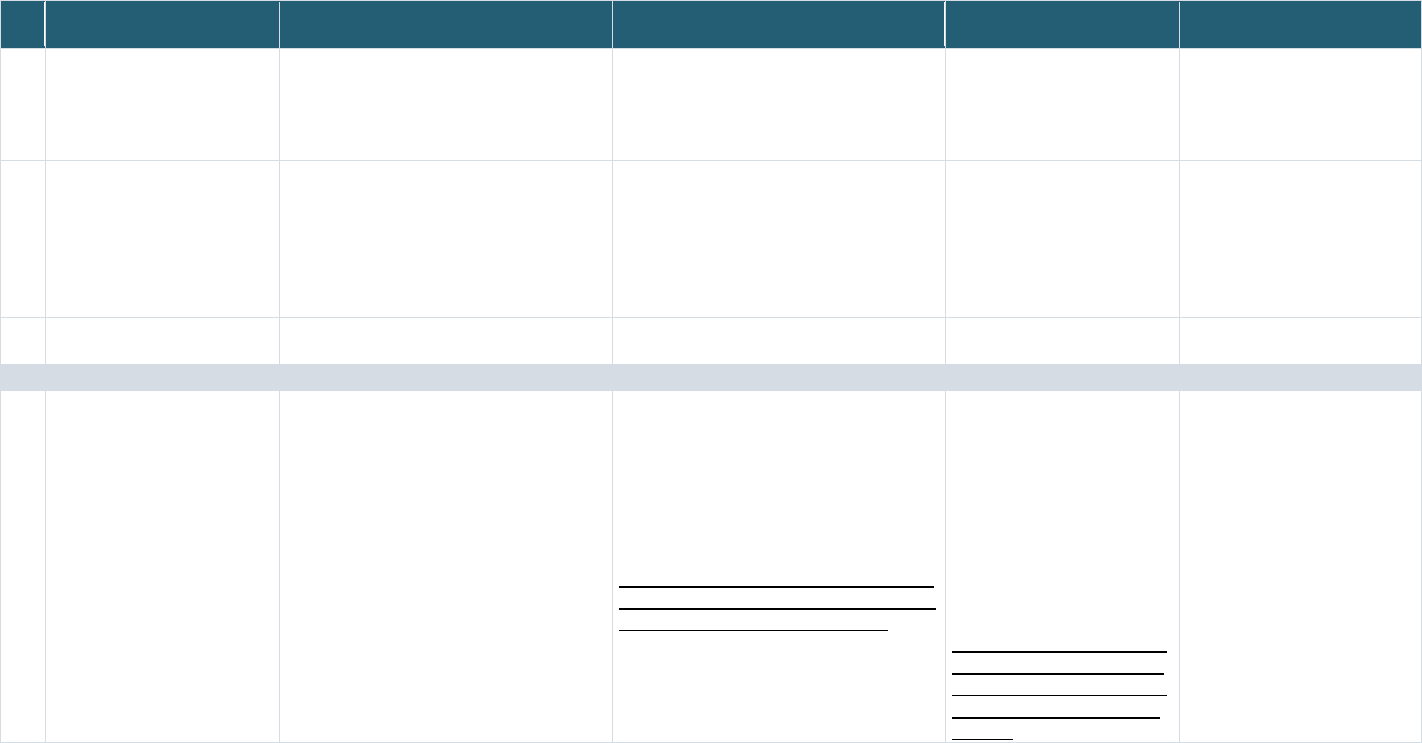
Page 118 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementations
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
3-5 N3C EHR Linkage
N3C Class 0 Linkage N3C
Class 2 Linkage
Participating institutions do not obtain consent
from individual patients for the data they send
to the N3C. NCATS obtained a waiver of
consent from NIH IRB
Participating institutions do not obtain consent
from individual patients for the data they send
to the N3C. NCATS obtained a waiver of
consent from NIH IRB
Participating institutions do not
obtain consent from individual
patients for the data they send
to the N3C. NCATS obtained a
waiver of consent from NIH IRB
Participating institutions do not
obtain consent from individual
patients for the data they send to
the N3C. NCATS obtained a
waiver of consent from NIH IRB
6 PEDSnet Much of PEDSnet’s work involves large
observational studies and operates under
waiver of consent, but when consent is
obtained, the consent language is broad and
addresses sharing linked data.
Much of PEDSnet’s work involves large
observational studies and operates under
waiver of consent, but when consent is
obtained, the consent language is broad and
addresses sharing linked data.
Much of PEDSnet’s work
involves large observational
studies and operates under
waiver of consent, but when
consent is obtained, the
consent language is broad and
addresses sharing linked data.
All sites have policies to reobtain
consent when the participant
turns 18.
It is specified in the policy as to
whether the data cuts off or is
removed entirely if they do not
reconsent.
7 CDC/The Childhood Obesity
Data Initiative (CODI)
No consent found No consent found No consent found No consent found
NON-PPRL RECORD LINKING IMPLEMENTATIONS
1 dbGaP Based on submitter provided Subject Consent
file
“Genomic and phenotypic data, and any other
data relevant for the study (such as exposure
or disease status) will be generated and may
be shared broadly and used for future
research in a manner consistent with the
participant’s informed consent and all
applicable federal and state laws and
regulations.”
https://sharing.nih.gov/sites/default/files/flmng
r/NIH_Guidance_on_Elements_of_Consent_u
nder_the_GDS_Policy_07-13-2015.pdf
“Participants may withdraw
consent for research use of
genomic or phenotypic data at
any time without penalty or loss
of benefits to which the
participant is otherwise entitled.
