
UNITED STATES DEPARTMENT OF DEFENSE
Data Validation
Guidelines Module 4:
Data Validation
Procedure for Organic
Analysis by GC
Environmental Data Quality Workgroup
03/09/2021

Data Validation Guidelines
Module 4
Digitally signed by
JORDAN.BRIAN.D.1141739820
JORDAN.BRIAN.D.1141739820
Date: 2021.03.11 07:09:34 -06'00'
Brian Jordan Date
Army Principal
GILLETTE.JOHN.S.1123328350
Digitally signed by GILLETTE.JOHN.S.1123328350
Date: 2021.03.10 11:09:04 -06'00'
Seb Gillette, Ph.D. Date
Air Force Principal
ADELSON.JORDAN.M.1268693137
Digitally signed by ADELSON.JORDAN.M.1268693137
Date: 2021.03.11 07:44:17 -05'00'
Jordan Adelson, Ph.D. Date
Navy Principal, EDQW Chair

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 1 of 40
Table of Contents
1.0 Purpose ........................................................................................................................................... 2
2.0 Procedure ........................................................................................................................................ 2
2.1 Introduction ................................................................................................................................. 2
2.2 Deliverables ................................................................................................................................. 3
2.3 Validation Stages ......................................................................................................................... 3
3.0 Stage 1 Validation ........................................................................................................................... 3
3.1 Sample Results ............................................................................................................................. 4
3.2 Chain of Custody (CoC) ................................................................................................................ 5
3.3 Field QC ........................................................................................................................................ 7
4.0 Stage 2A Validation ....................................................................................................................... 11
4.1 Surrogate Spikes ........................................................................................................................ 11
4.2 Laboratory Control Sample/Laboratory Control Sample Duplicate (LCS/LCSD) ........................ 13
4.3 Matrix Spike/Matrix Spike Duplicate (MS/MSD) ....................................................................... 13
4.4 Method Blanks ........................................................................................................................... 15
4.5 Sample Dilutions and Reanalysis ............................................................................................... 16
5.0 Stage 2B Validation ....................................................................................................................... 16
5.1 Sequence and Preparation Logs ................................................................................................ 16
5.2 Instrument Performance Checks (Method 8081: Organochlorine Pesticides) .......................... 17
5.3 Initial Calibration ........................................................................................................................ 18
5.4 Initial (Secondary Source) and Continuing Calibration Verification .......................................... 20
5.5 Internal Standards (Optional) .................................................................................................... 21
5.6 Cleanup Procedures for Methods 8081, 8082, 8141, and 8151 ................................................ 21
5.7 Second Column Confirmation/Dissimilar Detector Confirmation ............................................. 23
6.0 Stage 3 Validation ......................................................................................................................... 24
6.1 Samples and Field QC ................................................................................................................ 24
6.2 Method QC ................................................................................................................................. 25
6.3 Instrument QC ........................................................................................................................... 26
6.4 Standards Traceability ............................................................................................................... 28
6.5 Detection/Quantitation Limit Studies (Optional) ...................................................................... 29
7.0 Stage 4 Validation ......................................................................................................................... 29
7.1 Target Compound Identification ................................................................................................ 29
7.2 Retention Time Windows .......................................................................................................... 31
7.3 Manual Integrations .................................................................................................................. 32
Appendix A: Method QC Tables .......................................................................................................... 33
Appendix B: Formulas used in Stage 3 and 4 Data Validation ............................................................ 37

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 2 of 40
Module 4: Data Validation Procedure for Organic Analysis
by GC (SW-846 8000 Series)
1.0 Purpose
This document provides guidance on the validation of organic contaminants by SW-846
Series 8000 methods when analyzed on a Gas Chromatograph (GC):
• 1,2-Dibromoethane and 1,2-Dibromo-3-Chloropropane by Microextraction and GC
Method 8011;
• Nonhalogenated Organics by GC Method 8015;
• Aromatic and Halogenated Volatiles by GC Using Photoionization and/or Electrolytic
Conductivity Detectors Method 8021;
• Organochlorine Pesticides by GC Method 8081;
• Polychlorinated Biphenyls (PCBs) by GC Method 8082;
• Organophosphorus Compounds by GC Method 8141; and
• Chlorinated Herbicides by GC Using Methylation or Pentafluorobenzylation
Derivatization Method 8151;
Note: With the exception of EPA 8000D, this document does not identify specific Series
8000 method versions. Consult the project UFP-QAPP to determine specific
requirements for analysis and validation. The language within this document is only to be
used as guidance and the QAPP shall always supersede this document.
The objective of this procedure is to provide the end user with a clear understanding of
the quality and limitations of the data through documented validation procedures and to
encourage consistency in the validation technique and reporting for data generated for
Department of Defense (DoD) projects for organic constituents when analyzed on GC.
This document assumes the user is familiar with data validation conventions and
qualifiers used in the DoD General Data Validation Guidelines Version 1 (2019). This
document is also not intended to obviate the need for professional judgment during the
validation process.
This document references the Uniform Federal Policy for Quality Assurance Project Plans
(UFP-QAPP) Optimized Worksheets (March 2012). Other QAPP formats are equally
acceptable.
2.0 Procedure
This guidance can be applied to organic contaminant data generated in support of DoD
projects that was produced on GC. This guidance should be implemented by personnel
familiar with the methodology contained herein.
Data validation personnel are responsible for implementing this procedure for validation of
data and generation of data validation reports for GC organic contaminant data.
2.1 Introduction
This document was written with primary consideration to the latest SW-846 8000
series GC method versions with Quality Control (QC) criteria identified in the DoD
Quality Systems Manual (QSM). Actual validation should proceed using the acceptance
criteria for the method version specified in the laboratory data deliverable or in the QAPP.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 3 of 40
Appendix A summarizes the QC checks and the required frequency and acceptance criteria
for Method 8000D and the QSM version 5.3 requirements.
2.2 Deliverables
Laboratory data deliverables consist of a combination of forms and raw data. The manner in
which laboratories label their forms is not dictated nor specified. The labeling convention
below is used for simplicity.
• Cover Sheet
• Case Narrative
• Sample Receipt and Conditions Summary
• Sample Results Summary
• Surrogate Recovery Summary
• Laboratory Control Sample/Laboratory Control Sample Duplicate
• Matrix Spike/Matrix Spike Duplicate Recovery Summary
• Method Blank Summary
• Instrument Performance Check Summary (where applicable)
• Initial Calibration Summary
• Initial/Continuing Calibration Verification Summary
• Retention Time/Internal Standard Summary
• Sequence and preparation logs
2.3 Validation Stages
The types of laboratory data deliverables, staged data validation, and the relationship
between the two are outlined in the DoD General Data Validation Guidelines Version 1.
Stage 1 data validation consists of a review of sample results form, associated sample
receipt summaries (chain of custody), and field QC data.
Stages 2A and 2B data validation consist of review of summary forms only.
Stages 3 and 4 data validation require review of both summary forms and all associated raw
data.
Both the laboratory deliverable and the level of validation should be specified in the QAPP or
other planning documents. Data review guidelines and how they apply to the different
validation stages are indicated in the following sections.
Note: Any required stage of validation that reveals significant deviations from project
requirements may require a higher stage of validation to uncover the source. Data validators
are encouraged to communicate with their points of contact identified in the QAPP (such as
the UFP-QAPP Worksheet #6) to resolve discrepancies.
3.0 Stage 1 Validation
The following documents should be reviewed for representativeness (compliance with
required analytical protocols outlined in QAPP), completeness, and project sensitivity needs:
• Cover Sheet
• Table of Contents
• Case Narrative
• Sample results form or equivalent Laboratory Report

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 4 of 40
• Chain of Custody (CoC) forms, Laboratory Receipt Checklists, and other supporting
records
• Field QC forms and supporting records
Stage 1 is the validation of investigative and field QC samples.
3.1 Sample Results
Examine the Laboratory Report sample results summary (can be called by many names,
such as Form I) and verify the following information:
• Holding times have been met, as applicable
• All sample identification labels are unique, and match the CoC
• All project GC analytes have been analyzed and are reported
• Second (dissimilar) Column results are consistent with QAPP requirements
• All laboratory reported Limits of Detection (LODs) and Limits of Quantitation (LOQs)
are equal to or less than QAPP required LODs/LOQs
• All project required LOQs have been met and achieved LOQs are less than the
project required action levels
• All reported units (e.g., mg/kg) are accurate and reflect the requirements of the
project, and units are consistent with the type of sample matrix
• All required field QC samples (such as trip blanks, equipment blanks, reagent blanks,
and field duplicates) have been included in the Laboratory Report at the frequency
specified in the QAPP
• Soil samples have been reported on a dry weight basis, unless specified by the QAPP
to report on a wet weight basis
• Each laboratory report has a case narrative that explains non-conformities with the
data
• All sample collection date/time information matches the CoC or any inconsistency is
appropriately documented
The following statements apply to sample results (assuming no other qualifications due to
data quality issues):
Qualification of data is based upon the reporting requirements of the QAPP.
The QSM requires reporting non-detects as U-qualified at the LOD and requires reporting
detects between the DL and LOQ with a J qualification. There are several ways that a project
team may change these reporting requirements for project-specific reasons which are
outlined in the QAPP. Though not recommended for typical projects, these changes include
reporting non-detects as U-qualified at the DL; reporting non-detects and detects below the
LOD as non-detects with U qualification at the LOD; or reporting non-detects and detects
below the LOQ as non-detects with U qualification at the LOQ. These varying reporting
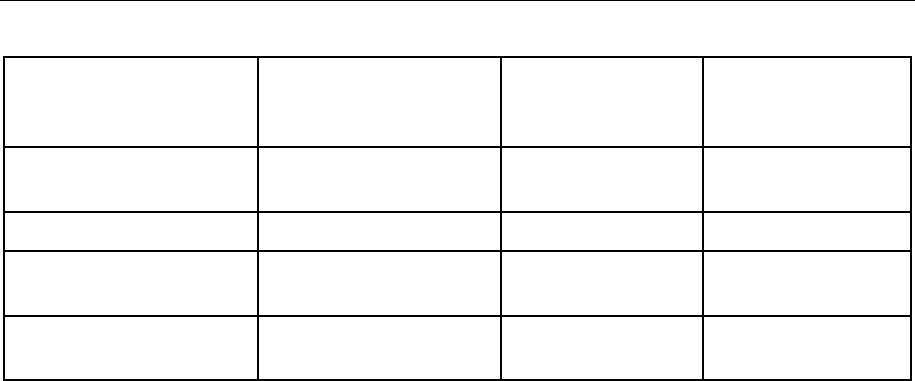
conventions are summarized in the following table:
Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 5 of 40
Table I: Reporting Requirements
Reporting
Requirements (listed
below)
Non-detects or
results Below (<) DL
Results Below
(<) LOD
Results Below (<)
LOQ
Standard QSM
Reporting
LOD value U
Reported Result J
Reported Result J
*Reporting results to DL
DL value U
Reported Result J
Reported Result J
Reporting results to
LOD
LOD value U
LOD value U
Reported Result J
Reporting results to
LOQ
LOQ value U
LOQ value U
LOQ value U
*Note: non-detects reported at the DL have a 50% false negative rate. For further discussion
please see Fact Sheet: Detection and Quantitation – What Project Managers and Data Users
Need to Know, DoD Environmental Data Quality Workgroup, October 2017.
Evaluation of the Laboratory Report
Any samples received for analysis that were not analyzed should be noted in the data
validation report, along with the reason(s) for failure to analyze the samples, if the reason(s)
can be determined; conversely, samples that were analyzed by GC but were not requested
should also be noted.
Analytes that have project action levels less than the laboratory’s LOD may reveal a severe
deficiency in the data and a failure to meet project goals, and such instances should be noted
in the data validation report. Errors in reported units and case narrative non-conformities that
call into question the quality of the data should also be discussed in the data validation
report.
Errors in quantitation limits or missing or misidentified samples may require a higher than
Stage 1 validation. Data validators are encouraged to reach out to their point of contact
(QAPP Worksheet #6) when preparing the data validation report.
3.2 Chain of Custody (CoC)
Examine the CoC form (some information may be included on Laboratory Receipt Checklists)
for legibility and check that all GC analyses requested on the CoC have been performed by
the laboratory. Ensure that the CoC Sample Identification on the laboratory sample results
summary (Form I [or equivalent]) matches the Sample Identification on the CoC. Ensure the
CoC was signed and dated during transfers of custody. Read the laboratory case narrative
for additional information.
Evaluation of the CoC
Any discrepancies in sample identification between the CoC and sample results form should
be noted in the data validation report with the correct sample identification in the report and
on the appropriate summary form, if the correct sample identification can be determined.
These edit corrections should also be verified in any associated electronic data deliverables
(EDDs).