In this event, data will be
withdrawn from any repository,
if possible, but data already
distributed for research use will
not be retrieved.”
https://sharing.nih.gov/sites/def
ault/files/flmngr/NIH_Gui2danc
e_on_Elements_of_Consent_u
nder_the_GDS_Policy_07-13-
2015.pdf
No explicit reconsent language

Page 119 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementations
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
2 All of Us (AoU) “If you join, we will gather data about you. We
will combine it with data from other people
who join. Researchers will use this data for
lots of studies. By looking for patterns,
researchers may learn more about what
affects people’s health.”
“If you decide to join All of Us, we will gather
data about you. We will gather some of the
data from you directly. We will gather some of
the data from elsewhere.”
"Data about your health from other sources:
We will add data from other sources to the
data you give us. For example, environmental
data and pharmacy records. This will give
researchers more data about factors that
might affect your health. ... We will use data
that identifies you like your name and date of
birth to add data that is specific to you. For
example, we may add data from pharmacy
records or health insurance records. If you
have had cancer, we may add data from
cancer registries...
These other sources can contain sensitive
data. For example, they may tell us about
your mental health, or use of alcohol or drugs.
They may contain sexual or infection data,
including HIV status. Because of this, we will
ask the All of Us ethics committee to review
and approve each data source before we add
it.”
.
https://allofus.nih.gov/sites/default/files/Conse
nt_to_Join_AoU_English.pdf
“Researchers can also ask to study your
samples or DNA directly. We may send them
a small amount of your samples or DNA so
that they can do this. Before we send
researchers your samples or DNA, they will
have to take special training and sign a
contract stating that they will not try to find out
who you are. They will have to tell us what
they want to study. All of Us will have to
approve it. Their research may be on nearly
any topic. They may look for patterns in DNA.
This may help them discover
different ways that DNA affects people. These
researchers may be from anywhere in the
world. They may work for commercial
companies, like drug companies. They may
be citizen or community scientists. Citizen
and community scientists are people who do
science in their spare time.”
“The data researchers get from studying your
samples and DNA may be added to the All of
Us scientific database.”
“In order to work with your health data,
researchers must sign a contract stating they
will not try to find out who you are.”
“Once your information is shared with All of
Us, it may no longer be protected by patient
privacy rules (like HIPAA). However, it will still
be protected by other privacy rules. These
include the rules that researchers must follow
to access the All of Us scientific database.”
https://allofus.nih.gov/sites/default/files/Conse
nt_to_Join_AoU_English.pdf
“If you decide to join All of Us,
you can change your mind at
any time. If you
decide you want to withdraw
(quit), you need to tell us. You
can tell us
through the app or website or
use the contact information at
the end of this
form to call or write to us.
If you withdraw, your samples
will be destroyed. Your data will
not be used
for new studies. However, if
researchers already have your
data or samples for their
studies, we at All of Us cannot
get it back. Also, we will let
researchers check the results
of past studies. If they need
your old data to do this work,
we will give it to them.”
https://allofus.nih.gov/sites/defa
ult/files/Consent_to_Join_AoU_
English.pdf
No explicit reconsent language
(currently not enrolling children)

Page 120 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linkage
Implementations
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of majority)
3 PCORnet-DS Connect/DS-
DETERMINED study
“You will be asked to share your information
from your Electronic Health Record with the
KUMC study team. We will only access data
on diagnosis, procedures, condition,
medications, vital signs, and provider.
We will combine your survey results (DS-
Connect completed IHQ and the SDI results)
with your medical information.
[DS-DETERMINED
consent form from ODSS]
“You will be asked to share your information
from your Electronic Health Record with the
KUMC study team. We will only access data
on diagnosis, procedures, condition,
medications, vital signs, and provider.
This information w
ill be shared with members
of the KUMC research team.”
No explicit dynamic consent
language
No explicit reconsent language
4 Georgetown Federal Statistical
Research Data Center
(FSRDC) – Census
No explicit consent language on sharing
linked data
No explicit consent language on sharing
linked data
No explicit dynamic consent
language
No explicit reconsent language
5 National Center for Health
Statistics (NCHS) with National
Death Index (NDI)
“We take your privacy very seriously. We
combine your answers with other people’s
answers in a way that keeps everyone’s
identity secret. As required by federal law,
your identity can be seen only by those NCHS
employees and specially designated agents
(such as the U.S. Census Bureau) who need
that information for a specific reason.”
“Once you complete the survey, your
confidential data will be combined with
information from thousands of other survey
participants. The combined data are then
analyzed and shared with health
professionals and the public to raise
awareness about progress and needs, and as
a means for inspiring positive change.”
https://www.cdc.gov/nchs/nhis/participants/im
ages/English-Letter-Generic-508.pdf
“Everything you tell us is confidential. Your
information is ONLY used for statistical
purposes. We remove all personally
identifying information from the data. After
that, the data is posted on the NHIS website
for future research or to guide public health
decisions.”
https://www.cdc.gov/nchs/nhis/participants/im
ages/English-Letter-Generic-508.pdf
No explicit dynamic consent
language
No explicit reconsent language
6 The Administration for Children
and Families (ACF) -- The
Child Maltreatment Incidence
(CMI) Data Linkages project-
Alaska Department of Health
and Social Services/Oregon
Health Sciences University
(ADHHS/OHSU)
No explicit consent language for data linkage No explicit consent language on sharing
linked data
No explicit dynamic consent
language
No explicit reconsent language

Page 121 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
11.2.5
Consent Language for the Record Linkage Examples Not Used in the Project
Appendix Table 5: Consent Language from Record Linkage Examples Not Used in the Project
Record Linking
Examples
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of
majority)
1 National Survey of
Child and
Adolescent Well-
Being NSCAW
46
Legal Guardian/Caregiver Permission for Child
Data Linkage form language:
“The NSCAW interview data we collect from the
child can be even more valuable to researchers
when combined with other data. This includes
information that exists now as well as future
information. We also ask the child’s current
caregiver to offer permission to combine interview
data with other information about them. This form
requests your approval to add other sources to the
child’s interview data.
We would add three types of information to the
child’s interview data.
• We would add records collected from child
welfare services agencies taking part in the
study. Adding this information will help us to
learn about foster care, adoption, or other
services the child receives.
• We would add information about the child’s
household, such as wages and other benefits
from the Social Security Administration and from
data available to the Administration for Children
and Families—the agency funding the study.
• Researchers interested in NSCAW data may
wish to add other types of information in the
future. For example, in the past, data on county
or state child welfare policies have been added
to NSCAW interviews.“
Youth Age 13-17 Data Linkage Assent Forms:
“The NSCAW interview with you can be even
more useful when we combine your answers with
other data. This includes information available now
as well as future information. We want to ask for
your okay to add other types of information to your
survey answers.
Legal Guardian/Caregiver Permission for Child
Participation Consent form language:
“This research is covered by a federal protection
called a Certificate of Confidentiality. This means
the researchers cannot share the information they
gather that may identify the child. The Certificate
prevents researchers from revealing this
information even if it is subpoenaed by a court.”
“However, the Certif
icate does allow researchers
to share information in some situations. For
example, researchers must follow reporting laws
about child and adult abuse. Also, as a part of
agreeing to be in this study, you are giving
permission for researchers to share information in
the rare circumstance that it is needed to prevent
serious risk to the child or others. In addition, the
agency that funds this research (the
Administration for Children and Families) is
permitted to access information to confirm that the
research is being conducted properly.”
“In the future, informat
ion from this study may be
securely shared with qualified individuals to help
learn more about the experiences of children and
families with the child welfare system. The
information that is shared will only include a study
ID number and not the child’s name.”
Youth Age 13-17 Data Linkage Assent Forms:
“We do many things to make sure your answers
stay private. I am going to enter your answers into
a laptop computer. We have a paper from the
government that promises that we do not have to
give your information to anyone. We will not tell
anyone your answers unless we are worried about
you or someone else’s safety. For example, if you
tell us you might hurt yourself or someone else,
Legal Guardian/Caregiver
Permission for Child Data Linkage
form language:
“If in the future should you decide you
no longer want the child’s interview data
combined with other records, you can
opt out of this request and stop further
collection from outside sources. Please
contact Jennifer Keeney at RTI
International (toll-free at 800-334-8571
extension 23525) or RTI's Office of
Human Research Protections at 1-866-
214-2043 (a toll-free number) to record
your request.”
Youth Age 13-17 Data Linkage
Assent Forms:
“You have the right to say yes or no to
this request. Your okay also allows us
to share it with researchers for other
approved research studies. If you
change your mind, please call Jennifer
Keeney at RTI International (RTI) (toll-
free at 800-334-8571, extension 23525)
or RTI's Office of Human Research
Protections at 1-866-214-2043 (a toll-
free number) to record this request.”
https://www.reginfo.gov/public/do/PRAV
iewDocument?ref_nbr=202109-0970-
003
Legal Guardian/Caregiver
Permission for Child Data
Linkage form language:
“Your permission allows us to
add these other types of
information to the child’s
interview data. We will add
this information to the child’s
interview data until the child
becomes an adult (turns 18
years old). We can add some
information about the child
now. Some of the information
we will add in the future.
“Your permission to add other
information will stop when the
child turns 18 years old. When
the child turns 18 years old,
we will ask the child’s
permission to add new
information. The child’s
permission will allow us to add
information about the child’s
adult life. “
Youth Age 13-17 Data
Linkage Assent Forms:
If you give your okay now, we
will collect these other kinds of
information to combine with
your interview data. Your okay
to link your interview data to
other information will last until
you turn 18 years old unless
you change your mind before
then. Your okay means we
can start adding information
about you now or in the future.

Page 122 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linking
Examples
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of
majority)
We would add three types of information to your
interview data.
• We would add records collected from child
welfare services agencies taking part in the
study. Adding this information will help us learn
about foster care, adoption, or other services
you receive.
• We would add information about your
household’s income, such as wages and
disability benefits from the Social Security
Administration and from data available to the
Administration for Children and Families—the
agency funding the study.
• Researchers interested in NSCAW data may
wish to add other information in the future. This
could include data on county or state child
welfare policies. “
https://www.reginfo.gov/public/do/PRAViewDocum
ent?ref_nbr=202109-0970-003
we may tell someone. If you tell us someone hurts
you, we may tell authorities to keep people safe.”
https://www.reginfo.gov/public/do/PRAViewDocum
ent?ref_nbr=202109-0970-003
When you turn 18 years old,
we will ask if it is okay to
collect information about you
as an adult. If you say no
then, we will stop adding
information about you as an
adult and only keep and share
the information collected
before you turned 18 years
old.
https://www.reginfo.gov/public
/do/PRAViewDocument?ref_n
br=202109-0970-003
2 LunaPBC “By sharing any Personal Data or Shared Data
into this platform, you agree to and consent to the
use of that data as set forth in this LunaDNA
Agreement and in our Privacy Policy. The Privacy
Policy contains further detail on many of the
concepts in this LunaDNA Consent, and we urge
you to read it in full.”
https://support.lunadna.com/support/solutions/artic
les/43000076335-lunadna-consent
“To protect your privacy, we separate your Shared
Data from your Personal Data. We refer to the
separation of your Personal Data from your
Shared Data as de-identifying your Shared Data.