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 6 of 40
If the receiving laboratory transferred the samples to another laboratory for analysis, both the
original CoCs and transfer CoCs should be present. Document in the data validation report if
the transfer CoCs are not present or if there is missing information (such as location of the
laboratory). Make note in the data validation report when signatures of relinquish and receipt
of custody were not present.
3.2.1 Sample Preservation, Handling, and Transport
Evaluate sample handling, transport, and laboratory receipt from the CoC and laboratory
receipt checklists to ensure that the samples have been properly preserved and handled.
The project QAPP (such as UFP-QAPP Worksheet #19) should provide specific preservation
requirements. The following are general guidance if project specifications were not stipulated.
Typically, organic samples do not require chemical preservation. An exception is 3 mL 10%
sodium thiosulfate solution is added per gallon (or 0.008%) to aqueous samples with residual
chlorine present. Addition of sodium thiosulfate solution to sample container may be
performed in the laboratory prior to field use. Reference the QAPP for specific preservation
requirements.
• Organic contaminant samples are to be shipped in amber bottles with PTFE-lined lids
• All samples are to be shipped in coolers that are maintained at ≤ 6 degrees Celsius
(°C)
Evaluation of Preservation, Handling, and Transport
If the temperature of receipt is > 6°C but ≤ 15⁰C, detects should be flagged as estimated J-
and non-detects as estimated UJ.
If the temperature of receipt is > 15°C, detects should be flagged as estimated J- and non-
detects as X, exclusion of data recommended.
On occasion, the samples may be delivered to the laboratory within a few hours of collection
and before the temperature of the cooler is able to reach 6⁰C. For those instances, if cooling
has begun, but the temperature is > 6⁰C, special note should be made but no qualification
should be required.
If the temperature is below 0°C, special note should be made but no qualification should be
required.
In the event that both a cooler temperature and a temperature blank were measured, the
temperature blank should be evaluated for temperature compliance as it best represents the
condition of the samples; however, both temperatures shall be noted in the data validation
report.
If the temperature upon receipt at the laboratory was not recorded, note this in the data
validation report and assume that a temperature non-conformance occurred. Detects should
be flagged as estimated J- and non-detects flagged X, exclusion of data recommended.
If the receiving laboratory transferred the samples to another laboratory for analysis, apply
the same temperature criteria to both the transfer CoC and the original CoC.
If aqueous samples known to contain chlorine were not chemically preserved with sodium
thiosulfate, apply professional judgment to qualify the sample results.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 7 of 40
3.2.2 Holding Times
Holding times for organics are measured from the time of collection (as shown on the CoC) to
the time of sample analysis as shown on the sample results summary (Forms I or equivalent)
or extraction log (if applicable). Based on input from the DoD Environmental Data Quality
Workgroup (EDQW) holding time exceedances are calculated as follows:
Total holding time is based on the timeframe (i.e., hours, days, or months) of the
requirement. The following example gives guidance on how holding time exceedances are
measured:
For a test with a recommended maximum holding time measured in days, the holding time is
tracked by the day.
• An exceedance of holding time for a sample with a 14-day holding time will occur
when the 15th day is reached. Therefore, a sample with a 14-day holding time
collected at 8:30 AM on April 4th must be analyzed or extracted before 12:00 AM April
19
th
(midnight, the start of the 15
th
day), or an exceedance has occurred.
Samples and extracts must be stored refrigerated to ≤ 6°C until the time of analysis. The
holding time for aqueous samples is 7 days from the collection date to the beginning of
extraction, and 40 days from extraction to analysis. The holding time for solid samples is 14
days from the collection date to the beginning of extraction, and 40 days from extraction to
analysis. The holding time for aqueous waste samples is 7 days from collection to leaching, 7
days from leaching to the beginning of extraction, and 40 days from extraction to analysis.
The holding time for solid waste samples is 14 days from collection to leaching, 7 days from
leeching to the beginning of extraction and 40 days from extraction to analysis.
There is no specified holding time for PCB Aroclor samples. The QAPP should specify the
holding time requirements.
Evaluation of Holding Time
If the holding time is exceeded, qualify all associated detects as estimated J- and all
associated non-detects as estimated UJ and document that holding times were exceeded.
If holding times are grossly exceeded (defined as > 14 days to extraction for aqueous
samples and > 28 days for solid samples), detects should be qualified as estimated J- and
non-detects as X, exclusion of data recommended.
For PCB Aroclor samples, the above holding times for organic contaminants can be used for
guidance if specific holding times are not listed in the QAPP. Exceedances do not require
qualification of the data but should be noted in the data validation report.
3.3 Field QC
Field QC can consist of various blanks, field duplicates, and field replicates. The purpose of
blanks is to identify potential cross-contamination at different stages of sampling and
cleaning of equipment for reuse. Duplicates and replicates help a project identify
reproducibility among samples at the project site.
3.3.1 Field Blanks
Not every field blank type may be utilized during any given sampling event and there may be
more blank types than described in this document. Field blanks may be varied throughout the

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 8 of 40
sampling events of a project. The types of blanks and their collection frequency should be
stipulated in the QAPP. Generally, the blanks are collected once a day or one per twenty field
investigative samples, by each sampling team, and may be matrix dependent.
Below are the common types of field blanks utilized in the collection of organic contaminants
analyzed by GC.
Trip blanks are included for aqueous volatile analytes only. Volatile organic compounds
detected in trip blanks indicate the possibility of contamination of site samples or cross-
contamination between site samples due to sample handling and transport while in the
cooler. A trip blank is usually included for every cooler that transports volatile samples.
An ambient blank is sample collected on site, without the need of equipment, filled directly
into a sample container. Ambient blanks are included for volatile analysis only. Analytes
detected in ambient blanks indicate the possibility of cross-contamination between the air
matrix and the matrix being collected for testing.
An equipment blank (also called a rinse or rinsate blank) is an aliquot of reagent water
subjected to all aspects of sample collection. Analytes detected in equipment blanks indicate
the possibility of cross-contamination between samples due to improper equipment
decontamination. Equipment blanks are usually collected at a frequency of one per twenty
investigative samples, or as specified in the QAPP.
A source blank (also called a reagent blank) may be collected from each source of water
used during each sampling event. This type of field blank may be analyzed to assess
whether the chemical nature of the water used in decontamination may have affected the
analytical results of site samples. A source blank is usually collected once per source prior to
sample collection.
Evaluation of Field Blanks
Check that all coolers containing samples to be analyzed for volatile organic contaminants by
GC contained a trip blank. If a cooler requiring a trip blank did not have an associated trip
blank, no qualification of the samples transported in the cooler is necessary, but the incident
should be discussed in the data validation report along with other required types of field
blanks that were found missing. The point of contact (QAPP Worksheet #6) should be
notified within the required time frame as required by the QAPP.
Determine which field blanks apply to samples in the sample delivery group (SDG) from the
CoC. If the applicability of multiple field blanks cannot be determined, communicate with the
point of contact (QAPP Worksheet #6) to inquire if applicability can be determined.
Note: SDGs can be called by different names such as SEDD Lab Reporting Batch,
depending on the project.
Ensure that units are correct when applying field blank qualifications.
Note: it may not be appropriate to make a direct quantitative comparison for aqueous field
blanks (such as equipment blanks reported as µg/mL) to a solid parent sample (such as a
soil sample reported as mg/kg). At best, only a qualitative comparison can be made.
Generally, when multiple blank type contaminations are present, the evaluation should not
involve a ‘hierarchy’ of one blank type over another. Each blank is evaluated separately and
independently. The final validated result should be assessed on the blank with the highest
value (i.e., greatest effect on sample analyte concentration).

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 9 of 40
The source blank water should be analyte free (undetected or as defined by QAPP) and
provided with the sample bottle kit by the contracted laboratory performing the analysis. To
ensure the origin of the water used, consult with the field sampling team leader via
appropriate channels identified in the QAPP (such as UFP-QAPP Worksheet #6). If source
blank water is used as equipment blank water and both are contaminated, the affected
samples are qualified by either the source blank or equipment blank results, whichever has
the higher contaminant concentration.
If analytes (as appropriate) are detected in the field blanks, the procedure for the qualification
of associated sample results is summarized below.
Compare the results of each type of field blank with the associated sample results. The
reviewer should note that the blank analyses may not involve the same units, volumes, or
dilution factors as the associated samples. These factors should be taken into consideration
when applying the 5X and 10X criteria discussed below, such that a comparison of the total
amount of contamination is actually made. Care should be taken to factor in any dilution
factors when doing comparisons between detects in the sample and the blank.
• If an analyte is detected in the field blank, but not in the associated samples, no
action is taken.
• If field blank contamination includes those analytes listed in Table I as common lab
contaminants, then 10X (in lieu of 5X) should be used to determine the qualification of
the sample.
• If field blanks were not collected at the proper frequency required by the QAPP, then
use professional judgment to qualify the data, and make note of this in the data
validation report.
• If an analyte is detected in the field blank (at any concentration) and in the associated
samples, the action taken depends on both the blank and sample concentrations
(Table III).
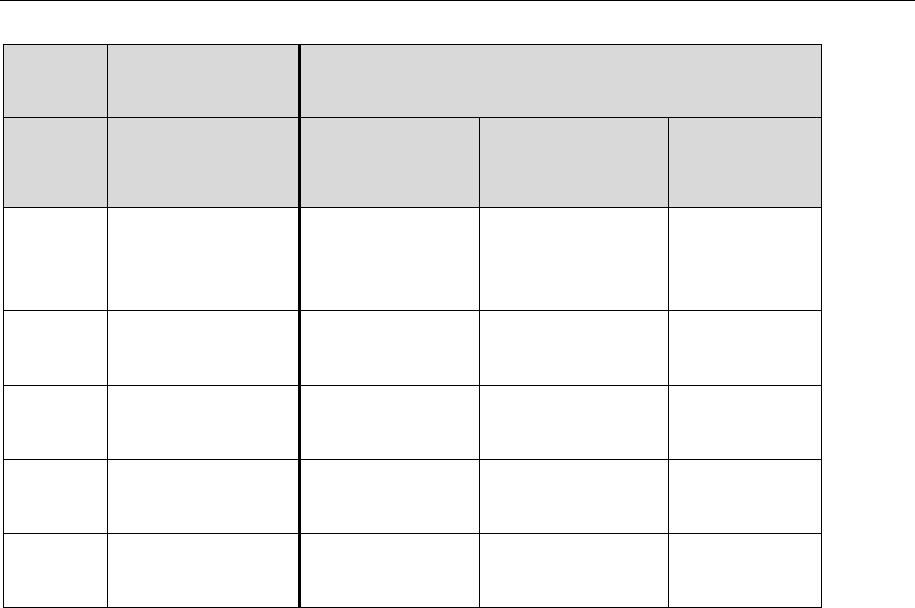
Table II: Common Lab Contaminants
Methylene chloride
Acetone
2-Butanone (MEK)
Phthalate Esters
Toluene
Hexane
2-Propanol
Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 10 of 40
Table III: Blank Qualifications
Blank
Sample
Row
Number
Result
Result
Validated
Result
Validation
Qualifier
1
≤ DL or LOD
≤ DL or LOD
Report as
required by QAPP
(at DL or LOD)
U
2
> DL or LOD
≤ DL or LOD
Report at DL or
LOD
U
3
> DL or LOD
> DL or LOD but
≤ LOQ
Report at LOQ
U
4
> DL or LOD
> LOQ but ≤ 5x
blank
Report at Sample
Result
U
5
> DL or LOD
> LOQ and > 5x
blank
Report at Sample
Result
None
LOD = Limit of Detection LOQ = Limit of Quantitation DL = Detection Limit
Note: The QAPP should specify reporting at either the DL, LOD or both
3.3.2 Field Duplicates (can also be called replicates)
Field duplicates consist of either collocated or subsampled (split) samples. Field duplicates
for groundwater and surface water samples are generally considered to be collocated
samples. Soil duplicate samples may be split samples or collocated, as specified in the
QAPP. Field duplicate results are an indication of both field and laboratory precision; the
results may be used to evaluate the consistency of sampling practices.
Evaluation of Field Duplicates
Check to ensure that field duplicates were collected and analyzed as specified in the QAPP.
If the sampling frequency is less than the frequency stated in the QAPP, no qualification of
the associated sample results is necessary, but the incident should be discussed in the data
validation report.
Relative Percent Differences (RPDs) should be calculated when detected results are
reported for the duplicate(s) and at least one of those results is greater than or equal to the
LOQ. For field duplicate results, if the RPDs or absolute differences are greater than those
stated in the QAPP, qualify the associated sample results as estimated J, and any non-
conformities should be noted in the data validation summary.
Professional judgment may be required in instances where the sample and field duplicate
results are less than the LOQ or project Reporting Limits (RLs). RPD results can be elevated
when low (e.g., <5x the LOQ) or estimated concentrations in the samples and duplicates are
reported. If one or both results in a duplicate pair are <5x the LOQ, the absolute difference

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 11 of 40
between the two results can be used as an alternative acceptance criterion, if approved by
the QAPP or project point of contact (QAPP Worksheet #6).
Some sampling schemes, such as Incremental Sampling Methodology (ISM) require specific
replicate calculations, which should be specified in the QAPP.
It should be noted that RPDs or absolute differences for field duplicates are generally not
calculated or reported by the laboratory and should be calculated by the validator.
There are instances where an RPD is not calculable (for example, when one result is a non-
detect and the other is greater than the LOQ). In those cases, the RPDs are not calculated
but the non-conformity should be noted in the data validation report. The reported
concentrations should be carefully examined to determine what conditions would permit one
result to be reported at or above the LOQ/RL and the other to be reported below the LOQ/RL
or as a non-detect.
4.0 Stage 2A Validation
Note: Stage 2A includes all of Stage 1
Stage 2A requires the review and qualification of the following summary documents.
• Surrogate Recovery Summary
• Laboratory Control Sample/Laboratory Control Sample Duplicate
• Matrix Spike/Matrix Spike Duplicate Recovery Summary
• Method Blank Summary Form
• Sample Dilution/Reanalysis Summaries
Stage 2A is the validation of preparation batch specific QC data in addition to any sample
specific parameters included in Stage 1.
Generally, a “preparation batch” of samples consists of up to twenty field samples (maximum)
along with duplicate/replicate (laboratory or field), method blank, and control/matrix type QC
samples. They are meant to be analyzed together on a single instrument. However,
laboratories may choose to split up a batch over multiple instruments to save time. In this
case, if the use of multiple instruments is uncovered in a Stage 2A validation, the validator
should request from their point of contact (QAPP Worksheet #6) a Stage 2B validation to
review sequence logs. The use of multiple instrumentation should be noted in the data
validation report.
4.1 Surrogate Spikes
Extraction efficiency on individual samples is established by means of surrogate spikes. All
samples are spiked with surrogates prior to sample extraction. The evaluation of the results
of these surrogate spikes is not necessarily straightforward. The sample itself may produce
effects due to such factors as interference and high concentrations of analytes. Because the
effects of the sample matrix are frequently outside the control of the laboratory and may
present relatively unique problems, the review and validation of data based on specific
sample results is frequently subjective and demands analytical experience and professional
judgment.
Verify that surrogate percent recoveries and acceptance limits were reported for all field and
batch QC samples.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 12 of 40
Sample and batch QC surrogate recoveries should be within control limits established in the
QAPP or the QSM. Verify that no samples or batch QC have surrogate percent recoveries
outside their criteria.
If any surrogate recovery is out of specification, then a re-extraction (if applicable) and
reanalysis should have been performed and reported. Re-extraction is not required for
surrogates with high bias associated with non-detect sample results. The laboratory should
have reported both runs if the first was unsuccessful.
The laboratory does not have to reanalyze a sample if a matrix spike/matrix spike duplicate
(MS/MSD) was performed on the sample with out-of-control surrogate results showing the
same matrix effects, as long as the batch QC display acceptable surrogate percent
recoveries.
Evaluation of Surrogates
If surrogate percent recoveries are out of specification with no evidence of re-extraction (if
applicable) and reanalysis, justification should be noted in the laboratory case narrative (e.g.,
limited sample volume prevented reanalysis). If justification is not noted, the point of contact
(QAPP Worksheet #6) should be reached for further guidance.
If the surrogate percent recovery control criteria displayed in the deliverable are not the same
ranges stipulated in the QAPP or the DoD QSM, reference the required control ranges for
evaluation instead of the summarized ranges in the deliverable. The project team should be
informed to implement changes to the current deliverables or those to be created in the
future. Please follow the notification protocols outlined in the QAPP (such as the UFP-QAPP
Worksheet #6).
GC Organic Contaminants
If any surrogate percent recovery is < 10%, qualify detects as estimated J-, and non-detects
as X, exclusion of data recommended for all associated target analytes in the sample.
If any surrogate percent recovery is greater than the upper acceptance limit, qualify
associated detects in the sample as estimated with a positive bias J+ and non-detects should
not be qualified.
If any surrogate percent recovery is less than the lower acceptance limit but ≥ 10%, qualify all
associated detects as estimated with a negative bias J- and non-detects as estimated UJ.
For samples that require dilution, surrogates may be reported as “diluted out”, if dilution is
such that the surrogate can no longer be detected above the LOD. If this is the case, note in
the data validation report that surrogate evaluation could not be performed due to a high
dilution factor. A full evaluation (Stage 4 validation) of the sample chromatogram and
quantitation report may be necessary to determine that surrogates are truly “diluted out.”
In the special case of blank analysis with surrogates out of specification, the reviewer should
give special consideration to the validity of associated sample data. The primary concern is
whether the blank failures represent an isolated incident with the blank alone, or whether
there is a systemic problem with the analytical process. For example, if the samples in the
batch show acceptable surrogate recoveries, the reviewer may determine the blank failure to
be an isolated occurrence for which no qualification of the data is required. However, if
surrogate failures occur throughout the field and QC samples, then consideration should be
given to communicate with the QAPP point of contact (QAPP Worksheet #6) to receive a

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 13 of 40
revised report. If this cannot be done, then consideration should be given to qualifying all
associated data as X, exclusion of data recommended.
4.2 Laboratory Control Sample/Laboratory Control Sample Duplicate (LCS/LCSD)
An LCS is a sample matrix free from the analytes of interest, spiked with known amounts of
the analytes and taken through all sample preparation, cleanup and analytical steps. LCSs
establish the method precision and bias for a specific batch of samples. Analysis of LCSDs
may be required by the QAPP or may be used as an indication of batch precision in
instances where MSD analysis is not possible (e.g., a limited volume of sample).
LCS (sometimes called a “Blank Spike”) and, if analyzed, LCSD recoveries should be within
the QC limits specified in the QAPP or as listed in the QSM. If an LCSD was analyzed, the
RPDs should be within the QC limits specified in the QAPP or as listed in the QSM.
Evaluation of LCS/LCSD
Verify that results (from appropriate summary form), percent recoveries, RPDs (if applicable)
and acceptance limits were reported for all target analytes and surrogates.
If the LCS/LCSD was not spiked with all target analytes, notify the project team by following
the notification protocols outlined in the QAPP (such as UFP-QAPP Worksheet #6) and
qualify all detects and non-detects for those analytes not spiked as X, exclusion of data
recommended.
If the spike percent recovery control criteria displayed in the deliverable are not the same
range (i.e., outside or wider than) as those stipulated in the QAPP or the DoD QSM,
reference the required control ranges for evaluation instead of the summarized ranges in the
deliverable. The project team should be informed to implement changes to the current
deliverables or those to be created in the future.
In-house control limits are acceptable for any analytes not specified in the QAPP or DoD
QSM. No qualification is necessary for any reported in-house control limit that is within (i.e.,
same or less than) those specified in the QAPP or DoD QSM. If the laboratory’s in-house
control limits are wider than those in the QSM and the results are outside of the DoD QSM
limits, qualify the appropriate data as X, exclusion of data recommended.
If the LCS percent recoveries were greater than the upper control limit, qualify detects for the
analyte in associated samples as estimated with a positive bias J+. Non-detects should not
be qualified.
If the LCS percent recoveries were less than the lower control limit, qualify detects for the
analyte in associated samples as estimated with a negative bias J- and non-detects as X,
exclusion of data recommended.
If the LCS/LCSD RPDs were greater than the acceptance limits, qualify detects for the
analyte in the associated sample(s) as estimated J. Non-detects should not be qualified.
Professional judgment should be utilized in qualifying data for circumstances other than
those listed above.
4.3 Matrix Spike/Matrix Spike Duplicate (MS/MSD)
MS/MSD data are used to determine the effect of the matrix on a method’s recovery
efficiency and precision for a specific sample matrix.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 14 of 40
Generally, qualifying action is taken only on the parent sample based on MS/MSD non-
conformities. In instances where it can be determined that the results of the MS/MSD affect
only the sample spiked, then qualification should be limited to that sample alone. Using
informed professional judgment, the data reviewer may use the MS/MSD results in
conjunction with other QC criteria (i.e., surrogates and LCS) and determine the need for
additional qualification beyond that applied to the parent sample when the laboratory is
having a systemic problem in the analysis of one or more analytes, which affects all
associated samples.
If a field blank was used for the MS/MSD, this fact should be included in the data validation
report. Sample matrix effects may not be observed with field blanks; therefore, the recoveries
and precision do not reflect the extraction or analytical impact of the site matrix.
The laboratory should spike and analyze an MS/MSD from the specific project site as
required by the QAPP for each matrix type and analytical batch. The MS and MSD should be
spiked per QSM requirements with all target analytes. If the parent sample for the MS/MSD
was from another site or project (for example, not enough sample collected, or multiple site
samples analyzed within a single batch), the reason should be documented in the data
validation report, and sample results should not be qualified due to any non-conformities
noted in non-site-specific matrices.
Evaluation of MS/MSD
MS/MSD data should be reported on a MS/MSD summary form (or equivalent). Verify that
the MS/MSD were spiked with all target analytes, and that percent recoveries were reported
for all target analytes.
Compare the percent recovery and RPD for each analyte with LCS control limits established
by the QAPP. If the spike percent recovery control criteria displayed in the deliverable are not
the same range (i.e., outside or wider than) as those or stipulated in the QAPP or the DoD
QSM, reference the required control ranges for evaluation instead of the summarized ranges
in the deliverable. The project team should be informed to implement changes to the current
deliverables or those to be created in the future. Please follow the notification protocols
outlined in the QAPP (such as UFP-QAPP Worksheet #6).
If the MS/MSD was not spiked with all target analytes, notify the project team by following the
notification protocols and qualify all detects and non-detects in the parent sample for those
analytes in each batch not spiked as X, exclusion of data recommended.
If the MS/ MSD percent recoveries were greater than the upper control limit, qualify detects
for the analyte in the associated parent sample as estimated J+. Non-detects should not be
qualified.
If the MS/MSD percent recoveries were less than the lower acceptance limit but ≥ 10%,
qualify detects for the analyte in the associated parent sample as estimated J- and non-
detects as estimated UJ. If the percent recoveries were < 10%, qualify detects for the analyte
in the associated parent sample as estimated J- and non-detects as X, exclusion of data
recommended.
If the MS/MSD RPDs were greater than the acceptance limits, qualify detects for the analyte
in the associated sample(s) as J. Non-detects should not be qualified.
If the MS/MSD fail due to the presence of target analytes in the parent sample at > 4X the
spike concentration or if matrix spikes are diluted to less than the LOQ, then MS non-