Your Shared Data is then combined, or
aggregated, with the entire pool of de-identified
Shared Data of our community, to create a
searchable database to power research and
discovery. LunaDNA and researchers (including
our manager, LunaPBC) may perform population-
level searches based on a pre-defined and
approved study design. We refer to these
searches as queries. The results of a study-linked
query will include a list containing de-identified
data file identification numbers of members whose
Shared Data is a match for the query. Based on
these results, a subset of aggregated, de-identified
Shared Data is populated in a private, secured
compute environment controlled by LunaDNA,
which we refer to as a sandbox, in order to
“Your data is owned by you. You may
revoke your consent at any time. At any
time after you provide Shared Data or
Personal Data to LunaDNA, you can
decide to revoke your consent, purge
some or all of your data, and even
delete your account completely from our
databases. If you revoke your consent
or delete your account, your data will be
permanently removed, or purged, from
our database (subject to retaining an
archival copy if, and for so long as,
required by law), and this Consent
Agreement relating to that data will
automatically terminate. If you choose
to purge some or all of your data,
LunaDNA will immediately prevent that
data from being found in any new
queries for researchers. LunaDNA will
also determine if your Shared Data is at
No explicit reconsent
language

Page 123 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
Record Linking
Examples
Consent for Linking Data Consent for Sharing (Linked) Data Dynamic Consent Language
(unenroll/withdraw)
Reconsent Language
(for reaching age of
majority)
complete the analysis required by the study
design.”
https://support.lunadna.com/support/solutions/artic
les/43000076335-lunadna-consent
that time available to any researchers in
a sandbox.”
https://support.lunadna.com/support/sol
utions/articles/43000076335-lunadna-
consent
3 Health and
Retirement Survey
– CMS data
linkage (NIA)
“We would like to understand how people’s
medical history affects their financial status, and
how use of health care may change as people
age. To do that, we need to obtain information
about health care costs and diagnoses for
statistical purposes. The best place to get this
information without taking up a lot more of your
time is in the (Medicaid/State name for Medicaid)
files.) Could you give me your Medicaid number
for this purpose?”
https://g2aging.org/?section=item&itemid=361791

Page 124 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
12 REFERENCES
1
White House Executive Order on Ensuring a Data-Driven Response to COVID-19 and Future High-
Consequence Public Health Threats: https://www.whitehouse.gov/briefing-room/presidential-
actions/2021/01/21/executive-order-ensuring-a-data-driven-response-to-covid-19-and-future-high-
consequence-public-health-threats/
2
Patel, J. M. (2022). Multisystem Inflammatory Syndrome in Children (MIS-C). Current Allergy and Asthma
Reports, 1-8. https://doi.org/10.1007/s11882-022-01031-4
3
NIH RePORTER search results of active projects using terms with terms (“COVID” or “SARS-CoV-2”) and
(child or children or pediatric)/(“COVID” or “SARS-CoV-2”) and (child or children or pediatric) and (MIS-C or
“multisystem inflammatory syndrome”) in project title or key terms performed on 8/30/2022
4
Dunn, H. L. (1946). Record linkage. American Journal of Public Health and the Nations Health, 36(12), 1412-
1416. https://doi.org/10.2105/ajph.36.12.1412
5
HIPAA: https://aspe.hhs.gov/reports/health-insurance-portability-accountability-act-1996
6
Schmidlin, K., Clough-Gorr, K. M., & Spoerri, A. (2015). Privacy preserving probabilistic record linkage
(P3RL): a novel method for linking existing health-related data and maintaining participant
confidentiality. BMC medical research methodology, 15(1), 1-10.https://doi.org/10.1186/s12874-015-0038-6
7
Dusetzina, S. B., Tyree, S., Meyer, A. M., Meyer, A., Green, L., & Carpenter, W. R. (2014). Linking data for
health services research: a framework and instructional guide. Available from:
https://www.ncbi.nlm.nih.gov/books/NBK253312/
8
Census/Data Linkage Infrastructure: https://www.census.gov/about/adrm/linkage.html
9
CDC/NCHS Data Linkage Activities: https://www.cdc.gov/nchs/data-linkage/index.htm
10
Linking Data for Health Services Research: A Framework and Instructional Guide:
https://effectivehealthcare.ahrq.gov/products/registries-linking-records- methods/research
11
HHS/Linking Administrative Data to Improve Understanding of Child Maltreatment Incidence and Related
Risk and Protective Factors: A Feasibility Study https://www.acf.hhs.gov/opre/report/linking-administrative-
data-improve-understanding-child-maltreatment-incidence-and
12
HHS/Federal Policy for the Protection of Human Subjects ('Common Rule'):
https://www.hhs.gov/ohrp/regulations-and-policy/regulations/common-rule/index.html
13
DOJ/PRIVACY ACT OF 1974: https://www.justice.gov/opcl/privacy-act-1974
14
Emily C. O’Brien, Ana Maria Rodriguez, Hye-Chung Kum, Laura E. Schanberg, Marcy Fitz-Randolph, Sean M.
O’Brien, Soko Setoguchi, Patient perspectives on the linkage of health data for research: Insights from an
online patient community questionnaire, International Journal of Medical Informatics, Volume 127, 2019,
Pages 9-17, ISSN 1386-5056, https://doi.org/10.1016/j.ijmedinf.2019.04.003
15
Mozersky J, Parsons M, Walsh H, Baldwin K, McIntosh T, DuBois JM. Research Participant Views regarding
Qualitative Data Sharing. Ethics Hum Res. 2020 Mar;42(2):13-27. doi: 10.1002/eahr.500044. PMID:
32233117; PMCID: PMC7418215.
16
Goodman D, Johnson CO, Bowen D, Smith M, Wenzel L, Edwards K. De-identified genomic data sharing:
the research participant perspective. J Community Genet. 2017 Jul;8(3):173-181. doi: 10.1007/s12687-017-
0300-1. Epub 2017 Apr 5. PMID: 28382417; PMCID: PMC5496839.
17
Mello MM, Lieou V, Goodman SN. Clinical Trial Participants' Views of the Risks and Benefits of Data
Sharing. N Engl J Med. 2018 Jun 7;378(23):2202-2211. doi: 10.1056/NEJMsa1713258. PMID: 29874542;
PMCID: PMC6057615.