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 15 of 40
conformities should not result in any qualifications. Note the incident in the data validation
report.
4.4 Method Blanks
A method blank is used to identify systemic contamination originating in the laboratory that
may have a detrimental effect on project sample results. The validator should identify
samples associated with each method blank using a method blank summary form (or
equivalent). Verify that the method blank has been reported per batch.
Compare the results of each method blank with the associated sample results. The reviewer
should note that the blank analyses may not involve the same weights, volumes, percent
moistures, or dilution factors as the associated samples.
These factors should be taken into consideration when applying the 5X and 10X criteria
(discussed in section 3.3.1), such that a comparison of the total amount of contamination is
actually made. Care should be taken to factor in the percent moisture or dilution factor when
doing comparisons between detects in the sample and the method blank. If available, raw
data should be used for comparison and evaluation.
Evaluation of Method Blanks
If no method blank was analyzed, qualify detects in samples with no associated method
blank X, exclusion of data recommended. Non-detects do not require qualification.
If gross contamination exists (defined as greater than a Project Action Limit) in the method
blanks, all analytes affected should be qualified X, exclusion of data recommended. due to
interference in all affected samples and this should be noted in the data validation report.
If target analytes other than common laboratory contaminants (see Table II) are found at low
levels in the method blank(s), it may be indicative of a problem at the laboratory and should
be noted in the data validation report.
If an analyte is detected in the method blank, but not in the associated samples, no action is
taken.
If an analyte is detected in the method blank and in the associated samples, the action taken
depends on both the blank and sample concentrations. Table III (Blank Qualifications) and
section 3.3.1 discussions on evaluations of results from the DL/LOD to LOQ is also
applicable to the method blank.
Additionally, there may be instances where little or no contamination was present in the
associated method blanks, but qualification of the sample was deemed necessary.
Contamination introduced through dilution water is one example. Although it is not always
possible to determine instances of this occurring can be detected when contaminants are
found in the diluted sample result but are absent in the undiluted sample result. It may be
impossible to verify this source of contamination. However, if the reviewer determines that
the contamination is from a source other than the sample, the data should be qualified. In this
case, the 5X or 10X rule does not apply. The reason should be documented in the data
validation report. Qualification of the data should be performed as given in Table III.
Multiple blank contaminations (such as a batch with field blanks and a method blank) does
not establish a ‘hierarchy’ of one blank over another. Each blank must be evaluated
individually. Blanks should not be qualified due to the results of other blanks.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 16 of 40
4.5 Sample Dilutions and Reanalysis
Laboratories may dilute samples due to high analyte concentrations or reanalyze samples
due to QC non-conformities, and document both sets of results. Generally, the laboratory will
report the “best” value for a given analyte in the official laboratory report (or equivalent form).
In these instances, the validator should evaluate both the reported and the initial analysis
result. The validator should consider the application of appropriate qualifiers to the reported
results within the scope of the project due to elevated LODs/LOQs or other QC non-
conformities. Qualifiers apply only to the reported results in the official laboratory report.
Evaluation of Sample Dilutions and Reanalysis
When sample results are reported at more than one dilution due to analyte concentrations
exceeding the calibration curve, the lowest LODs are generally used for the non-detects
unless a QC criterion has been exceeded.
Results reported from dilutions leads to elevated LODs for non-detects. The data validation
report should indicate the reason for all reported dilutions (including cases where the
laboratory did not perform an undiluted analysis) resulting in elevated sensitivity limits for
non-detected results.
When reanalysis has occurred due to QC non-conformities, the validator should ensure that
the non-conformity was corrected during the reanalysis. If that is not the case, then the
appropriate qualifier should be placed on the reported results.
In some cases, using professional judgment, the validator may determine that an alternate
result was more appropriate than the one reported. In those cases, explain the rationale for
accepting the alternate result in the data validation report.
In some cases, reanalysis may lead to exceedances of holding time. Use professional
judgment to evaluate the results and apply the appropriate qualifiers (if required).
5.0 Stage 2B Validation
Note: Stage 2B includes all of Stage 1, and Stage 2A
Stage 2B requires the review and qualification of the following summary documents.
• Sequence and Preparation Logs (or equivalent)
• Instrument Performance Check Summary (any equivalent to include Degradation
Checks)
• Initial Calibration Summary (any equivalent to include Initial Calibration, Average
Response Factors, and Regression)
• Initial/Continuing Calibration Verification Summary (any equivalent to include Initial
and Continuing Calibration Verifications)
• Internal Standard Summary (any equivalent to include Internal Standards)
• Cleanup Procedure Summary (any equivalent to include Cleanup Recovery Checks)
• Second Column Summary (or equivalent to show analysis by a Second Column)
Stage 2B is the validation of instrument specific QC data.
5.1 Sequence and Preparation Logs
Sequence logs are reviewed by the data validator to ensure all QC samples (both batch and
instrument specific) were analyzed within a specific batch, in the correct order. Preparation