Page 125 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
18
HHS/Guidance Regarding Methods for De-identification of Protected Health Information in Accordance
with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule:
https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html
19
Gkoulalas-Divanis, A., Vatsalan, D., Karapiperis, D., & Kantarcioglu, M. (2021). Modern Privacy-Preserving
Record Linkage Techniques: An Overview. IEEE Transactions on Information Forensics and Security.
https://doi.org/10.1109/TIFS.2021.3114026
20
Ranbaduge, T., Christen, P., Schnell, R. (2020, May). Secure and Accurate Two-Step Hash Encoding for
Privacy-Preserving Record Linkage. In: Lauw, H., Wong, RW., Ntoulas, A., Lim, EP., Ng, SK., Pan, S. (eds)
Advances in Knowledge Discovery and Data Mining. PAKDD 2020. Lecture Notes in Computer Science(), vol
12085. Springer, Cham. https://doi.org/10.1007/978-3-030-47436-2_11
21
Randall, S. M., Ferrante, A. M., Boyd, J. H., Bauer, J. K., & Semmens, J. B. (2014). Privacy-preserving record
linkage on large real-world datasets. Journal of biomedical informatics, 50, 205-212.
https://doi.org/10.1016/j.jbi.2013.12.003
22
Wellcome/Sharing research data and findings relevant to the novel coronavirus (COVID-19) outbreak:
https://wellcome.org/press-release/sharing-research-data-and-findings-relevant-novel-coronavirus-ncov-
outbreak
23
NIH calls on clinical researchers to swiftly share COVID-19 results: https://www.nih.gov/about-nih/who-
we-are/nih-director/statements/nih-calls-clinical-researchers-swiftly-share-covid-19-results
24
NIH Genomic Data Sharing: https://osp.od.nih.gov/scientific-sharing/genomic-data-sharing/
25
NIH/ Workshop on the Policy and Ethics of Record Linkage: Workshop Summary:
https://datascience.nih.gov/nih-policy-and-ethics-of-record-linkage-workshop-summary
26
Johnson, S. B., Whitney, G., McAuliffe, M., Wang, H., McCreedy, E., Rozenblit, L., & Evans, C. C. (2010).
Using global unique identifiers to link autism collections. Journal of the American Medical Informatics
Association, 17(6), 689-695. https://doi.org/10.1136/jamia.2009.002063
27
American Academy of Pediatrics. (n.d.). Children and covid-19: State-level data report. (Data as of
8/25/2022). Retrieved August 30, 2022, from https://www.aap.org/en/pages/2019-novel-coronavirus-covid-
19-infections/children-and-covid-19-state-level-data-report/
28
CDC/Health Department-Reported Cases of Multisystem Inflammatory Syndrome in Children (MIS-C) in
the United States: https://covid.cdc.gov/covid-data-tracker/#mis-national-surveillance Accessed 8/30/2022
29
Jesse Aronson et al. Landscape Analysis of Privacy Preserving Patient Record Linkage Software (P3RLS)
Final Report. Cancer Moonshot Task Order Phase I, prepared by Synectics for the National Cancer Institute,
August 15, 2019. Available from: https://surveillance.cancer.gov/reports/
30
NIH Pharmacokinetics, Pharmacodynamics, and Safety Profile of Understudied Drugs Administered to
Children Per Standard of Care (POPS) (POPS or POP02): https://clinicaltrials.gov/ct2/show/NCT04278404
31
Truong, D. T., Trachtenberg, F. L., Pearson, G. D., Dionne, A., Elias, M. D., Friedman, K., ... & Chrisant, M.
(2022). The NHLBI Study on Long-terM OUtcomes after the Multisystem Inflammatory Syndrome In Children
(MUSIC): Design and Objectives. American heart journal, 243, 43-53.
https://doi.org/10.1016/j.ahj.2021.08.003
32
NIH/Pediatric Research Immune Network on SARS-CoV-2 and MIS-C (PRISM):
https://www.niaid.nih.gov/clinical-trials/pediatric-research-immune-network-sars-cov-2-and-mis-c
33
Release: NIH funds eight studies to uncover risk factors for COVID-19-related inflammatory syndrome in
children: https://www.nichd.nih.gov/newsroom/news/122120-prevail-kids
34
NIH/Completing an Institutional Certification Form: https://sharing.nih.gov/genomic-data-sharing-
policy/institutional-certifications/completing-an-institutional-certification-form

Page 126 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
35
RADx-rad Study Registration: Institutional Certification and Study Sharing and Submission Registration
documents (NIH): https://www.radxrad.org/resource/radx-rad-study-registration-institutional-certification-
and-study-sharing-and-submission-registration-documents-nih/
36
HHS/Report on Statistical Disclosure Limitation Methodology:
https://www.hhs.gov/sites/default/files/spwp22.pdf
37
Census/Researcher Handbook: https://www.census.gov/content/dam/Census/programs-
surveys/sipp/methodology/Researcher_Handbook_20091119.pdf
38
Vatsalan, D., Christen, P., & Verykios, V. S. (2013). A taxonomy of privacy-preserving record linkage
techniques. Information Systems, 38(6), 946-969.