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 17 of 40
logs are reviewed by the data validator to ensure that samples had the proper extraction
performed, within specified holding times. Non-conformities uncovered in the review of the
logs may point the validator to specific samples that require further review. Non-conformities
uncovered in preparation or sequence logs should be noted in the data validation report.
Sequence logs are helpful in identifying when multiple instrumentation is used to analyze a
batch of samples. For example, it is not uncommon to analyze a single batch of twenty
samples at the same time on two or more different instruments. At a minimum, each
instrument should be calibrated independently. Batch QC should be reviewed on each
instrument, as appropriate. Non-conformities involving the use of multiple instruments should
be noted in the data validation report.
5.2 Instrument Performance Checks (Method 8081: Organochlorine Pesticides)
4,4′-Dichlorodiphenyltrichloroethane (DDT) and Endrin are prone to degradation in the
injection port liner with the presence of high boiling residue from sample injection or when the
injector contains metal fittings. Degradation problems are checked by injecting a standard
containing only DDT and Endrin. Presence of DDE, DDD, Endrin Aldehyde, or Endrin Ketone
indicates breakdown. Unless otherwise specified in an approved project plan, this test should
be performed as a test of the inertness of the analytical system even when DDT and Endrin
are not target analytes for a given project.
Evaluation of Performance Checks
The breakdown of DDT and Endrin should be measured before samples are analyzed at the
beginning of each 12-hour shift. Professional judgment should be applied to qualify results for
samples that were analyzed more than 12 hours after the breakdown standard was analyzed.
Verify that the degradation does not exceed 15% for either DDT or Endrin on both GC
columns.
1. If DDT breakdown is > 15%:
• Flag all associated detects for DDT, DDD, and DDE as J, estimated.
• If DDT was not detected in the breakdown standard, then qualify all results for
DDT as X, exclusion of data recommended. Qualify DDD and DDE detects as
presumptively present at an estimated quantity NJ.
• If DDT is present and passes on one column, but not confirmed on the other
column that has > 15% breakdown, qualify the associated DDT data X,
exclusion of data recommended. Qualify DDE and DDE detects as
presumptively present at an estimated quantity NJ.
2. If Endrin breakdown is > 15%:
• Flag all associated detects for Endrin, Endrin Aldehyde, and Endrin Ketone as
J, estimated.
• If Endrin was not detected, but Endrin Aldehyde or Endrin Ketone are detected
in the breakdown standard, then qualify the Endrin result as X, exclusion of
data recommended. Qualify Endrin Ketone or Endrin Aldehyde detects as
presumptively present at an estimated quantity NJ.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 18 of 40
• If Endrin is present on one column but not confirmed on the other column that
has > 15% breakdown, qualify the associated data X, exclusion of data
recommended. In this case, the reviewer may determine that the compound
needs to be reported as a tentative identification N.
5.3 Initial Calibration
The objective of initial calibration is to ensure that the instrument is capable of producing
acceptable qualitative and quantitative data. Initial calibration demonstrates that the
instrument is capable of acceptable performance in the beginning of the analytical run and of
producing an acceptable calibration curve. The GC system can be calibrated using the
external standard technique or internal standard technique. Because of the difficulty in
selecting suitable internal standards, the external standard technique will most often be the
method of choice.
Evaluation of External Calibration
A minimum of five standards is required for a linear calibration. The lowest calibration
standard concentration should be at or below the LOQ. If the laboratory used more than the
minimum number of standards and must exclude calibration points, only exclusion of the high
or low standard is allowed. The calibration points in between should not be excluded without
sound technical justification.
If reported target analytes were not properly calibrated, make note of this in the data
validation report and qualify the associated data as X, exclusion of data recommended.
If the concentration of the lowest standard in the initial calibration was greater than the LOQ,
qualify all detects between the DL and the lowest standard as X, exclusion of data
recommended. Detects above the low standard do not require qualification. Non-detects do
not require qualification.
Inform the point of contact (QAPP Worksheet #6) for further instruction in those instances of
unwarranted manipulation of calibration curves. As an example, calibration curves that were
run with excessive calibration points that are misapplied to achieve passing criteria (without
any technical justification) require prompt notification of the project team. If the issue cannot
be resolved with the laboratory, make note of this in the data validation report and qualify all
affected data as X, exclusion of data recommended.
Calibration Factor (CF): External standard calibration involves a comparison of instrument
responses from the sample to the target compound responses in the calibration standards.
The ratio of the detector response to the amount of analyte in the calibration standard is
defined as the CF. The instrument should have been calibrated for all target analytes and
surrogates.
Evaluate the percent relative standard deviation (%RSD) for all target compounds. If any
analyte has a %RSD greater than 20%, qualify detects for the affected compounds as J and
non-detects as UJ in the associated samples that correspond to that initial calibration.
If the %RSD is excessively high (defined as > 40%) qualify associated target analyte sample
results as X, exclusion of data recommended.
Linear Regression: The laboratory may employ a linear or weighted linear least squares
regression curve. Evaluate the Correlation Coefficients (r) for all applicable target analytes.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 19 of 40
The r value should be ≥ 0.995. Some instrumentation reports Coefficient of Determination
(r
2
). If the instrument reports r
2
, the value should be ≥ 0.99.
If the r value for any target analyte is < 0.995 (or the r
2
value is < 0.99), qualify detects for the
affected analytes J and non-detects as UJ in the associated samples.
If the r value is excessively low (defined as < 0.95) or the r
2
value is excessively low (defined
as < 0.90), qualify all associated non-detects as X, exclusion of data recommended and
detects as estimated J.
Non-Linear Regression: The laboratory may also generate a higher order curve for the
calibration. The calibration curve should not be more than second order (Quadratic) in
accordance with QSM requirements.
A minimum of six standards is required for a second order (quadratic) curve.
Evaluate the correlation coefficients(r) for all applicable target analytes. The r value should
be ≥ 0.995. Some instrumentation reports coefficient of determination (r
2
). If the instrument
reports r
2
, the value should be ≥ 0.99.
If the required number of calibration standards was not used, qualify detects J. Apply
professional judgment to qualify non-detects based on the concentrations of the standards
used.
If the r value for any target analyte is < 0.995 (or the r
2
value is < 0.99), qualify detects for the
affected analytes J and non-detects UJ in the associated samples.
If the r value is excessively low (defined as < 0.95) or the r
2
value is excessively low (defined
as < 0.90), qualify all associated non-detects as X, exclusion of data recommended and
detects as estimated J.
Calibration curves that are higher than second order (such as a third order polynomial fit) are
not allowed in accordance with QSM requirements. Qualify X, exclusion of data
recommended all associated data based on third order (or higher) calibration curves.
5.3.1 Method 8081: Organochlorine Pesticides & Method 8082: Polychlorinated
Biphenyls (Aroclors)
For Organochlorine Pesticides with multicomponent analytes such as Toxaphene and
Chlordane, quantitation must be performed using a five-point calibration, in accordance with
QSM requirements. Results may not be quantitated using a single point calibration. If
Toxaphene and Chlordane results are reported without a multipoint calibration, then inform
the QAPP point of contact (QAPP Worksheet #6). If the situation cannot be resolved with a
revised laboratory report, then qualify all associated detects as X, exclusion of data
recommended and make note in the data validation report.
For PCB Aroclors, a multipoint calibration employing a mixture of Aroclors 1016 and 1260 at
five different concentrations is sufficient to demonstrate detector linearity because it will
usually include many of the peaks of some of the other Aroclors. Although the method may
be used to demonstrate that a sample does not contain peaks for some of the other Aroclors,
the qualitative identification of the Aroclor is subject to the professional judgment of the
analyst after comparison to standard(s) of that respective Aroclor. If any Aroclor other than
1016 or 1260 is detected, the result must be quantified against a multipoint calibration for the
specific Aroclor mixture of interest prior to reporting a quantitative result for that Aroclor.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 20 of 40
If any Aroclor other than 1016 or 1260 is detected in the associated sample and is not
quantified against a multipoint curve of the identified Aroclor, then inform the QAPP point of
contact (QAPP Worksheet #6). If the situation cannot be resolved with a revised laboratory
report, then qualify all associated detects as X, exclusion of data recommended and make
note in the data validation report.
5.3.2 Method 8151: Chlorinated Herbicides
Herbicide samples undergo a hydrolysis step during extraction and may undergo a
derivatization step. If the calibration standards have undergone these steps as well, then the
calibration curve is directly comparable to the samples. However, if the calibration standards
did not undergo these steps, then then calculation of concentration (quantitation of the
results) should have included a correction of the molecular weight of the methyl ester versus
the acid herbicide.
This calculation may require a Stage 3 validation to determine if the correction factor was
applied appropriately. Inform the point of contact (QAPP Worksheet #6) if sufficient
information cannot be obtained for a Stage 2B validation.
5.4 Initial (Secondary Source) and Continuing Calibration Verification
The initial calibration curve should be verified with a standard that has been purchased or
prepared from an independent source each time initial calibration is performed. This standard
is called the secondary source or Initial Calibration Verification (ICV). The ICV should contain
all of the GC target analytes. Note that multiple ICVs may be analyzed to encompass all of
the target analytes.
After the initial calibration has been verified with a second source, samples may be run
continuously until the initial calibration fails. To verify this, a Continuing Calibration
Verification (CCV) containing all GC target compounds should be analyzed before sample
analysis, after every 10 field samples, and at the end of the analysis sequence. Continuing
calibration checks satisfactory performance of the instrument on a day-to-day basis.
The CCVs for Pesticide multicomponent mixtures Toxaphene and Chlordane by method
8081 and Aroclors other than 1016/1260 by Method 8082 are only required before sample
analysis.
Evaluating the ICV and CCV
Verify the ICV was analyzed following the initial calibration and contained all target analytes.
Verify the CCVs have been run at the proper frequency. When a new initial calibration is
performed, the ICV can serve as the first CCV if samples are being run afterwards. The
CCVs after the first ICV are not required to be a second source.
The ICV percent difference (%D) or percent drift for each target analyte and surrogate should
be ≤ 20%. The CCV %D for each target analyte should also be ≤ 20%.
If the ICV (second source) has not been performed successfully after an initial calibration or if
samples have been analyzed prior to a valid ICV, qualify X, exclusion of data recommended
all associated data. No samples should have been analyzed in accordance with QSM
requirements.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 21 of 40
If the CCV has not been analyzed (either continuing or end-of-run), qualify X, exclusion of
data recommended all associated data. No samples should have been analyzed without a
valid CCV.
Verify that the %Ds are within the acceptance criteria. If any target analytes do not meet the
acceptance criteria for the CCV, qualify detects for that analyte as estimated J+ when the
%D is higher than acceptance criteria and J- when below acceptance criteria. Non-detects
are qualified as UJ in all associated samples for %D outside of acceptance criteria.
For gross exceedances of %D (defined as > 50%) qualify all associated data as X, exclusion
of data recommended.
5.5 Internal Standards (Optional)
Internal standard (IS) calibration involves comparison of instrument responses from the target
compounds in the sample to responses of internal standards added to the sample or extract
before injection. A constant amount of the IS is added to all samples extracts and calibration
standards. The peak response ratio of the target compound to the IS in the sample extract is
compared with the same ratio for each calibration standard. This ratio indicates that the
target compound response is calculated relative to that of the IS.
Evaluation of Internal Standards
Each IS area should be within 50-200% (same as QSM criteria of -50 to +100) of the area of
the mid-point standard in the ICAL for associated standards. On days when ICAL is not
performed, the daily initial CCV is used.
The IS retention times for all field and QC samples should be within 30 seconds of the
retention time of the midpoint standard in the ICAL, or on days when ICAL is not performed,
the initial CCV is used.
Detects for analytes quantitated using an IS area count > 200% should be qualified estimated
with a J. Non-detects should not be qualified.
Analytes quantitated using an IS area count < 50% but ≥ 20% should be qualified estimated
with a J for detects. Non-detects should be qualified estimated UJ.
If extremely low area counts are reported (< 20% of the area for associated standards),
detects and non-detects should be qualified X, exclusion of data recommended.
Large retention time variations may call into question peak identifications. If an IS retention
time varies by more than 30 seconds, detects and non-detects should be qualified X,
exclusion of data recommended.
5.6 Cleanup Procedures for Methods 8081 (Organochlorine Pesticides), 8082 (PCBs),
8141 (Ortho phosphorous Compounds), and 8151 (Chlorinated Herbicides)
Cleanup techniques are used to eliminate or minimize chemical and chromatographic
interferences arising from the samples themselves. Most environmental or waste samples
may require one or more cleanup techniques after extraction and prior to analysis. The
specific cleanup performed will be dependent on the nature of the samples.
A summary of recommended cleanup procedures can be found in Table IV. SW-846 method
3600 provides more in-depth guidance on cleanup method selection.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 22 of 40
All associated batch QC samples must undergo the same cleanup procedure(s) as the
samples.
A description of the different cleanup approaches follows:
• Adsorption Chromatography - Florisil (Method 3620), and Silica Gel (Method 3630)
are useful for separating analytes of a relatively narrow polarity range away from
extraneous, interfering peaks of a different polarity. These are primarily used for
cleanup of a specific chemical group of relatively non-polar analytes such as
pesticides and PCBs.
• Gel Permeation Chromatography (GPC) (Method 3640) - This cleanup technique
applies to a broad range of pesticides and is capable of separating high molecular-
weight, high boiling point material from the target analytes. GPC may not be
applicable to elimination of extraneous peaks on a chromatogram which interfere with
the analytes of interest. It is, however, useful for the removal of high boiling point
materials which would contaminate injection ports and improve continuing calibration.
• Acid-base Partitioning (Method 3650) – This technique is useful for separating acidic
or basic organics from neutral organics. It has been applied to analytes such as the
Chlorinated Herbicides.
• Sulfur cleanup (Method 3660) – This technique is useful in eliminating known sulfur
from sample extracts, which may cause chromatographic interference with analytes of
interest.
• Sulfuric Acid/Permanganate Cleanup (Method 3665) – This technique improves
elevated baselines for PCB sample extracts prior to analysis. This method cannot be
used to cleanup extracts for other target analytes, as it will destroy most organic
chemicals, including the pesticides.
Table IV: Cleanup Methods
Analyte Group
Analytical Method
Cleanup Methods
Organochlorine Pesticides
8081
3620, 3640, 3660
PCBs
8082
3620, 3630, 3665
Organophosphorus Pesticides
8141
3620
Chlorinated Herbicides
8151
3620, 3650
Evaluation of Cleanup Methods (Recovery Checks)
The analyst must demonstrate that the compounds of interest are quantitatively recovered
before applying this method to actual samples. This test applies to both the column cleanup
and cartridge cleanup procedures. A recovery check needs to be performed using standards
of the target analytes at a known concentration near the LOQ for the target analyte. Only lots
of cartridges/columns from which the spiked analytes are quantitatively recovered may be
used to process the samples.
When using Florisil (3620) for pesticides, the lot of Florisil cartridges is acceptable if all
pesticides are recovered at 80 to 110%, the recovery of Trichlorophenol is < 5%, and no
peaks interfere with the target analytes. For Chlorophenoxy acid herbicides, the lot of Florisil
is acceptable if the target analytes are quantitatively recovered, the recovery of
trichlorophenol is < 5%, and no peaks interfere with the target analytes.
For Silica Gel (3630) the recovery of all analytes in the recovery check must be between 85-
115%.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 23 of 40
The GPC (3640) should be calibrated at least once per week. The retention time shift must
be < 5% when compared to retention times in the last calibration UV trace. The UV trace
requirements are as follows: corn oil and Phthalate peaks must exhibit > 85% resolution;
Phthalate and Methoxychlor peaks must exhibit > 85% resolution; Methoxychlor and
Perylene peaks must exhibit > 85% resolution; Perylene and Sulfur peaks must not be
saturated and must exhibit > 90% baseline resolution. GPC elution should continue until after
Perylene has eluted, or long enough to recover at least 85% of the analytes, whichever time
is longer.
Acid-base Partition (3650), Sulfur Cleanup (3660), and Sulfuric acid/Permanganate (3665)
requires only that the batch QC pass the QC limits outlined in the QAPP or the laboratory
Standard Operating Procedure (SOP).
If it is determined that the QC samples were not treated with the same cleanup procedures
as the field samples, inform the point of contact (QAPP Worksheet #6) to receive a revised
laboratory report. If this is not possible, qualify all associated data as, exclusion of data
recommended and make note of this in the data validation report.
If the recovery check standard fails high, qualify detects J+ and no qualification is necessary
for non-detects.
If the recovery check standard fails low, qualify associated positive field sample results as
estimated J and non-detects as estimated UJ.
If the recovery standard fails unusually low (defined as < 10%), qualify positive field sample
results as estimated J and non-detects as X, exclusion of data recommended.
If there is recovery for negative test analytes (Trichlorophenol > 5%) or peak resolution is
less than required, inspect the field sample chromatograms and use professional judgment to
qualify associated results.
5.7 Second Column Confirmation/Dissimilar Detector Confirmation
Second column confirmation of all detects above the QAPP stated DL must be performed for
all GC work, unless an alternate detector was utilized for confirmation. The only exceptions
are for single column methods such as Total Petroleum Hydrocarbon (TPH) by Method 8015
where confirmation is not required. For the purposes of reporting, both columns are
considered equivalent, provided QC evaluations are within acceptance limits on both
columns. Barring chromatographic problems (overlapping peaks, baseline shifts) or QC
anomalies on one column or the other, the result from the column specified in the QAPP
should be reported. Some projects may require the reporting of results from both columns.
Evaluation of Second Column Confirmation
The RPD between columns should be ≤ 40%. The concentrations of both analytical column
peaks must be greater than the stated DL. If one column has a peak that corresponds to a
concentration that is less than the associated DL, then the result must be reported as a non-
detect, and the RPD is noted in the data validation report as “non-calculable”.
If the RPD between columns is > 40%, qualify the results as estimated J.
If second column confirmation is not performed, qualify any reported detect as presumptive
and estimated, NJ. The validator should inform the QAPP point of contact (QAPP Worksheet
#6) to obtain a revised laboratory report, if possible.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 24 of 40
6.0 Stage 3 Validation
Note: Stage 3 validation includes all of Stage 1, Stage 2A and Stage 2B
Stage 3 requires the review of the following documents
• Raw Data (including any laboratory forms, instrument outputs, spreadsheets, or
handwritten calculations necessary for recalculation and re-quantification)
• Standards Traceability forms and worksheets
• DL studies (optional)
Stage 3 validation includes the recalculation and re-quantification of selected samples, and
method and instrument QC. The types of results that should be recalculated and re-
quantified include target analytes, analytes with detects above the LOQ, second column
results, and field QC samples (blanks and duplicates). For method QC results, spiked
recoveries and method blanks should be considered. For instrument QC, calibrations
(including CFs and regressions), calibration verifications, and internal standards should be
recalculated and re-quantified. Some calculations may include the need to review standards
preparation and serial dilutions.
6.1 Samples and Field QC
When choosing samples, field QC and analytes for re-quantification and recalculation,
consideration should be given to the laboratory’s batching scheme to ensure a
representative subsample of recalculations is performed. Additionally, if priority
contaminants or contaminants of concern are identified in the QAPP, those analytes should
be selected for re-quantification and recalculation. Other circumstances that should be
prioritized for re-quantification and recalculation are diluted samples, manual integrations,
re-runs of samples, and field QC blank failures.
Re-quantification and recalculation should be performed on the designated percentage of the
samples per SDG (or however defined in the QAPP, such as percentage of total project
samples) per analytical suite. As a minimum, it is recommended that 10% of the data should
be re-quantified and recalculated unless specific instructions are given in the QAPP.
Sample recalculations should include the raw instrument result, re-quantified from the
instrument response against the calibration function, and the final reported sample result,
including any dilution, preparation factor, or percent moisture (if applicable). The equations
in Appendix B can be used to calculate a sample result from the corresponding reported
calibration or regression function, as appropriate.
Verify that one or more of the laboratory’s RLs (such as LOQ) are calculated correctly for the
non-detects and reported accordingly. If a DL study was identified by the QAPP, recalculate
one or more analyte DLs.
Re-quantitate all detected target analytes in the 10% sample data chosen. For some
samples, all results may be non-detects, therefore recalculation would not be necessary.
Verify that sample-specific results have been adjusted correctly to reflect percent solids,
original sample mass/volume, and any applicable dilutions.
Re-quantitate all detects found in the field QC blanks (such as trip blanks, source blanks, or
equipment blanks). Field QC sample replicate calculations should include re-quantification of
the same detected sample/replicate sets and determination of RPD or RSD.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 25 of 40
When recalculations require rounding of data, that rounding should be completed only once
at the end of all calculations to minimize rounding errors. Calculations should be rounded to
the significant figures of the underlying criteria. For example, an LCS criteria of 80-117%
would still be considered acceptable if the recalculation was 117.4%.
Evaluation of Sample and field QC recalculations
If the laboratory’s quantitation, or RLs (however defined) are calculated incorrectly, then
continue to recalculate limits until it is determined whether the problem is systemic (such as
incorrect equations used) or isolated (such as a transcription or rounding errors).
For systemic (defined as widespread and major in nature) issues that cannot be corrected
through a revised laboratory report, qualify all results as X, exclusion of data recommended.
For isolated cases, use professional judgment. It may be necessary to engage the point of
contact (QAPP Worksheet #6) to communicate with the laboratory, so they can provide
revised (corrected) results. In all cases, if calculation errors affect project target analytes, the
point of contact (QAPP Worksheet #6) should be notified, and all affected results noted in
the data validation report, including listing the calculation errors.
6.2 Method QC
Re-quantification of batch QC sample results should use raw instrument response in
tandem with the reported CF and regressions, the preparation information, and percent
moisture for solid samples to recreate the reported result.
Verify the concentrations of surrogates from the raw data. Verify that the surrogate result and
percent recovery were calculated and reported correctly by recalculating all surrogates in the
10% of chosen sample data and method QC that were originally selected.
To check that the spike percent recovery was calculated and reported correctly, using the
equation in Appendix B, re-quantitate and then recalculate all contaminants of concern as
outlined in the UFP-QAPP Worksheet #12 or #15. Use a random 10% of the analytes in the
LCS/LCSD if contaminants of concern (target analytes) have not been specifically identified.
Recalculate RPDs (if applicable) from LCS/LCSD pairs that would result in the qualification of
a sample.
Re-quantitate 10% of the contaminants of concern as listed in the UFP-QAPP Worksheet #12
or #15 for both the MS and the MSD. Use a random 10% of the analytes in the MS and MSD
if contaminates of concern have not been identified. The RPDs of the recalculated MS/MSD
pairs should be calculated from the MS/MSD concentrations, not from the recoveries.
Method Blank (MB) analytical results are assessed to determine the existence and
magnitude of contamination problems associated with sample extraction and analysis. If
problems with any method blank exist, all associated data should be carefully evaluated to
determine whether there is any bias associated with the data, or if the problem is an isolated
occurrence not affecting other data. Results may not be corrected by subtracting any blank
values.
Re-quantitate one or more detects found in the method blank (if applicable) per each batch of
samples.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 26 of 40
Evaluation of all Surrogate Spike, LCS, MS, and MB Recalculations
If transcription errors (or other minor issues such as rounding errors) are found in method
QC results, use professional judgment to qualify the data. It may be necessary to engage
the point of contact (QAPP Worksheet #6) to communicate with the laboratory, so they can
provide revised (corrected) results. In all cases, if method QC calculation errors affect project
target analytes, the point of contact (QAPP Worksheet #6) should be notified, and all
affected results noted in the data validation report, including listing the calculation errors.
For systemic (defined as widespread and major in nature) problems with LCS/LCSD, method
blanks, surrogate spikes, or MS/MSD calculations that cannot be corrected by the laboratory
in a revised report, qualify all affected analytes in associated samples as X, exclusion of data
recommended.
6.3 Instrument QC
6.3.1 Instrument Performance Checks
Verify by recalculation at least one of the reported DDT or Endrin breakdown degradation
checks per SDG were calculated correctly, if Method 8081 was required by the QAPP.
6.3.2 Initial (Calibration Factors and Regressions) and Continuing Calibration
Verifications
Initial calibration (ICAL) recalculations should use the raw instrument response for the
target analytes and associated internal standards (if used) to recreate the calibration curve
from the individual calibration standards. If multiple types of calibration curves are
employed in an analytical suite, then one analyte per curve type should be recalculated.
Re-quantitate and recalculate the individual and average CFs, %RSDs, and regression
function (if used) and r values reported for at least 10% of the target analytes per each IS,
(preferably analytes of concern which were identified in the QAPP), per initial calibration
curve type.
Re-quantitate and recalculate the ICV and CCV CF result and %D for at least 10% of the
target analytes, proportionally selecting analytes based on each calibration curve type.
The laboratory may employ a linear or weighted linear least squares regression. The low
standard should be recalculated using the calibration curve and evaluated. CFs should not
be evaluated for analytes with linear or higher order regression curves. Recalculation of the
low calibration standard is not required for higher order (Quadratic) calibration curves.
The analyte quantitation should be evaluated for all detects by evaluating the raw data (for
example by visual inspection of chromatograms). The analyte quantitation should be based
on the CF (%RSD) or regression function from the appropriate ICAL.
Verify all internal standards (if used) reported from the raw data for at least one sample per
batch of samples and verify IS areas for samples that were qualified due to out-of-control IS
areas.
For methods 8081 and 8082, re-quantitate and recalculate at least one multicomponent
analyte (Toxaphene, Chlordane, or Aroclor) per SDG ensuring that a multipoint calibration
was used for each.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 27 of 40
For Method 8151, re-quantitate and recalculate at least 10% of the target analytes.
Determine if the hydrolysis/derivatization steps were included in the calibration standards. If
not, verify the quantitation included a molecular weight correction factor.
Evaluation of Instrument Performance Checks, ICAL, CF, Regressions, ICV/CCV, and IS
Recalculations
If degradation breakdown checks are calculated incorrectly, use professional judgment to
qualify the data based on the actual correct calculations. The QSM states that no data should
have been collected without DDT and Endrin breakdown of ≤ 15%.
If the files provided do not match the quantitation report, the CFs reported are likely to be
from another initial calibration and the laboratory report should be revised. The point of
contact (UFP-QAPP Worksheet #6) should be reached to get a revised (corrected) report
from the laboratory. For calculation errors for CFs or any other regression equations that
cannot be corrected in a revised report, qualify all the data as X, exclusion of data
recommended.
The reprocessed low standard of a regression curve should be within 30% of the true value.
If the recalculated concentration is not within 30% of the true value, qualify detects (at the
LOQ and above) for the affected analytes J and non-detects UJ in the associated samples.
Qualify all associated data as X, exclusion of data recommended if the corresponding
ICV/CCV %D has been calculated incorrectly by the laboratory and cannot be corrected in a
revised laboratory report.
If the issue cannot be corrected in a revised laboratory report, qualify all data as X, exclusion
of data recommended when the corresponding IS (if used) has been calculated incorrectly by
the laboratory or when more than one IS was used and the analyte has been assigned to the
wrong IS.
For multicomponent analytes such as Aroclors, Toxaphene, and Chlordane with detects that
were calculated with single point calibrations, if a multipoint curve cannot be obtained with a
revised laboratory report, then qualify all associated detects as X, exclusion of data
recommended and make note in the data validation report.
For Chlorinated Herbicides that were quantitated without the proper correction factor (if
applicable) and cannot be corrected with a revised laboratory report, then qualify all
associated detects as X, exclusion of data recommended and make note in the data
validation report.
All instrument QC must be analyzed on both the primary and any secondary columns, such
as the Instrument Performance Checks, ICAL, ICV/CCVs, and sample specific QC analyses
such as surrogates. Consideration should be given to recalculating and re-quantifying target
analyte detects on both the primary and secondary columns, to include the instrument QC of
both as a minimum.
In all cases where instrument QC are calculated incorrectly, the UFP-QAPP point of contact
(QAPP Worksheet #6) should be notified and noted in the data validation report.
6.3.3 Cleanup Recovery Checks
For samples that require a cleanup step, verify at least one recovery check per cleanup
method has been calculated correctly. For some cleanup methods this may require review of
the laboratory SOP. For samples with negative test analyte (Trichlorophenol < 5%) verify the
absence of the analyte.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 28 of 40
In all cases where cleanup recovery checks were not performed or calculated incorrectly,
notify the UFP-QAPP point of contact (QAPP Worksheet #6) and make note in the data
validation report. Use professional judgment to evaluate the effects on the associated
samples and qualify accordingly.
6.3.4 Second Column Confirmation
Recalculate the RPD between at least one positive detect and the second column
confirmation per each SDG if detects were found.
If the RPD was calculated incorrectly, notify the UFP-QAPP point of contact (QAPP
Worksheet #6) and make note in the data validation report. Use professional judgment to
evaluate the effects on the associated samples and qualify accordingly.
6.4 Standards Traceability
Evaluate the calibration standards used for the analytes of concern. From the Certificate of
Analysis (however named), verify that the “true values” of each analyte of concern were
correctly applied to create the calibration curve, and that all analytes of concern were in the
calibration mix.
All initial instrument calibrations should be verified with a standard obtained from a second
manufacturer prior to analyzing any samples. From the standard Certificate of Analysis verify
that a second source was used for the ICV. The use of a standard from a second lot obtained
from the same manufacturer (independently prepared from different source materials) is
acceptable for use as a second source standard.
Check that the stock standards were diluted properly into working standards by recalculating
the dilutions of one or more calibration standards. Recalculate one or more surrogate
dilutions. Recalculate one or more method QC sample dilutions (such as LCS or MS/MSD)
from the stock to the working standard.
Note: It is not the role of the data validator to evaluate the Certificate of Analysis for
compliance with the ISO-17034 Standard, but to verify that stock and working standards were
correctly applied in the creation of calibration curves.
Evaluation of Standards
Professional judgment should be used when evaluating errors in standards preparation. For
minor issues, the point of contact (QAPP Worksheet #6) identified in the QAPP (UFP-QAPP
Worksheet #6) should be reached to get a revised (corrected) report from the laboratory.
Issues that do not affect the results of any target analytes should be noted in the data
validation report.
For systemic (widespread) issues that cannot be corrected by the laboratory, or issues that
affect the results of target analytes, the data should be qualified as X, exclusion of data
recommended.
For ICV standards that were not verified to be from a second source, qualify X, exclusion of
data recommended all affected data. No samples should have been run without a valid
second source standard (per QSM requirements).
Per QSM requirements, a laboratory cannot use a standard beyond its expiration date. All
associated data should be qualified as X, exclusion of data recommended if expired

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 29 of 40
standards were used. The expiration date of any working standard is based on the expiration
date of the primary or stock standard.
6.5 Detection/Quantitation Limit Studies (Optional)
In some cases, a QAPP may specify the review and validation of a detection/quantitation limit
study. This could include studies such as Detection Limit (DL) studies (for instance MDL),
quarterly LOD verification, or LOQ verifications. The QAPP should specify the criteria for
evaluating the study. As a minimum, at least 10% of the raw data in the study should be
recalculated.
Evaluation of Detection Limit Studies
The criteria for evaluating a detection/quantitation limit study should be listed in the QAPP.
The following guidance should be enacted if the QAPP does not specify the evaluation
criteria.
If transcription errors (or other minor issues such as rounding errors) are found in
detection/quantitation limit studies, use professional judgment to qualify the data. It may be
necessary to engage the point of contact (QAPP Worksheet #6) as identified in the QAPP to
communicate with the laboratory, so they can provide revised (corrected) results. In all
cases, if calculation errors affect project detection or quantitation limits, the point of contact
(QAPP Worksheet #6) should be notified, and all affected results noted in the data validation
report, including listing the calculation errors.
When calculation errors are uncovered that cannot be corrected by the laboratory and that
affect detection/quantitation results, consideration should be given to qualify the affected data
as X, exclusion of data recommended. Points to consider are whether effects are limited to
data below the LOQ or extend to data above the LOQ and whether calculation errors are
limited to the detection limit study or if other calculation errors may be present.
7.0 Stage 4 Validation
Stage 4 requires the review of the following documents
Note: Stage 4 validation includes all of Stage 1, Stage 2A, Stage 2B and Stage 3
Raw Data (including any instrument outputs, spectra, or chromatograms)
Stage 4 is a qualitative review of all sample results from instrument outputs. Chromatograms
are checked for peak integration (10% of automated integration and 100% of manual
integrations), baseline, and interferences; chromatographic spectra are checked for minimum
signal to noise; retention times or relative retention times (RRTs) are checked to ensure they
are within method requirements for analyte identification. Raw data quantitation reports and
chromatograms are required to perform review of the instrument outputs.
7.1 Target Compound Identification
The objective of the criteria for GC qualitative analysis is to minimize the number of
erroneous identifications of target compounds. An erroneous identification can either be
false positive (reporting a compound present when it is not) or a false negative (not
reporting a compound that is present).