39
HHS/45 CFR 46: https://www.hhs.gov/ohrp/regulations-and-policy/regulations/45-cfr-46/index.html
40
HHS/NIH Genomic Data Sharing Policy: NIH Request for Public Comment:
https://www.hhs.gov/ohrp/sachrp-committee/recommendations/tab-c-nih-genomic-data-sharing-policy-
nih-request-for-public-comments.html
41
Cho, M. K., Magnus, D., Constantine, M., Lee, S. S., Kelley, M., Alessi, S., Korngiebel, D., James, C., Kuwana,
E., Gallagher, T. H., Diekema, D., Capron, A. M., Joffe, S., & Wilfond, B. S. (2015). Attitudes Toward Risk and
Informed Consent for Research on Medical Practices: A Cross-sectional Survey. Annals of internal medicine,
162(10), 690–696. https://doi.org/10.7326/M15-0166
42
Sanderson SC, Brothers KB, Mercaldo ND, et al. Public Attitudes toward Consent and Data Sharing in
Biobank Research: A Large Multi-site Experimental Survey in the US. Am J Hum Genet. 2017;100(3):414-427.
doi:10.1016/j.ajhg.2017.01.021
43
NIH/Request for Information: Developing Consent Language for Future Use of Data and Biospecimens:
https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-131.html
44
All of Us Research Program HIPAA Authorization for Research EHR/Part 2 Supplement:
https://allofus.nih.gov/sites/default/files/f2_hipaa_ehr_part_2_supplement-eng-sample.pdf
45
NHGRI/Studies Involving Children: https://www.genome.gov/about-genomics/policy-issues/Informed-
Consent/Special-Considerations-for-Genome-Research#children
46
OMB Office of Information and Regulatory Affairs/ICR Documents:
https://www.reginfo.gov/public/do/PRAViewDocument?ref_nbr=202109-0970-003
47
NIH/Informed Consent for Secondary Research with Data and Biospecimens: Points to Consider and
Sample Language for Future Use and/or Sharing: https://osp.od.nih.gov/wp-content/uploads/Informed-
Consent-Resource-for-Secondary-Research-with-Data-and-Biospecimens.pdf
48
HHS/2018 Revised Common Rule – General Requirements for Informed Consent:
https://www.hhs.gov/ohrp/regulations-and-policy/regulations/45-cfr-46/revised-common-rule-regulatory-
text/index.html#46.116
49
NYSPI/National Data Archive Data Sharing Standards: https://nyspi.org/sites/default/files/inline-
files/NYSPI-NIMH_NDAR-DataSharing.pdf
50
NIH/Genomic Data Sharing Policy: https://grants.nih.gov/grants/guide/notice-files/NOT-OD-14-124.html
51
NIH/Request for Information on Proposed Updates and Long-Term Considerations for the NIH Genomic
Data Sharing Policy https://grants.nih.gov/grants/guide/notice-files/NOT-OD-22-029.html
52
NIH/Request for Public Comments on DRAFT Supplemental Information to the NIH Policy for Data
Management and Sharing: Protecting Privacy When Sharing Human Research Participant Data:
https://grants.nih.gov/grants/guide/notice-files/NOT-OD-22-131.html
53
Harvard/Identifying Participants in the Personal Genome Project by Name:
https://arxiv.org/ftp/arxiv/papers/1304/1304.7605.pdf

Page 127 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
54
CMU/k-Anonymity: A Model For Protecting Privacy: https://epic.org/wp-
content/uploads/privacy/reidentification/Sweeney_Article.pdf
55
AMIA/Re-Identification of DNA through an Automated Linkage Process:
https://dataprivacylab.org/dataprivacy/projects/genetic/dna2.pdf
56
NIH/Supplemental Information to the NIH Policy for Data Management and Sharing: Responsible
Management and Sharing of American Indian/Alaska Native Participant Data:
https://grants.nih.gov/grants/guide/notice-files/NOT-OD-22-214.html
57
Vatsalan, D., Sehili, Z., Christen, P., Rahm, E. (2017). Privacy-Preserving Record Linkage for Big Data:
Current Approaches and Research Challenges. In: Zomaya, A., Sakr, S. (eds) Handbook of Big Data
Technologies. Springer, Cham. https://doi.org/10.1007/978-3-319-49340-4_25
58
Patient Matching, Aggregation, and Linking (PMAL) Project – Final Report (2019). Office of the National
Coordinator for Health Information Technology. https://www.healthit.gov/sites/default/files/page/2019-
09/PMAL%20Final%20Report-08162019v2.pdf
59
McClure, R. C., Macumber, C. L., Kronk, C., Grasso, C., Horn, R. J., Queen, R., ... & Davison, K. (2022).
Gender harmony: improved standards to support affirmative care of gender-marginalized people through
inclusive gender and sex representation. Journal of the American Medical Informatics Association, 29(2),
354-363.
ttps://doi.org/10.1093/jamia/ocab196
60
NCHS/Life Tables: https://www.census.gov/topics/population/migration/guidance/calculating-migration-
expectancy.html#:~:text=Using%202007%20ACS%20data%2C%20it,one%20move%20per%20single%20year.
61
HealthIT/United States Core Data for Interoperability (USCDI) – Patient demographics:
https://www.healthit.gov/isa/uscdi-data/mothers-maiden-name
62
University of Michigan/Data Sharing for Demographic Research:
https://www.icpsr.umich.edu/web/pages/DSDR/disclosure.html
63
Notice of Special Interest (NOSI) - Emerging and Existing Issues of Coronavirus Disease 2019 (COVID-19)
Research Related to the Health and Well-Being of Women, Children and Individuals with Physical and/or
Intellectual Disabilities: https://grants.nih.gov/grants/guide/notice-files/NOT-HD-22-002.html
64
IJARET/Privacy Preserving Record Linkage Using Phonetic and Bloom Filter Encoding :
https://iaeme.com/MasterAdmin/Journal_uploads/IJARET/VOLUME_11_ISSUE_7/IJARET_11_07_035.pdf
65
N3C/N3C Privacy-Preserving Record Linkage: https://covid.cd2h.org/PPRL
66
NIH/Data Use Certification Agreement: https://osp.od.nih.gov/wp-content/uploads/Model_DUC.pdf
67
NIH/Emergency Award: Rapid Acceleration of Diagnostics Tribal Data Repository (RADx TDR):
https://grants.nih.gov/grants/guide/rfa-files/RFA-OD-22-011.html
68
HHS/Explore the OS-PCORTF Project Profiles: https://aspe.hhs.gov/collaborations-committees-advisory-
groups/os-pcortf/explore-portfolio
69
BRICS/Catalyzing Team Science: https://brics.cit.nih.gov/
70
Navale, V., Ji, M., Vovk, O., Misquitta, L., Gebremichael, T., Garcia, A., ... & McAuliffe, M. (2019).