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 30 of 40
The identification criteria can be applied more easily in detecting false positives than false
negatives. More information is available for false positives because of the requirement for
submittal of data supporting positive identifications. Negatives, or non-detects, on the
other hand represent an absence of data and are therefore more difficult to assess.
Target analyte detections should display a signal to noise of 3:1, have proper peak
integration and have a stable baseline.
For internal standard calibration, Relative Retention Times (RRTs) should be within ± 0.06
RRT units of the midpoint standard of the ICAL curve or, on days when ICAL is not
performed, the initial CCV. When not employing internal standard calibration, the
Retention Time window width is determined at method set-up and is ± 3 times the
standard deviation for each analyte RT from the 72-hour study or equal to 0.03 minutes,
whichever is greater. The midpoint position is set using the midpoint standard of the ICAL
curve or, on days when ICAL is not performed, the initial CCV.
Check a minimum of 10% of the reported target analyte detects for RRT or RT. RRT or
RT performance in samples with only non-detects can be evaluated by reviewing the
surrogate retention times. Both primary and secondary columns should be reviewed for
target compound identification of target analyte detects.
Evaluation of Target Compound Identification
The application of qualitative criteria for GC analysis of target analytes requires professional
judgment. It is up to the reviewer's discretion to obtain additional information from their point
of contact (QAPP Worksheet #6) identified in the QAPP, if qualitative identification problems
are uncovered. The point of contact (QAPP Worksheet #6) should arrange with the laboratory
to obtain a revised (corrected) laboratory report. All qualitative identification problems should
be discussed in the data validation report. If it is determined that incorrect identifications were
made, or if a confirmed positive detect was made on one column but not found on the
dissimilar column (without baseline or interference issues) and the laboratory cannot correct
the problem, then all affected data should be qualified as X, exclusion of data recommended.
In all other cases it is understood that evaluation of the confirmation results can be
subjective, and qualification requires professional judgment when the results do not meet the
criteria.
Evaluate the chromatogram for signal to noise requirement for positive detections (3:1).
Positive results require this minimum response above baseline. Verify that positive results
are based on concentrations greater than the QAPP provided DL on both analytical columns
from peaks that elute within their established retention times. Professional judgment should
determine if reported results are usable.
Professional judgment should also be used to qualify the data if it is determined that
cross-contamination has occurred, or if interferences found in one column conflicts with
the result of the secondary column. Any changes made to the reported analytes or
concerns regarding target analyte identifications should be clearly indicated in the data
validation report.
If the spectra for a detected target analyte is considered invalid (such as interference or
baseline issues), confer with the point of contact (QAPP Worksheet #6) identified in the
QAPP to consider changing the reported detect to a non-detect for the affected analyte.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 31 of 40
7.2 Retention Time Windows
Retention Time Windows (RTWs) are critical to GC systems, and can have a direct impact on
compound identification. RTWs are established to compensate for minor shifts in absolute
retention times as a result of variability in sample injections, gas flows, or normal
chromatographic variability. The width of the RTW is carefully established to minimize the
occurrence of both false positive and false negative results and improve reproducibility.
RTWs are required to be established for both primary and secondary columns.
RTW width is required to be determined at method setup and after major maintenance.
RTWs are established by analyzing a minimum of three standards over a 72- hour period and
calculating the standard deviation (± 3 SD). As an alternative, the laboratory can establish a
0.03-minute retention time width, whichever is greater. RTW position is established once per
initial calibration and at the beginning of each analytical sequence. Position should be set
using the midpoint standard of the Initial Calibration curve when an Initial Calibration is
performed. On days when the Initial Calibration is not performed, the retention times
displayed in the initial CCV are used to set the position.
Multicomponent analytes such as Technical Chlordane, Toxaphene, Strobane, Aroclors and
Fuels are distinguished based on the ranges of retention times for characteristic components.
Separate calibration standards are necessary for each multi-component target analyte like
Toxaphene. Typically, an alkane standard containing a homologous series of n-alkanes is
used for establishing retention times for fuels like diesel. When the whole response (including
the hump-o-gram) is used for quantitation, appropriate RTWs are established for the boiling
point range or carbon number range used to define each multicomponent analyte. According
to 8015, two specific gasoline components are used to establish the GRO range (2-
methylpentane and 1,2,4-trimethylbenzene) or the DRO range (C10 and C 28 alkanes).
Refer to the method or the QAPP for specific requirements. Retention time windows are
established similarly as described above for the two components that bracket the multi-
component analyte. The retention time range is then calculated based on the lower limit of
the RTW for the first eluting component and the upper limit of the RTW for the last eluting
component. When three to five unique peaks are used in the identification of Technical
Chlordane, Toxaphene, Strobane, and/or Aroclors, these individual quantitation peaks shall
have established RTWs as described above.
Evaluation of Retention Time Windows
Verify from the raw data that the RTWs were established and are calculated correctly. The
CCV can be used to verify RTWs, and at least two analytes per calibration curve should be
recalculated (if a standard deviation was used) to verify a correct window width and
placement. If deviations are discovered, the laboratory should be contacted to correct and re-
produce the report.
Verify that all reported analytes and surrogates are within their established RTWs for both the
primary and secondary column. If positive results (detects) were reported from peaks outside
of the established RTW, contact the laboratory to evaluate for false positives and re-issue a
revised report.
Retention times of the peaks for detects must fall within the calculated window for both
chromatographic columns, and the pattern of the peaks should match the pattern in the
CCVs on both columns for multicomponent analytes. If the peaks fall outside of the RTW on