Development of an informatics system for accelerating biomedical research. F1000Research, 8.
https://doi.org/10.12688/f1000research.19161.2
71
Introducing BRICS: https://brics.cit.nih.gov/intro
72
Information gathered during BRICS stakeholder interview
73
Slides from BRICS stakeholder
74
NINDS/Centralized GUID Server: https://pdbp.ninds.nih.gov/ninds-centralized-guid-server
75
NIH/GUID: https://eyegene.nih.gov/how-to/guid

Page 128 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
76
NIH/NCATS Global Rare Diseases Patient Registry Data Repository (GRDR®): https://avillach-
lab.hms.harvard.edu/nihncats-global-rare-diseases-patient-registry-data-repository-grdr%C2%AE
77
NIH/Data Submission Request: https://cdrns.nih.gov/policies/intramural/data-submission-request
78
NIH/Global Unique Identifier: https://fitbir.nih.gov/content/global-unique-identifier
79
BRICS/Data Sharing Policy for NINR Funded P20 and P30 Pilot
Studies:https://cdrns.nih.gov/sites/default/files/inline-files/NINR_Data_Sharing_Policy_0.pdf
80
NIH/FITBIR Data Sharing Policy: https://fitbir.nih.gov/sites/default/files/inline-
files/FITBIR_Data_Sharing_Policy_final.pdf
81
Information from NINDS/PDBP stakeholder
82
NIH/BRICS GUID: https://brics.cit.nih.gov/sites/default/files/inline-files/GUID_MANUAL_1.pdf
83
FITBIR/Data Access Request: https://fitbir.nih.gov/sites/default/files/inline-
files/FITBIR_Data_Access_Request_DUC.pdf
84
NIH/NDA About Us: https://nda.nih.gov/about/about-us.html
85
NIH/NDA FAQ: https://nda.nih.gov/about/faq.html
86
NIH/Notice of Data Sharing Policy for the National Institute of Mental Health:
https://grants.nih.gov/grants/guide/notice-files/NOT-MH-19-033.html
87
NIMH Data Archive Data Submission Agreement:
https://nda.nih.gov/ndapublicweb/Documents/NDA+Submission+Request.pdf
88
NDA/Policy for the NIMH Data Archive (NDA):
https://s3.amazonaws.com/nda.nih.gov/Documents/NIMH+Data+Archive+Policy.pdf
89
NDA/Using the NDA GUID: https://nda.nih.gov/contribute/using-the-nda-guid.html
90
NDA/pseudoGUIDs: https://nda.nih.gov/contribute/using-the-nda-guid.html#pseudoGUID
91
NDA/SOP-08 GUID Generation Permission Request: https://nda.nih.gov/about/standard-operating-
procedures.html#sop8
92
NDA GUID Tool: https://nda.nih.gov/binaries/content/documents/ndacms/resources/nda-guid-tool-user-
manual/nda-guid-tool-user-manual/ndacms%3Aresource
93
NDA/Security Controls: https://nda.nih.gov/contribute/using-the-nda-guid.html#securitycontrols
94
NDA/What is the GUID and why is it so important to the NDA: https://nda.nih.gov/abcd/s/nda/about-
us/faq.html#nc.3
95
NDA/NIMH Data Archive Data Use Certification:
https://nda.nih.gov/ndapublicweb/Documents/NDA+Data+Access+Request+DUC+FINAL.pdf
96
NCATS/About the National COVID Cohort Collaborative: https://ncats.nih.gov/n3c/about
97
NCATS/N3C Data Overview: https://ncats.nih.gov/n3c/about/data-overview
98
NCATS/Linkage Honest Data Broker: https://ncats.nih.gov/files/NCATS_LHBA-508.pdf
99
NCATS/N3C Data Contribution Forms and Resources: https://ncats.nih.gov/n3c/resources/data-
contribution
100
N3C Privacy-Preserving Record Linkage and Linked Data Governance:
https://doi.org/10.5281/zenodo.5165212
101
Information gathered from N3C stakeholders
102
NCATS/Access Requirements for Researchers by Data Level: https://ncats.nih.gov/n3c/about/data-
overview#access-requirements

Page 129 of 130
103
NCATS/What data does the N3C have and where does it come from?:
https://ncats.nih.gov/n3c/about#where
104
N3C External Dataset Process Document- FINAL
https://docs.google.com/document/d/1QJi_sNi0wnZFV3ghTBubI7d3kFLtdkVfQwsLQJOFil8/edit#
105
NCATS/N3C Viral Variants wiki: https://github.com/National-COVID-Cohort-Collaborative/variants/wiki
106
Pedsnet Home Page: https://pedsnet.org/
107
Pedsnet/Institutions: https://pedsnet.org/about/institutions/
108
Pedsnet/Data Domains in PEDSnet Database: https://pedsnet.org/data/data-domains/
109
Information gathered during PEDSnet stakeholder interview
110
PEDSnet Policy, Version 2020; accessed 04/2022 https://pedsnet.org/documents/303/PEDSnet-Policies-
May-2020_.pdf
111
Kraus, E. M., Scott, K. A., Zucker, R., Heisey-Grove, D., King, R. J., Carton, T. W., ... & Davidson, A. J. (2021).