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 32 of 40
either analytical column, then the result must be reported as a non-detect, unless a known
baseline shift has occurred.
When laboratory reports cannot be revised to correct retention time issues, use professional
judgment to qualify the data. Since retention time is critical for analyte identification, strong
consideration should be given to qualifying the data as X, exclusion of data recommended.
Other qualifications should be explained within the data validation report. The RPD between
the primary and secondary column and the chromatograms should be evaluated to determine
if interference is indicated. Consideration should also be given to the magnitude of the
detects on both columns (such as results less than the LOQ) and baseline shifts and
interferences due to target or non-target analytes present in the sample when qualifying the
data.
7.3 Manual Integrations
For Stage 4, the reviewer should examine and verify the validity of all manual integrations.
Performing improper manual integrations, including peak shaving, peak enhancing, or
baseline manipulation to meet QC criteria or to avoid corrective actions is unwarranted
manipulation and misrepresents the data. All manual integrations should be reviewed by the
data validator. When manual integrations are performed, raw data records should include a
complete audit trail for those manipulations (i.e., the chromatograms obtained before and
after the manual integration should be retained to permit reconstruction of the results). This
requirement applies to all analytical runs including calibration standards and QC samples.
The person performing the manual integration should sign and date each manually integrated
chromatogram and record the rationale for performing manual integration (electronic
signature is acceptable). Any manual integration should be fully discussed in the case
narrative, including the cause and justification.
Evaluation of Manual Integrations
Some level of manual integrations is considered necessary for the normal operation of
chromatographic systems. Instances of properly integrated peaks do not require qualification
but should be noted in the validation report. However, excessive manual integrations may
show a lack of routine maintenance by the laboratory, a rush to complete samples, or the
results of analyzing excessively ‘dirty’ samples. Excessive manual integrations may also be
the result of faulty software peak/baseline integration.
The data validator should use professional judgment in the review of manual integrations. All
instances of manual integrations should be noted in the data validation report. Instances of
incomplete information for manual integrations (such as failure to provide justification) should
be reported to the point of contact (QAPP Worksheet #6) to obtain a revised (corrected)
laboratory report. Instances of excessive manual integrations that cannot be corrected by the
laboratory (such as ‘dirty’ samples that cannot undergo further cleanup procedures) should
be qualified as X, exclusion of data recommended.
If, in the professional judgment of the validator, there are instances of unwarranted
manipulation of data (such as multiple manual integrations used to ‘pass’ QC criteria) then
those cases should be reported to the project team as soon as practical (UFP-QAPP
Worksheet #6).
Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 33 of 40
Appendix A: Method QC Tables
Note: The following Table is based on the QSM Standard, with the Methods associated with
8000D for comparison. The Table does not include all the QC elements from the methods or
as listed in this guidance document.
QC Check
QSM Ver. 5.3 Frequency & Acceptance
Criteria
8000D Methods Frequency &
Acceptance Criteria
Breakdown Check
(Method 8081)
Before sample analysis and at the beginning
of each 12-hour shift.
Degradation of DDT and Endrin must be
≤ 15%.
Before sample analysis and at
the beginning of each 12-hour
shift.
Degradation of DDT and Endrin
must be ≤ 15%.
Presence of 4,4’-DDE, 4,4’-DDD,
or Endrin Ketone indicates
breakdown.
Initial Calibration
(ICAL)
For all analytes and surrogates
At instrument set-up and after ICV or CCV
failure, prior to sample analysis.
Each analyte should meet one of the
options below:
Option 1: RSD for each analyte ≤ 20%;
Option 2: linear least squares regression for
each analyte: r
2
≥ 0.99;
Option 3: non-linear least squares
regression (quadratic) for each analyte: r
2
≥
0.99.
Minimum 5 levels for linear and 6 levels for
quadratic.
Quantitation for multipoint analytes such as
Chlordane, Toxaphene, and Aroclors must
be performed using a 5-point calibration.
At instrument set-up and after
ICV or CCV failure, prior to
sample analysis.
Each analyte should meet one
of the options below:
Option 1: RSD for each analyte
≤ 20%;
Option 2: linear least squares
regression for each analyte: r
2
≥
0.990 or r ≥ 0.995;
Option 3: non-linear least
squares regression (quadratic)
for each analyte: r
2
≥ 0.99 or r ≥
0.995;
Option 4: Relative Standard
Error (RSE) ≤ 20%.
Minimum 5 levels for linear and 6
levels for quadratic.
Single point calibration for
multicomponent analytes.
Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 34 of 40
QC Check
QSM Ver. 5.3 Frequency & Acceptance
Criteria
8000D Methods Frequency &
Acceptance Criteria
Retention Time
Window Width
Establishment
Once per ICAL, and at the beginning of the
analytical sequence.
Position shall be set using the midpoint
standard of the ICAL curve when the ICAL is
performed. Use initial CCV when ICAL not
performed.
Once per ICAL, and at the
beginning of the analytical
sequence.
Position shall be set using the
midpoint standard of the ICAL
curve when the ICAL is
performed. Use initial CCV when
ICAL not performed.
Retention Time
(RT) Window
Width
Method setup and after major maintenance
RT width is ± 3 times standard deviation
from 72-hour study or 0.03 minutes,
whichever is greater.
Method setup and after major
maintenance
RT width is ± 3 times standard
deviation from 72-hour study or
0.03 minutes, whichever is
greater.
Initial Calibration
Verification (ICV)
Once after each ICAL, analysis of a second
source standard prior to sample analysis.
All reported analytes within ± 20% of true
value.
Once after each ICAL, analysis of
a second source standard prior to
sample analysis.
All reported analytes within ±
20% of true value.
Continuing
Calibration
Verification (CCV)
Before sample analysis; after every 10 field
samples; and at the end of the analytical
batch run.
Exception: Multicomponent analytes
Toxaphene, Chlordane, Aroclors (other than
1016 and 1260) are only required before
sample analysis.
All reported analytes and surrogates within ±
20% of true value for opening CCV.
After every 10 field samples;
more frequent verification of
calibration (i.e., after every 10
samples) may be necessary for
some types of detectors.
Multicomponent analytes are only
required before sample analysis.
If ≤ 10% of the analytes exceed
the calibration verification criteria,
then the initial calibration may still
be used, but any detected
analytes exceeding the limit must
be reported as estimated.
All reported analytes and
surrogates within ± 20% of true
value for CCV.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 35 of 40
QC Check
QSM Ver. 5.3 Frequency & Acceptance
Criteria
8000D Methods Frequency &
Acceptance Criteria
Internal Standards
(if used)
Every field sample, standard, and QC
sample.
Retention time within ± 30 seconds from
retention time of the midpoint standard in the
ICAL; EICP area within - 50% to +100% of
ICAL midpoint standard.
Every field sample, standard, and
QC sample.
Retention time within ± 30
seconds from retention time of
the midpoint standard in the
ICAL; EICP area within - 50% to
+100% of ICAL midpoint
standard.
RRT range of each analyte within
0.80-1.20 units.
Method Blank
(MB)
One per preparatory batch.
No analytes detected > ½ LOQ or > 1/10 the
amount measured in any sample or 1/10 the
regulatory limit, whichever is greater.
Method blanks should be
prepared at a frequency of at
least 5%: one method blank for
each group of up to 20 samples
prepared at the same time.
Results of the method blank
should be less than the lower
limit of quantitation (LLOQ) for
the analyte or less than the level
of acceptable blank
contamination specified in the
approved QAPP.
Laboratory
Control Sample
(LCS);
Matrix Spike (MS);
Matrix Spike
Duplicate (MSD)
Relative percent
difference (RPD)
One each per preparatory batch.
A laboratory should use the QSM Appendix
C Limits for batch control and matrix spikes if
project limits are not specified.
If the analyte(s) are not listed, use in-house
LCS limits if project limits are not specified.
MSD or MD: RPD of all analytes ≤ 20%
(between MS and MSD or sample and MD).
Should contain all of the target
analytes. One each per batch of
20 samples.
The laboratory should use 70 -
130% as interim acceptance
criteria for recoveries of spiked
analytes, until in-house LCS
limits are developed.
Duplicate RPD will be
established for the field samples
through the DQOs contained in a
written QAPP.
Surrogate Spikes
All field and QC samples.
QC acceptance criteria specified by the
project if available; otherwise use QSM
Appendix C limits or in-house LCS limits if
analyte(s) are not listed.
All field and QC samples.
Compared to developed in-house
surrogate recovery limits. Data is
reported as “estimated” if any re-
analysis is not within limits.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 36 of 40
QC Check
QSM Ver. 5.3 Frequency & Acceptance
Criteria
8000D Methods Frequency &
Acceptance Criteria
Confirmation of
positive results
(second column)
All results > the DL must be confirmed
(except for single column methods such as
TPH by method 8015).
Results between primary and secondary
column RPD ≤ 40%.
When confirmation is made on a
second column, that analysis
should meet all of the QC criteria
(calibrations, retention times, and
performance checks) of the first
column.
No RPD criteria given in 8000D.

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 37 of 40
Appendix B: Formulas used in Stage 3 and 4 Data Validation
Note: Not all formulas that may be necessary for validation are included herein.
Calibration:
Calibration Factor (CF): A
s
C
S
Note: A CF is calculated for each individual peak.
Response Factor (RF): A
S
*C
IS
A
IS
*C
S
A
S
= Area, Standard
C
IS
= Concentration, Internal Standard
A
IS
= Area, Internal Standard
C
S
= Concentration, Standard
Note: a RF is calculated if an internal standard is used.
Average CF or RF: ∑ RF
n
∑ RF = Sum of CFs or RFs for the standards
n= Number of standards
Percent Relative Standard Deviation: S
RF
*100
ARF
S
RF
= Standard Deviation of CFs or RFs
ARF= Average CF or RF
Relative Retention time (RRT): RT
A
RT
IS
RT
A
= Retention time of Analyte
RT
IS
= Retention time of Internal Standard
Percent Difference: %D= C
S
-C
K
*100
C
K
C
s
= Concentration, reported
C
K
= Concentration, known

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 38 of 40
Sample Concentration (regression):
Raw Values
CF: C
p
= A
p
RF
RF (IS): C
p
= A
p
*C
IS
A
IS
*ARF
C
s
= ∑C
p
n
C
p
= Concentration of peak, sample
A
p
=Area of peak, Sample
C
IS
= Concentration, Internal Standard
A
IS
= Area, Internal Standard
RF= CF or RF for each peak
C
s
= Concentration, sample
n= number of peaks
Linear Regression (External calibration): y= mx + b
C
p
= A
p
-b
m
C
p
= Concentration of peak, sample
A
p
=Area of peak, Sample
b= Intercept
m= Slope
Linear Regression (Internal calibration): y= mx + b
C
p
=[(A
p
/A
IS
)-b]*C
IS
m
C
p
= Concentration of peak, sample
A
p
=Area of peak, Sample
A
IS
= Area, Internal standard
C
IS
= Concentration, Internal Standard
b= Intercept
m= Slope

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 39 of 40
Quadratic Regression (External Calibration): y=ax2+bx+c
C
p
= -b+√[b
2
-4a*(c-A
p
)]
2a
C
p
= Concentration of peak, sample
A
p
=Area of peak, Sample
a= Quadratic Coefficient
b= Linear Coefficient
c= Constant Term
Quadratic Regression (Internal Calibration): y=ax2+bx+c
C
p
= -b+√[b
2
-4a*(c-A
p
/A
IS
)]
2a*C
IS
C
p
= Concentration of peak, sample
A
p
=Area of peak, Sample
A
IS
= Area, Internal standard
C
IS
= Concentration, Internal Standard
a= Quadratic Coefficient
b= Linear Coefficient
c= Constant Term
For linear and quadratic regressions:
C
s
= ∑C
p
n
C
s
= Concentration, sample
n= number of peaks
Reported Values:
Waters
Concentration µg/L = R*V
f
*D
f
/V
i
R= Raw value from above in micrograms per liter (ug/L)
V
f
= Final Volume of extract in liters (L)
V
i
= Initial Volume extracted in liters (L)
D
f=
Dilution Factor

Department of Defense
Module 4 Data Validation Guidelines: Data Validation Procedure for Organic Analysis by GC
March 2021
Page 40 of 40
Solids
Concentration µg/Kg (Dry weight basis) = (R x V
f
× 1,000 × D
f
)
/
( Ws × D)
R = Raw value from above in micrograms per liter (ug/L)
V
t
= Final volume of extract in liters (L)
W
s
= Weight of soil/sediment extracted, in grams (g)
D
f
= Dilution factor.
D = 100 – % moisture
100
LCS or Surrogate Percent Recovery:
Percent Recovery: C
s
*100
C
K
C
s
= Concentration, Reported
C
K
= Concentration, Known
MS or MSD percent recovery:
Percent Recovery: (C
M
-C
S
) *100
C
K
C
M
= Concentration, MS or MSD
C
S
= Concentration, Sample
C
K
= Concentration, Known
MS/MSD or Duplicate Relative Percent Difference (RPD):
RPD: |(C
S
- C
d
)| *100
[(C
S
+ C
d
)/2]
C
s
= Concentration, Sample
C
d
= Concentration, Duplicate