A Governance Framework to Integrate Longitudinal Clinical and Community Data in a Distributed Data
Network: The Childhood Obesity Data Initiative. Journal of Public Health Management and Practice, , 28(2),
E421-E429.
doi:10.1097/PHH.0000000000001408
112
CODI/Clinical and Community Data Initiative: https://www.cdc.gov/obesity/initiatives/codi/community-
and-clinical-data-initiative.html
113
CODI/Data Owner Tools: https://github.com/mitre/data-owner-tools
114
CODI/Linkage Agent Tools: https://github.com/mitre/linkage-agent-tools
115
CODI/Master Data Sharing and Use Agreement:
https://www.coloradohealthinstitute.org/sites/default/files/file_attachments/CODI%40CHORDS_MSUA_Ap
pendix_I.pdf
116
dbGaP/Home page: https://www.ncbi.nlm.nih.gov/gap/
117
Information gathered during dbGaP stakeholder interview
118
dbGaP/dbGaP Study Submission Guide:
https://www.ncbi.nlm.nih.gov/gap/docs/submissionguide/#astart
119
dbGaP/Individual-level Data: General Questions: https://www.ncbi.nlm.nih.gov/books/NBK570260/
120
dbGaP/Software: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/Software.cgi
121
dbGaP/Study Meta DS and DD Files: Subject Consent, Subject Sample Mapping (SSM), and Pedigree:
https://www.ncbi.nlm.nih.gov/gap/docs/submissionguide/#9-how-do-i-create-subject-consen
122
dbGaP/What files do I need to submit to dbGaP?:
https://www.ncbi.nlm.nih.gov/gap/docs/submissionguide/#astart
123
dbGaP/Versioning dbGaP Datasets: https://osp.od.nih.gov/wp-
content/uploads/Versioning%20plan%20for%20dbGaP%20Datasets%202008.pdf
124
NIH/Data Access Committee Review: https://osp.od.nih.gov/scientific-sharing/data-access-request-dar-
approvals-and-disapprovals-by-data-access-committee-dac
125
dbGaP/How to Request and Access Datasets from dbGaP: https://sharing.nih.gov/accessing-
data/accessing-genomic-data/how-to-request-and-access-datasets-from-dbgap#step-2
126
All of Us/
Home Page: https://allofus.nih.gov/
127
All of Us/
FAQ: https://allofus.nih.gov/about/faq
128
All of Us/
Participation: https://allofus.nih.gov/get-involved/participation
129
Information gathered
during All of Us stakeholder interview
NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report (September 2022)

Page 130 of 130 NICHD Office of Data Science and Sharing (ODSS)
PPRL for Pediatric COVID Studies – Final Report
(September 2022)
130
NIH/Progress Towards Developing Data Infrastructure for COVID-19:
https://datascience.nih.gov/jumpstart-executive-summary
131
All of Us/Protocol v1 Summary:
https://allofus.nih.gov/sites/default/files/all_of_us_protocol_v1_summary.pdf
132
All of Us/The All of Us Consent Process: https://allofus.nih.gov/about/protocol/all-us-consent-process
133
All of Us/All of Us Research Program Operation Protocol:
https://allofus.nih.gov/sites/default/files/aou_operational_protocol_v1.7_mar_2018.pdf
134
All of Us Research Program HIPAA Authorization for Research EHR/Part 2 Supplement:
https://allofus.nih.gov/sites/default/files/f2_hipaa_ehr_part_2_supplement-eng-sample.pdf
135
All of Us/Framework for Access to All of Us Data Resources v1.1: https://www.researchallofus.org/wp-
content/themes/research-hub-wordpress-theme/media/data&tools/data-access-
use/AoU_Data_Access_Framework_508.pdf
136
PCORnet/Research We've Made Possible: https://www.pathnetwork.org/Research/DS-Determined.html
137
Information gathered during DS-DETERMINED study stakeholder interview
138
DS-DETERMINED Consent Form. IRB Approval Period 4/20/2021 – 5/6/2021: Obtained from Dr. Evan
Dean, Ph.D., Associate Director of Community Services, Kansas University Center on Developmental
Disabilities.
139
PCORnet/Include Data Hub: https://portal.includedcc.org
140
Census/Available Data: https://www.census.gov/about/adrm/fsrdc/about/available_data.html
141
Census/Research Data Centers: https://www.census.gov/about/adrm/fsrdc/locations.html
142
Information gathered during Census stakeholder interview
143
Census/The Person Identification Validation System (PVS): Applying the Center for Administrative Records
Research and Applications’ (CARRA) Record Linkage Software:
https://www.census.gov/content/dam/Census/library/working-papers/2014/adrm/carra-wp-2014-01.pdf
144
Census/How do I access and use data?: https://www.census.gov/about/adrm/linkage/guidance.html
145
Census/A History of the U.S. Census Bureau's Disclosure Review Board:
https://www.census.gov/library/working-papers/2019/adrm/history-DRB.html
146
CDC/The Linkage of National Center for Health Statistics Survey Data to the National Death Index – 2015
Linked Mortality File (LMF): Methodology Overview and Analytic Considerations:
https://www.cdc.gov/nchs/data/datalinkage/LMF2015_Methodology_Analytic_Considerations.pdf
147
CDC/The Linkage of National Center for Health Statistics Survey Data to the National Death Index – 2019
Linked Mortality File (LMF): Linkage Methodology and Analytic Considerations:
https://www.cdc.gov/nchs/data/datalinkage/2019NDI-Linkage-Methods-and-Analytic-Considerations-
508.pdf
148
CDC/National Health Interview Survey:
https://ftp.cdc.gov/pub/Health_Statistics/NCHS/Survey_Questionnaires/NHIS/2017/frmanual.pdf
149
CDC/Output Policies and Procedures: https://www.cdc.gov/rdc/b1datatype/rdc-Output.htm
150
CDC/Data User Agreement: https://www.cdc.gov/nchs/data_access/restrictions.htm
151
ACF/Linking Administrative Data to Improve Understanding of Child Maltreatment Incidence and Related
Risk and Protective Factors: A Feasibility Study:
https://www.acf.hhs.gov/sites/default/files/documents/opre/OPRE-Linking-Administrative-Data-to-
Improve-Understanding-Childhood-Maltreatment-Dec2021.pdf
